Abstract
Metabolomics and pharmacometabolomics are at the forefront of precision medicine, serving as powerful tools in drug discovery and development. These approaches help address critical challenges in the field, including high clinical trial failure rates, adverse drug reactions, and interindividual variability in drug response. Comprehensive metabolome profiling enables the elucidation of disease mechanisms, identification of drug targets, optimization of therapeutic strategies, and assessment of drug safety and efficacy. It also supports more informed clinical trial design. This review highlights the pivotal role of metabolomics in advancing precision medicine and aims to broaden the perspectives of emerging scientists entering this complex field. Key analytical techniques–namely mass spectrometry and nuclear magnetic resonance spectroscopy–are discussed for their respective strengths and limitations in metabolite identification, quantitation, and structural elucidation. Additionally, analytical separation technologies such as liquid and gas chromatography, ion mobility spectrometry, capillary electrophoresis, and supercritical fluid chromatography are explored for their potