Cytokine-Based Insights into Bloodstream Infections and Bacterial Gram Typing in ICU COVID-19 Patients
Abstract
1. Introduction
2. Materials and Methods
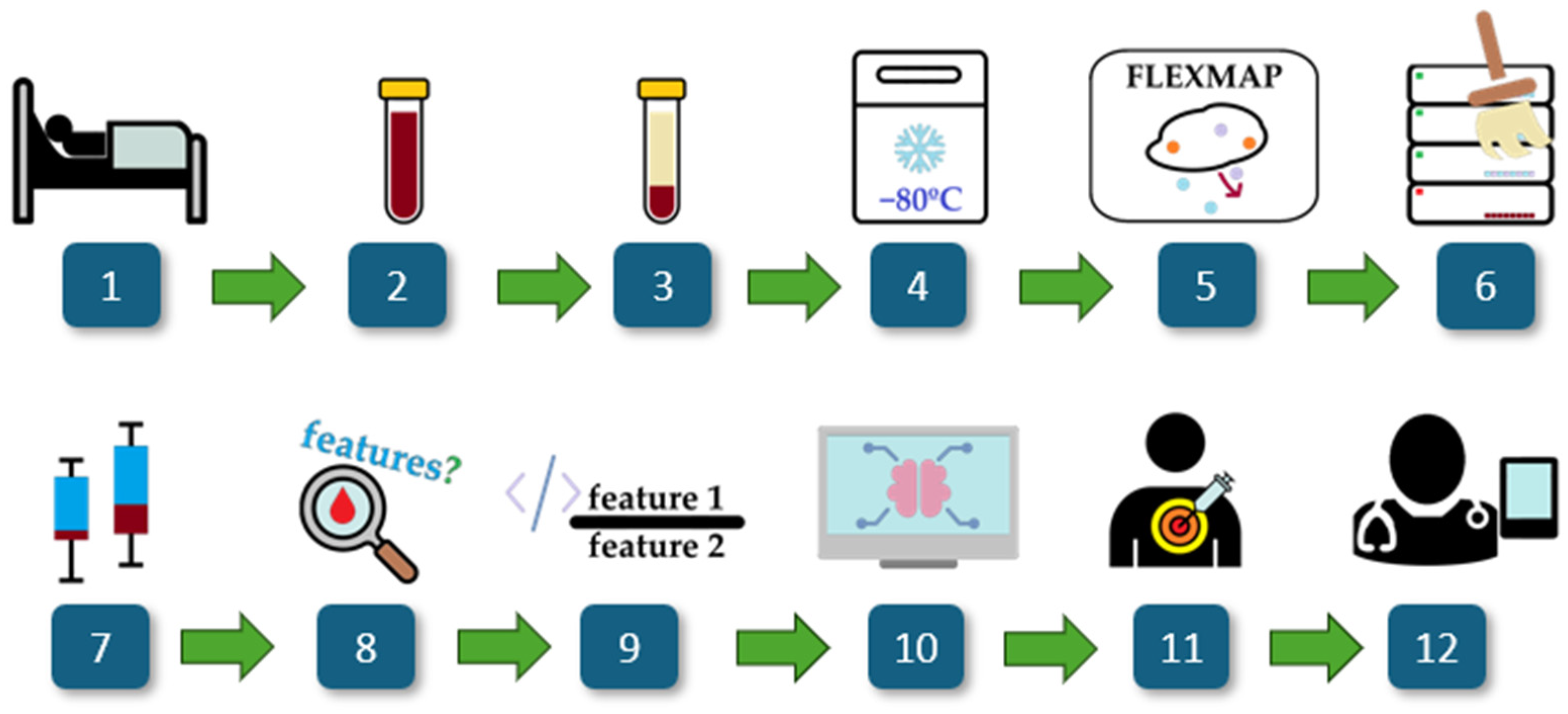
2.1. Workflow Overview
2.2. Study Population
2.3. Collection of Biological Samples
2.4. Cytokine Profiling
2.5. Data and Statistical Analysis
3. Results and Discussion
3.1. Study Population Characteristics
3.2. Commonly Reported Cytokines and Ratios
- BSIs: Presence vs. Absence
- Gram-Positive vs. Gram-Negative
- Convergence With and Divergence From the Literature
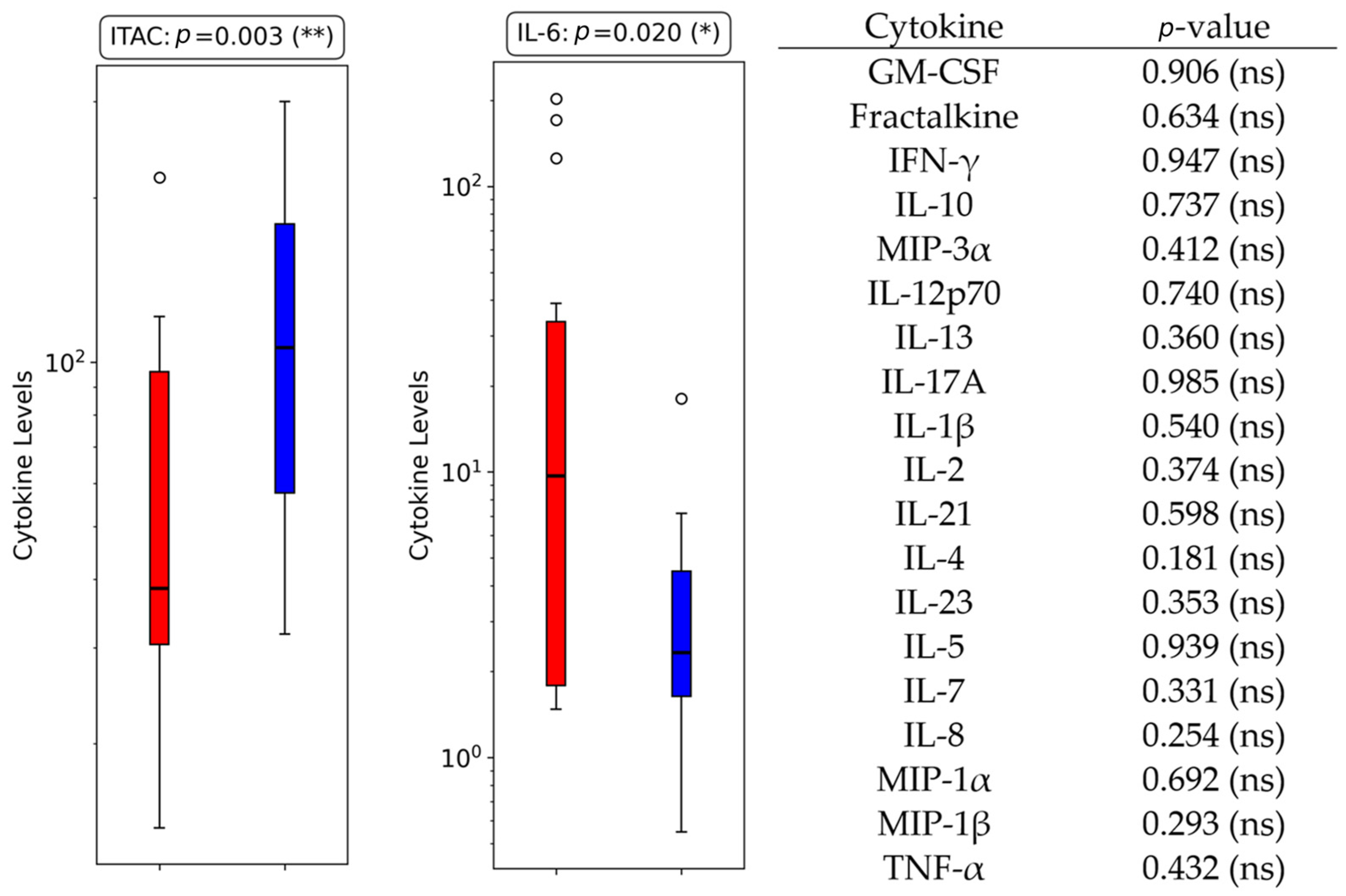
3.3. Univariate Cytokines Analysis
3.3.1. Bloodstream Infections
3.3.2. Gram Typing
3.4. Multivariate Cytokines Analysis
3.4.1. Individual Cytokine Analysis
3.4.2. Computationally Generated Cytokine Ratios
3.5. Key Considerations and Future Directions
- Data Quality versus Quantity
- Importance of Data Cleaning and Normalization
- Selecting the Right Evaluation Metrics
- No Sample Size is Big Enough for Universal Generalization
- The Path Forward: Reproducibility, Scalability, and Economic Considerations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vincent, J.-L.; Sakr, Y.; Singer, M.; Martin-Loeches, I.; Machado, F.R.; Marshall, J.C.; Finfer, S.; Pelosi, P.; Brazzi, L.; Aditianingsih, D.; et al. Prevalence and Outcomes of Infection Among Patients in Intensive Care Units in 2017. JAMA 2020, 323, 1478. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, R.; Bal, A.; Kapila, K.; Mave, V.; Gupta, A. Blood Stream Infections. BioMed Res. Int. 2014, 2014, 515273. [Google Scholar] [CrossRef] [PubMed]
- Bloch, N.; Rüfenacht, S.; Ludwinek, M.; Frick, W.; Kleger, G.-R.; Schneider, F.; Albrich, W.C.; Flury, D.; Kuster, S.P.; Schlegel, M.; et al. Healthcare-associated Infections in Intensive Care Unit Patients with and without COVID-19: A Single Center Pro-spective Surveillance Study. Antimicrob. Resist. Infect. Control 2023, 12, 147. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Cattaneo, E.; Florio, G. Secondary Infections in Critically Ill Patients with COVID-19. Crit. Care 2021, 25, 317. [Google Scholar] [CrossRef]
- Póvoa, P.; Coelho, L.; Dal-Pizzol, F.; Ferrer, R.; Huttner, A.; Conway Morris, A.; Nobre, V.; Ramirez, P.; Rouze, A.; Salluh, J.; et al. How to Use Biomarkers of Infection or Sepsis at the Bedside: Guide to Clinicians. Intensive Care Med. 2023, 49, 142–153. [Google Scholar] [CrossRef]
- Magrini, L.; Gagliano, G.; Travaglino, F.; Vetrone, F.; Marino, R.; Cardelli, P.; Salerno, G.; Di Somma, S. Comparison between White Blood Cell Count, Procalcitonin and C Reactive Protein as Diagnostic and Prognostic Biomarkers of Infection or Sepsis in Patients Presenting to Emergency Department. Clin. Chem. Lab. Med. 2014, 52, 1465–1472. [Google Scholar] [CrossRef]
- Losser, M.-R.; Damoisel, C.; Payen, D. Bench-to-Bedside Review: Glucose and Stress Conditions in the Intensive Care Unit. Crit. Care 2010, 14, 231. [Google Scholar] [CrossRef]
- Tsalik, E.L.; Jones, D.; Nicholson, B.; Waring, L.; Liesenfeld, O.; Park, L.P.; Glickman, S.W.; Caram, L.B.; Langley, R.J.; Van Velkinburgh, J.C.; et al. Multiplex PCR To Diagnose Bloodstream Infections in Patients Admitted from the Emergency De-partment with Sepsis. J. Clin. Microbiol. 2010, 48, 26–33. [Google Scholar] [CrossRef]
- Coelho, L.; Rabello, L.; Salluh, J.; Martin-Loeches, I.; Rodriguez, A.; Nseir, S.; Gomes, J.A.; Povoa, P. C-Reactive Protein and Procalcitonin Profile in Ventilator-Associated Lower Respiratory Infections. J. Crit. Care 2018, 48, 385–389. [Google Scholar] [CrossRef]
- Russell, C.D.; Parajuli, A.; Gale, H.J.; Bulteel, N.S.; Schuetz, P.; De Jager, C.P.C.; Loonen, A.J.M.; Merekoulias, G.I.; Baillie, J.K. The Utility of Peripheral Blood Leucocyte Ratios as Biomarkers in Infectious Diseases: A Systematic Review and Meta-Analysis. J. Infect. 2019, 78, 339–348. [Google Scholar] [CrossRef]
- Maves, R.C.; Enwezor, C.H. Uses of Procalcitonin as a Biomarker in Critical Care Medicine. Infect. Dis. Clin. N. Am. 2022, 36, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.A.; Aghaeepour, N.; Bhattacharyya, R.P.; Clish, C.B.; Gaudillière, B.; Hacohen, N.; Mansour, M.K.; Mudd, P.A.; Pasupneti, S.; Presti, R.M.; et al. Harnessing the Potential of Multiomics Studies for Precision Medicine in Infectious Disease. Open Forum Infect. Dis. 2021, 8, ofab483. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Kraft, R.; Herndon, D.N.; Finnerty, C.C.; Cox, R.A.; Song, J.; Jeschke, M.G. Predictive Value of IL-8 for Sepsis and Severe In-fections After Burn Injury: A Clinical Study. Shock 2015, 43, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Carlini, V.; Noonan, D.M.; Abdalalem, E.; Goletti, D.; Sansone, C.; Calabrone, L.; Albini, A. The Multifaceted Nature of IL-10: Regulation, Role in Immunological Homeostasis and Its Relevance to Cancer, COVID-19 and Post-COVID Conditions. Front. Immunol. 2023, 14, 1161067. [Google Scholar] [CrossRef]
- Dharra, R.; Kumar Sharma, A.; Datta, S. Emerging Aspects of Cytokine Storm in COVID-19: The Role of Proinflammatory Cytokines and Therapeutic Prospects. Cytokine 2023, 169, 156287. [Google Scholar] [CrossRef]
- Majeed, A.Y.; Zulkafli, N.E.S.; Ad’hiah, A.H. Serum Profiles of Pro-Inflammatory and Anti-Inflammatory Cytokines in Non-Hospitalized Patients with Mild/Moderate COVID-19 Infection. Immunol. Lett. 2023, 260, 24–34. [Google Scholar] [CrossRef]
- Montazersaheb, S.; Hosseiniyan Khatibi, S.M.; Hejazi, M.S.; Tarhriz, V.; Farjami, A.; Ghasemian Sorbeni, F.; Farahzadi, R.; Ghasemnejad, T. COVID-19 Infection: An Overview on Cytokine Storm and Related Interventions. Virol. J. 2022, 19, 92. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef]
- Song, J.; Park, D.W.; Moon, S.; Cho, H.-J.; Park, J.H.; Seok, H.; Choi, W.S. Diagnostic and Prognostic Value of Interleukin-6, Pentraxin 3, and Procalcitonin Levels among Sepsis and Septic Shock Patients: A Prospective Controlled Study According to the Sepsis-3 Definitions. BMC Infect. Dis. 2019, 19, 968. [Google Scholar] [CrossRef]
- Zhu, S.; Zeng, C.; Zou, Y.; Hu, Y.; Tang, C.; Liu, C. The Clinical Diagnostic Values of SAA, PCT, CRP, and IL-6 in Children with Bacterial, Viral, or Co-Infections. Int. J. Gen. Med. 2021, 14, 7107–7113. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Stow, J.L. Cytokine Release from Innate Immune Cells: Association with Diverse Membrane Trafficking Pathways. Blood 2011, 118, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Historical Insights into Cytokines. Eur. J. Immunol. 2007, 37, S34–S45. [Google Scholar] [CrossRef]
- Qin, Y.; Feng, M.; Wu, Y.; Wang, Y.; Zhao, X.; Liu, G.; Gao, C.; Luo, J.; Guo, H. Comprehensive Analysis of Multiple Cytokines and Blood Parameters for the Diagnosis of Bacterial Infections in Rheumatoid Arthritis. Cytokine 2020, 136, 155251. [Google Scholar] [CrossRef]
- Tan, K.; Minejima, E.; Lou, M.; Mack, W.J.; Nieberg, P.; Wong-Beringer, A. Cytokine Measurements Add Value to Clinical Variables in Predicting Outcomes for Staphylococcus aureus Bacteremia. BMC Infect. Dis. 2021, 21, 317. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-T.; Liu, Y.-H.; Lin, C.-Y.; Kuo, P.-L.; Yen, M.-C. Inflammatory Molecules Expression Pattern for Identifying Pathogen Species in Febrile Patient Serum. Exp. Ther. Med. 2016, 12, 312–318. [Google Scholar] [CrossRef]
- Yu, W.; Zeng, L.; Lian, X.; Jiang, L.; Xu, H.; Guo, W.; Zheng, B.; Xiao, Y. Dynamic Cytokine Profiles of Bloodstream Infection Caused by Klebsiella pneumoniae in China. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 79. [Google Scholar] [CrossRef]
- Taj-Aldeen, S.J.; Mir, F.A.; Sivaraman, S.K.; AbdulWahab, A. Serum Cytokine Profile in Patients with Candidemia versus Bacteremia. Pathogens 2021, 10, 1349. [Google Scholar] [CrossRef]
- Ye, Q.; Shao, W.-X.; Xu, X.-J.; Yang, Y. The Clinical Application Value of Cytokines in Treating Infectious Diseases. PLoS ONE 2014, 9, e98745. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, H.; Zheng, S.; Wang, B.; Li, M.; Zeng, W.; Zhou, L.; Guan, Z.; Wang, H.; Liu, Y.; et al. IL-6 and IL-10 Are Associated With Gram-Negative and Gram-Positive Bacteria Infection in Lymphoma. Front. Immunol. 2022, 13, 856039. [Google Scholar] [CrossRef]
- Araújo, R.; Bento, L.F.N.; Fonseca, T.A.H.; Von Rekowski, C.P.; da Cunha, B.R.; Calado, C.R.C. Infection Biomarkers Based on Metabolomics. Metabolites 2022, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Grankvist, K.; Gomez, R.; Nybo, M.; Lima-Oliveira, G.; Von Meyer, A. Preanalytical Aspects on Short- and Long-Term Storage of Serum and Plasma. Diagnosis 2019, 6, 51–56. [Google Scholar] [CrossRef]
- Valo, E.; Colombo, M.; Sandholm, N.; McGurnaghan, S.J.; Blackbourn, L.A.K.; Dunger, D.B.; McKeigue, P.M.; Forsblom, C.; Groop, P.-H.; Colhoun, H.M.; et al. Effect of Serum Sample Storage Temperature on Metabolomic and Proteomic Biomarkers. Sci. Rep. 2022, 12, 4571. [Google Scholar] [CrossRef]
- Vendrame, E.; Hussain, S.K.; Breen, E.C.; Magpantay, L.I.; Widney, D.P.; Jacobson, L.P.; Variakojis, D.; Knowlton, E.R.; Bream, J.H.; Ambinder, R.F.; et al. Serum Levels of Cytokines and Biomarkers for Inflammation and Immune Activation, and HIV-Associated Non-Hodgkin B-Cell Lymphoma Risk. Cancer Epidemiol. Biomark. Prev. 2014, 23, 343–349. [Google Scholar] [CrossRef]
- Gogos, C.A.; Drosou, E.; Bassaris, H.P.; Skoutelis, A. Pro- versus Anti-inflammatory Cytokine Profile in Patients with Severe Sepsis: A Marker for Prognosis and Future Therapeutic Options. J. Infect. Dis. 2000, 181, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Blanc, E.; Chaize, G.; Fievez, S.; Féger, C.; Herquelot, E.; Vainchtock, A.; Timsit, J.F.; Gaillat, J. The Impact of Comorbidities and Their Stacking on Short- and Long-Term Prognosis of Patients over 50 with Community-Acquired Pneumonia. BMC Infect. Dis. 2021, 21, 949. [Google Scholar] [CrossRef] [PubMed]
- Hockham, C.; Linschoten, M.; Asselbergs, F.W.; Ghossein, C.; Woodward, M.; Peters, S.A.E. Sex Differences in Cardiovascular Complications and Mortality in Hospital Patients with Covid-19: Registry Based Observational Study. BMJ Med. 2023, 2, e000245. [Google Scholar] [CrossRef]
- Perez, A.V.; Viana, M.V.; Dall’Orto Thomazini, L.; Loss, S.H.; De Machado, F.C.R.; Do Nascimento, A.G.; Kropidlofscky, A.P.; Gerchman, F.; Leitão, C.B.; Rech, T.H.; et al. BMI and Mortality in Critically Ill Patients with COVID-19: Another Brick in the Wall of the Obesity Paradox. Obesity 2024, 32, 1474–1482. [Google Scholar] [CrossRef]
- Simpson, A.; Puxty, K.; McLoone, P.; Quasim, T.; Sloan, B.; Morrison, D.S. Comorbidity and Survival after Admission to the Intensive Care Unit: A Population-Based Study of 41,230 Patients. J. Intensive Care Soc. 2021, 22, 143–151. [Google Scholar] [CrossRef]
- Chang, R.; Elhusseiny, K.M.; Yeh, Y.-C.; Sun, W.-Z. COVID-19 ICU and Mechanical Ventilation Patient Characteristics and Outcomes—A Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0246318. [Google Scholar] [CrossRef]
- Ling, R.R.; Ramanathan, K.; Sim, J.J.L.; Wong, S.N.; Chen, Y.; Amin, F.; Fernando, S.M.; Rochwerg, B.; Fan, E.; Barbaro, R.P.; et al. Evolving Outcomes of Extracorporeal Membrane Oxygenation during the First 2 Years of the COVID-19 Pandemic: A Systematic Review and Meta-Analysis. Crit. Care 2022, 26, 147. [Google Scholar] [CrossRef] [PubMed]
- van der Maaten, L.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Yu, L.; Liu, H. Feature Selection for High-Dimensional Data: A Fast Correlation-Based Filter Solution. In Proceedings of the Machine Learning, Proceedings of the Twentieth International Conference (ICML 2003), Washington, DC, USA, 21–24 August 2003. [Google Scholar]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Lustgarten, J.L.; Gopalakrishnan, V.; Visweswaran, S. Measuring Stability of Feature Selection in Biomedical Datasets. AMIA Annu. Symp. Proc. 2009, 2009, 406–410. [Google Scholar]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Lyle, P. The Construction of Nomograms for Use in Statistics: Part I. True and Empirical Nomograms. Appl. Stat. 1954, 3, 116. [Google Scholar] [CrossRef]
- Araújo, R.; Ramalhete, L.; Viegas, A.; Von Rekowski, C.P.; Fonseca, T.A.H.; Calado, C.R.C.; Bento, L. Simplifying Data Analysis in Biomedical Research: An Automated, User-Friendly Tool. Methods Protoc. 2024, 7, 36. [Google Scholar] [CrossRef]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the Pathophysiology of Severe Asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef]
- Bikker, A.; Erik Hack, C.; Lafeber, F.P.J.G.; Van Roon, J.A.G. Interleukin-7: A Key Mediator in T Cell-Driven Autoimmunity, Inflammation, and Tissue Destruction. Curr. Pharm. Des. 2012, 18, 2347–2356. [Google Scholar] [CrossRef]
- Yang, S.; Zeng, J.; Hao, W.; Sun, R.; Tuo, Y.; Tan, L.; Zhang, H.; Liu, R.; Bai, H. IL-21/IL-21R Promotes the Pro-Inflammatory Effects of Macrophages during C. muridarum Respiratory Infection. Int. J. Mol. Sci. 2023, 24, 12557. [Google Scholar] [CrossRef]
- Ye, D.; Wang, Z.; Ye, J.; Wang, M.; Liu, J.; Xu, Y.; Jiang, H.; Chen, J.; Wan, J. Interleukin-5 Levels Are Decreased in the Plasma of Coronary Artery Disease Patients and Inhibit Th1 and Th17 Differentiation in Vitro. Rev. Española De Cardiol. (Engl. Ed.) 2020, 73, 393–402. [Google Scholar] [CrossRef]
- Kim, H.K.; Waickman, A.T.; Castro, E.; Flomerfelt, F.A.; Hawk, N.V.; Kapoor, V.; Telford, W.G.; Gress, R.E. Distinct IL-7 Signaling in Recent Thymic Emigrants versus Mature Naïve T Cells Controls T-cell Homeostasis. Eur. J. Immunol. 2016, 46, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Jian, L.; Li, C.; Wang, X.; Sun, L.; Ma, Z.; Zhao, J. IL-21 Impairs pro-Inflammatory Activity of M1-like Macrophages Exerting Anti-Inflammatory Effects on Rheumatoid Arthritis. Autoimmunity 2022, 55, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.N.A.; Tauber, P.A.; Stieger, R.B.; Kratzer, B.; Pickl, W.F. The T-Cell Growth Factor Interleukin-2, Which Is Occa-sionally Targeted by Autoantibodies, Qualifies as Drug for the Treatment of Allergy, Autoimmunity, and Cancer: Collegium Internationale Allergologicum (CIA) Update 2024. Int. Arch. Allergy Immunol. 2024, 185, 286–300. [Google Scholar] [CrossRef]
- Klatzmann, D.; Abbas, A.K. The Promise of Low-Dose Interleukin-2 Therapy for Autoimmune and Inflammatory Diseases. Nat. Rev. Immunol. 2015, 15, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zeng, J.; Yu, X.; Wang, Z.; Wang, D.; Zhou, Q.; Bai, T.; Xu, Y. PCT, IL-6, and IL-10 Facilitate Early Diagnosis and Pathogen Classifications in Bloodstream Infection. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 103. [Google Scholar] [CrossRef]
- Schelonka, R.L.; Maheshwari, A.; Carlo, W.A.; Taylor, S.; Hansen, N.I.; Schendel, D.E.; Thorsen, P.; Skogstrand, K.; Hougaard, D.M.; Higgins, R.D. T Cell Cytokines and the Risk of Blood Stream Infection in Extremely Low Birth Weight Infants. Cytokine 2011, 53, 249–255. [Google Scholar] [CrossRef]
- Monastero, R.N.; Pentyala, S. Cytokines as Biomarkers and Their Respective Clinical Cutoff Levels. Int. J. Inflamm. 2017, 2017, 4309485. [Google Scholar] [CrossRef]
- Sun, J.; Ma, X.; Zhang, M.; Xie, M.; Zhang, X.; Han, X.; Li, X.; Zhou, E.; Wang, J.; Wang, J. Comparisons of Lymphocytes Profiles and Inflammatory Cytokines Levels in Blood of Patients with Differed Severity of Infection by Human Adenovirus Type 7. BMC Infect. Dis. 2023, 23, 174. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, J.C.; Plata-Menchaca, E.P.; Chiscano-Camón, L.; Ruiz-Sanmartin, A.; Ferrer, R. Blood Purification in Sepsis and COVID-19: What’s New in Cytokine and Endotoxin Hemoadsorption. J. Anesth. Analg. Crit. Care 2022, 2, 15. [Google Scholar] [CrossRef]
- Otto, G.; Braconier, J.; Andreasson, A.; Svanborg, C. Interleukin-6 and Disease Severity in Patients with Bacteremic and Nonbacteremic Febrile Urinary Tract Infection. J. Infect. Dis. 1999, 179, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Slaats, J.; Ten Oever, J.; Van De Veerdonk, F.L.; Netea, M.G. IL-1β/IL-6/CRP and IL-18/Ferritin: Distinct Inflammatory Programs in Infections. PLoS Pathog. 2016, 12, e1005973. [Google Scholar] [CrossRef] [PubMed]
- Picod, A.; Morisson, L.; De Roquetaillade, C.; Sadoune, M.; Mebazaa, A.; Gayat, E.; Davison, B.A.; Cotter, G.; Chousterman, B.G. Systemic Inflammation Evaluated by Interleukin-6 or C-Reactive Protein in Critically Ill Patients: Results From the FROG-ICU Study. Front. Immunol. 2022, 13, 868348. [Google Scholar] [CrossRef] [PubMed]
- Gamarra-Morales, Y.; Molina-López, J.; Santiago-Ruiz, F.-C.; Herrera-Quintana, L.; Vázquez-Lorente, H.; Gascón-Luna, F.; Planells, E. Efficiency of IL-6 in Early Prognosis and Follow-Up in Critically Ill Patients with Septic Shock. Diseases 2024, 12, 298. [Google Scholar] [CrossRef]
- Yamamoto, R.; Sasaki, J.; Shibusawa, T.; Nakada, T.; Mayumi, T.; Takasu, O.; Matsuda, K.; Shimazui, T.; Otsubo, H.; Teshima, Y.; et al. Accuracy for Mortality Prediction With Additive Biomarkers Including Interleukin-6 in Critically Ill Patients: A Mul-ticenter Prospective Observational Study. Crit. Care Explor. 2021, 3, e0387. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, C.; Agudelo Higuita, N.I.; Bhattacharya, R.; Chakrabarty, J.H.; Mukherjee, P. Evaluation of I-TAC as a Potential Early Plasma Marker to Differentiate between Critical and Non-Critical COVID-19. Cell Stress 2021, 6, 6–16. [Google Scholar] [CrossRef]
- Bi, Q.; Zhu, J.; Zheng, J.; Xu, Q.; Chen, J.; Zhang, L.; Mu, X. Blood Inflammatory Markers and Cytokines in COVID-19 Patients With Bacterial Coinfections. Immun. Inflam. Amp. Dis. 2024, 12, e70105. [Google Scholar] [CrossRef]
- Craig, A.; Mai, J.; Cai, S.; Jeyaseelan, S. Neutrophil Recruitment to the Lungs during Bacterial Pneumonia. Infect. Immun. 2009, 77, 568–575. [Google Scholar] [CrossRef]
- Skovbjerg, S.; Martner, A.; Hynsjö, L.; Hessle, C.; Olsen, I.; Dewhirst, F.E.; Tham, W.; Wold, A.E. Gram-Positive and Gram-Negative Bacteria Induce Different Patterns of Cytokine Production in Human Mononuclear Cells Irrespective of Tax-onomic Relatedness. J. Interferon Cytokine Res. 2010, 30, 23–32. [Google Scholar] [CrossRef]
- Nakada, T.; Russell, J.A.; Boyd, J.H.; Walley, K.R. IL17A Genetic Variation Is Associated with Altered Susceptibility to Gram-Positive Infection and Mortality of Severe Sepsis. Crit. Care 2011, 15, R254. [Google Scholar] [CrossRef]
- Kern, W.V.; Heiss, M.; Steinbach, G. Prediction of Gram-Negative Bacteremia in Patients with Cancer and Febrile Neutropenia by Means of Interleukin-8 Levels in Serum: Targeting Empirical Monotherapy versus Combination Therapy. Clin. Infect. Dis. 2001, 32, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Larsson, B.M.; Larsson, K.; Malmberg, P.; Palmberg, L. Gram Positive Bacteria Induce IL-6 and IL-8 Production in Human Alveolar Macrophages and Epithelial Cells. Cell. Mol. Neurobiol. 1999, 23, 217–230. [Google Scholar] [CrossRef]
- Sorrentino, R.; De Souza, P.M.; Sriskandan, S.; Duffin, C.; Paul-Clark, M.J.; Mitchell, J.A. Pattern Recognition Receptors and Interleukin-8 Mediate Effects of Gram-positive and Gram-negative Bacteria on Lung Epithelial Cell Function. Br. J. Pharmacol. 2008, 154, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Chetaille Nézondet, A.-L.; Poubelle, P.E.; Pelletier, M. The Evaluation of Cytokines to Help Establish Diagnosis and Guide Treatment of Autoinflammatory and Autoimmune Diseases. J. Leukoc. Biol. 2020, 108, 647–657. [Google Scholar] [CrossRef]
- Guo, W.; Lian, X.; Li, H.; Jiang, L.; Chen, Y.; Shen, P.; Yu, W. Characteristics of Immunocytes and Cytokines in Patients with Bloodstream Infections Caused by Carbapenem-Resistant Klebsiella pneumoniae in China. Infect. Drug Resist. 2024, 17, 719–725. [Google Scholar] [CrossRef]
- Ru, Y.; Ding, X.; Luo, Y.; Li, H.; Sun, X.; Zhou, M.; Zhou, Y.; Kuai, L.; Xing, M.; Liu, L.; et al. Adverse Events Associated With Anti-IL-23 Agents: Clinical Evidence and Possible Mechanisms. Front. Immunol. 2021, 12, 670398. [Google Scholar] [CrossRef]
- Jones, B.A.; Beamer, M.; Ahmed, S. Fractalkine/CX3CL1: A Potential New Target for Inflammatory Diseases. Mol. Interv. 2010, 10, 263–270. [Google Scholar] [CrossRef]
- Surbatovic, M.; Popovic, N.; Vojvodic, D.; Milosevic, I.; Acimovic, G.; Stojicic, M.; Veljovic, M.; Jevdjic, J.; Djordjevic, D.; Radakovic, S. Cytokine Profile in Severe Gram-Positive and Gram-Negative Abdominal Sepsis. Sci. Rep. 2015, 5, 11355. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Y.; Zhang, T.; Chen, J.; Hu, W.; Sun, G.; Zheng, P. Machine Learning Algorithms for the Early Detection of Bloodstream Infection in Children with Osteoarticular Infections. Front. Pediatr. 2024, 12, 1398713. [Google Scholar] [CrossRef]
- Murri, R.; De Angelis, G.; Antenucci, L.; Fiori, B.; Rinaldi, R.; Fantoni, M.; Damiani, A.; Patarnello, S.; Sanguinetti, M.; Valentini, V.; et al. A Machine Learning Predictive Model of Bloodstream Infection in Hospitalized Patients. Diagnostics 2024, 14, 445. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Liu, L.; Su, T.; Ji, B. Machine Learning Model for the Prediction of Gram-Positive and Gram-Negative Bacterial Bloodstream Infection Based on Routine Laboratory Parameters. BMC Infect. Dis. 2023, 23, 675. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Xu, E.; Zhang, T.; Zhu, Q.; Liu, Y.; Tian, Q. Cytokine-Based Nomogram for Discriminating Viral Pneumonia from Mycoplasma pneumoniae Pneumonia in Children. Diagn. Microbiol. Infect. Dis. 2025, 111, 116611. [Google Scholar] [CrossRef]
- Cheng, Q.; Zhao, F.; Zhang, B.; Zhang, Y.; Cai, L.; Qiao, B.; Hu, Y.; Sun, C. Prognostic Nomogram Incorporating Cytokines for Overall Survival in Patients with Newly Diagnosed Multiple Myeloma. Int. Immunopharmacol. 2021, 99, 108016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, T.; Chen, H.-J.; Chen, N.; Xing, Z.-X.; Fu, X.-Y. Risk Prediction Model for Distinguishing Gram-Positive from Gram-Negative Bacteremia Based on Age and Cytokine Levels: A Retrospective Study. World J. Clin. Cases 2023, 11, 4833–4842. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Zhao, W.; Shi, R.; Zhu, Y.; Wang, Z.; Pan, H.; Wang, D. Development and Validation of a Nomogram for Pre-dicting Gram-Negative Bacterial Infections in Patients with Peritoneal Dialysis-Associated Peritonitis. Heliyon 2023, 9, e18551. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.M.; Chambers, D.A.; Glasgow, R.E. Big Data and Large Sample Size: A Cautionary Note on the Potential for Bias. Clin. Transl. Sci. 2014, 7, 342–346. [Google Scholar] [CrossRef]
- Zhan, X.; Xu, Q.; Zheng, Y.; Lu, G.; Gevaert, O. Reliability-Enhanced Data Cleaning in Biomedical Machine Learning Using Inductiveconformal Prediction. PLoS Comput. Biol. 2025, 21, e1012803. [Google Scholar] [CrossRef]
- Van Steenkiste, T.; Ruyssinck, J.; De Baets, L.; Decruyenaere, J.; De Turck, F.; Ongenae, F.; Dhaene, T. Accurate Prediction of Blood Culture Outcome in the Intensive Care Unit Using Long Short-Term Memory Neural Networks. Artif. Intell. Med. 2019, 97, 38–43. [Google Scholar] [CrossRef]
- Cross, J.L.; Choma, M.A.; Onofrey, J.A. Bias in Medical AI: Implications for Clinical Decision-Making. PLOS Digit. Health 2024, 3, e0000651. [Google Scholar] [CrossRef]
- Miller, C.; Portlock, T.; Nyaga, D.M.; O’Sullivan, J.M. A Review of Model Evaluation Metrics for Machine Learning in Genetics and Genomics. Front. Bioinform. 2024, 4, 1457619. [Google Scholar] [CrossRef]
- Rockenschaub, P.; Hilbert, A.; Kossen, T.; Elbers, P.; Von Dincklage, F.; Madai, V.I.; Frey, D. The Impact of Multi-Institution Datasets on the Generalizability of Machine Learning Prediction Models in the ICU. Crit. Care Med. 2024, 52, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable Generalization Performance of a Deep Learning Model to Detect Pneumonia in Chest Radiographs: A Cross-Sectional Study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [PubMed]

| No Bloodstream Infections (n = 23) | Bloodstream Infections (n = 22) | p-Value | ||
|---|---|---|---|---|
| Age (years), median (IQR) | 58 (12) | 62 (22) | 0.641 # | |
| Gender, n (%) | Female | 5 (0.22) | 3 (0.14) | 0.699 + |
| Male | 18 (0.78) | 19 (0.86) | ||
| ECMO, n (%) | No | 17 (0.74) | 16 (0.73) | 1.000 * |
| Yes | 6 (0.26) | 6 (0.27) | ||
| IMV, n (%) | No | 6 (0.26) | 0 (0.00) | 0.022 + |
| Yes | 17 (0.74) | 22 (1.00) | ||
| Presence of comorbidities, n (%) | No | 2 (0.09) | 1 (0.05) | 1.000 + |
| Yes | 21 (0.91) | 21 (0.95) | ||
| BMI, median (IQR) | 29 (8) | 27 (6) | 0.716 # | |
| Feature | BSIs: Presence vs. Absence 1 | Gram: Negative vs. Positive 1 | ||
|---|---|---|---|---|
| p-Value | Elevated in 2 | p-Value | Elevated in 2 | |
| Traditional Laboratory Markers 3 | ||||
| CRP (mg/dL) | 0.799 (ns) 4 | BSIs | 0.006 (**) | Gram+ |
| PCT (ng/mL) | 0.508 (ns) | No BSIs | 0.953 (ns) | Gram− |
| Lymphocytes (×109/L) | 0.839 (ns) | No BSIs | 0.014 (*) | Gram− |
| Neutrophil % (NE%) | 0.383 (ns) | No BSIs | 0.179 (ns) | Gram+ |
| Cytokines | ||||
| IFN-γ | 0.895 (ns) | BSIs | 0.987 (ns) | Gram+ |
| IL-10 | 0.764 (ns) | No BSIs | 0.579 (ns) | Gram+ |
| IL-17A | 0.860 (ns) | No BSIs | 0.056 (ns) | Gram+ |
| IL-1β | 0.900 (ns) | BSIs | 0.235 (ns) | Gram+ |
| IL-2 | 0.209 (ns) | No BSIs | 0.943 (ns) | Gram− |
| IL-6 | 0.020 (*) | BSIs | 0.553 (ns) | Gram+ |
| IL-8 | 0.173 (ns) | BSIs | 0.391 (ns) | Gram− |
| TNF-α | 0.375 (ns) | BSIs | 1.000 (ns) | Gram− |
| Simple Ratios | ||||
| IL-2/IL-10 | 0.920 (ns) | None 5 | 0.758 (ns) | Gram− |
| IL-6/IL-10 | 0.000 (***) | BSIs | 0.888 (ns) | Gram+ |
| IL-10/TNF-α | 0.244 (ns) | No BSIs | 0.564 (ns) | Gram+ |
| Cytokine | BSI Median (IQR) | Non-BSI Median (IQR) | Abs. Diff. | % Diff. | Fold Change |
|---|---|---|---|---|---|
| ITAC | 59.42 (76.78) | 106.36 (121.28) | 46.93 | −78.98 | 0.56 |
| GM-CSF | 7.42 (3.91) | 8.62 (6.49) | 1.21 | −13.99 | 0.86 |
| Fractalkine | 15.52 (8.71) | 16.30 (6.82) | 0.77 | −4.75 | 0.95 |
| IFN-γ | 31.01 (16.41) | 28.77 (21.94) | 2.24 | 7.79 | 1.08 |
| IL-10 | 24.92 (59.17) | 32.99 (18.45) | 8.06 | −24.45 | 0.76 |
| MIP-3α | 14.34 (10.69) | 13.19 (8.04) | 1.15 | 8.72 | 1.09 |
| IL-12p70 | 5.64 (3.60) | 4.76 (3.04) | 0.88 | 18.56 | 1.19 |
| IL-13 | 2.44 (1.50) | 3.05 (3.17) | 0.61 | −19.90 | 0.80 |
| IL-17A | 25.50 (15.71) | 29.71 (20.33) | 4.20 | −14.15 | 0.86 |
| IL-1β | 1.22 (0.52) | 1.08 (0.67) | 0.14 | 12.64 | 1.13 |
| IL-2 | 3.31 (1.40) | 3.40 (2.87) | 0.09 | −2.64 | 0.97 |
| IL-21 | 10.55 (6.43) | 10.29 (6.71) | 0.26 | 2.49 | 1.02 |
| IL-4 | 8.31 (7.53) | 12.75 (7.90) | 4.44 | −34.81 | 0.65 |
| IL-23 | 232.51 (125.91) | 211.53 (134.44) | 20.98 | 9.92 | 1.10 |
| IL-5 | 6.16 (4.01) | 6.16 (6.44) | 0.00 | 0.02 | 1.00 |
| IL-6 | 21.50 (192.18) | 2.78 (10.84) | 18.71 | 672.65 | 7.73 |
| IL-7 | 17.41 (5.57) | 17.60 (7.41) | 0.19 | −1.10 | 0.99 |
| IL-8 | 16.74 (36.74) | 9.65 (12.54) | 7.09 | 73.52 | 1.74 |
| MIP-1α | 19.00 (11.97) | 15.09 (11.92) | 3.91 | 25.91 | 1.26 |
| MIP-1β | 9.49 (22.53) | 16.58 (15.05) | 7.09 | −42.75 | 0.57 |
| TNF-α | 9.19 (11.55) | 9.03 (4.26) | 0.16 | 1.83 | 1.02 |
| Cytokine | Gram− Median (IQR) | Gram+ Median (IQR) | Abs. Diff. | % Diff. | Fold Change |
|---|---|---|---|---|---|
| ITAC | 33.73 (39.99) | 90.97 (75.69) | 57.24 | −169.72 | 0.37 |
| GM-CSF | 7.32 (2.37) | 7.51 (4.01) | 0.19 | −2.58 | 0.97 |
| Fractalkine | 23.25 (19.38) | 13.38 (4.80) | 9.86 | 42.43 | 1.74 |
| IFN-γ | 28.98 (22.68) | 33.03 (15.15) | 4.05 | −13.96 | 0.88 |
| IL-10 | 21.88 (245.68) | 25.87 (53.37) | 3.99 | −18.21 | 0.85 |
| MIP-3α | 9.60 (34.20) | 14.84 (9.90) | 5.24 | −54.51 | 0.65 |
| IL-12p70 | 4.74 (2.86) | 5.85 (3.25) | 1.11 | −23.44 | 0.81 |
| IL-13 | 2.29 (1.11) | 3.33 (1.57) | 1.05 | −45.73 | 0.69 |
| IL-17A | 23.93 (8.84) | 26.60 (15.79) | 2.67 | −11.16 | 0.90 |
| IL-1β | 1.05 (0.51) | 1.27 (0.49) | 0.21 | −20.23 | 0.83 |
| IL-2 | 3.48 (1.66) | 3.22 (1.25) | 0.25 | 7.28 | 1.08 |
| IL-21 | 11.68 (4.47) | 9.48 (6.21) | 2.21 | 18.89 | 1.23 |
| IL-4 | 11.60 (8.22) | 6.35 (7.46) | 5.25 | 45.24 | 1.83 |
| IL-23 | 223.29 (110.93) | 249.41 (186.69) | 26.12 | −11.70 | 0.90 |
| IL-5 | 5.59 (4.07) | 7.21 (6.65) | 1.62 | −28.91 | 0.78 |
| IL-6 | 3.97 (12207.22) | 21.55 (157.65) | 17.58 | −442.75 | 0.18 |
| IL-7 | 14.47 (4.08) | 18.26 (6.09) | 3.79 | −26.19 | 0.79 |
| IL-8 | 30.58 (642.58) | 13.45 (27.51) | 17.12 | 56.00 | 2.27 |
| MIP-1α | 17.22 (5.03) | 19.99 (20.05) | 2.76 | −16.05 | 0.86 |
| MIP-1β | 8.24 (12.85) | 10.74 (27.35) | 2.51 | −30.45 | 0.77 |
| TNF-α | 13.60 (16.65) | 8.72 (10.74) | 4.89 | 35.92 | 1.56 |
| Cytokine | p-Value |
|---|---|
| ITAC | 0.058 (ns) 1 |
| GM-CSF | 0.704 (ns) |
| Fractalkine | 0.147 (ns) |
| IFN-γ | 0.987 (ns) |
| IL-10 | 0.579 (ns) |
| MIP-3α | 0.433 (ns) |
| IL-12p70 | 0.182 (ns) |
| IL-13 | 0.797 (ns) |
| IL-17A | 0.056 (ns) |
| IL-1β | 0.235 (ns) |
| IL-2 | 0.943 (ns) |
| IL-21 | 0.665 (ns) |
| IL-4 | 0.782 (ns) |
| IL-23 | 0.159 (ns) |
| IL-5 | 0.485 (ns) |
| IL-6 | 0.553 (ns) |
| IL-7 | 0.119 (ns) |
| IL-8 | 0.391 (ns) |
| MIP-1α | 0.633 (ns) |
| MIP-1β | 0.430 (ns) |
| TNF-α | 1.000 (ns) |
| Target Group | Model | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| BSI | kNN | 0.625 | 0.409 | 0.739 |
| Naïve Bayes | 0.620 | 0.455 | 0.652 | |
| Random Forest | 0.505 | 0.591 | 0.522 | |
| SVM | 0.690 | 0.273 | 0.478 | |
| Decision Tree | 0.520 | 0.500 | 0.565 | |
| BSI Ranked: FCBF | kNN | 0.525 | 0.409 | 0.739 |
| Naïve Bayes | 0.575 | 0.682 | 0.522 | |
| Random Forest | 0.610 | 0.545 | 0.522 | |
| SVM | 0.510 | 0.227 | 0.652 | |
| Decision Tree | 0.430 | 0.409 | 0.478 | |
| Gram | kNN | 0.517 | 0.143 | 0.667 |
| Naïve Bayes | 0.533 | 0.571 | 0.533 | |
| Random Forest | 0.567 | 0.286 | 0.667 | |
| SVM | 0.633 | 0.000 | 1.000 | |
| Decision Tree | 0.733 | 0.714 | 0.867 | |
| Gram Ranked: FCBF | kNN | 0.483 | 0.143 | 0.933 |
| Naïve Bayes | 0.700 | 0.571 | 0.667 | |
| Random Forest | 0.733 | 0.286 | 0.867 | |
| SVM | 0.300 | 0.000 | 1.000 | |
| Decision Tree | 0.750 | 0.714 | 0.800 |
| Target Group | Model | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| BSI | kNN | 0.635 | 0.455 | 0.783 |
| Naïve Bayes | n.a. 1 | 0.545 | 0.652 | |
| Random Forest | 0.625 | 0.500 | 0.696 | |
| SVM | 0.530 | 0.455 | 0.739 | |
| Decision Tree | 0.675 | 0.727 | 0.565 | |
| BSI Ranked: FCBF | kNN | 0.665 | 0.636 | 0.652 |
| Naïve Bayes | 0.970 | 0.909 | 0.913 | |
| Random Forest | 0.895 | 0.682 | 0.870 | |
| SVM | 0.520 | 0.500 | 0.652 | |
| Decision Tree | 0.830 | 0.727 | 0.870 | |
| Gram | kNN | 0.517 | 0.143 | 0.800 |
| Naïve Bayes | n.a. 1 | 0.714 | 0.133 | |
| Random Forest | 0.600 | 0.286 | 0.800 | |
| SVM | 0.533 | 0.000 | 1.000 | |
| Decision Tree | 0.567 | 0.429 | 0.733 | |
| Gram Ranked: FCBF | kNN | 0.800 | 0.714 | 0.667 |
| Naïve Bayes | 0.983 | 1.000 | 0.800 | |
| Random Forest | 0.900 | 0.857 | 0.867 | |
| SVM | 0.667 | 0.000 | 0.867 | |
| Decision Tree | 0.800 | 0.714 | 0.867 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, R.; Ramalhete, L.; Von Rekowski, C.P.; Fonseca, T.A.H.; Calado, C.R.C.; Bento, L. Cytokine-Based Insights into Bloodstream Infections and Bacterial Gram Typing in ICU COVID-19 Patients. Metabolites 2025, 15, 204. https://doi.org/10.3390/metabo15030204
Araújo R, Ramalhete L, Von Rekowski CP, Fonseca TAH, Calado CRC, Bento L. Cytokine-Based Insights into Bloodstream Infections and Bacterial Gram Typing in ICU COVID-19 Patients. Metabolites. 2025; 15(3):204. https://doi.org/10.3390/metabo15030204
Chicago/Turabian StyleAraújo, Rúben, Luís Ramalhete, Cristiana P. Von Rekowski, Tiago A. H. Fonseca, Cecília R. C. Calado, and Luís Bento. 2025. "Cytokine-Based Insights into Bloodstream Infections and Bacterial Gram Typing in ICU COVID-19 Patients" Metabolites 15, no. 3: 204. https://doi.org/10.3390/metabo15030204
APA StyleAraújo, R., Ramalhete, L., Von Rekowski, C. P., Fonseca, T. A. H., Calado, C. R. C., & Bento, L. (2025). Cytokine-Based Insights into Bloodstream Infections and Bacterial Gram Typing in ICU COVID-19 Patients. Metabolites, 15(3), 204. https://doi.org/10.3390/metabo15030204








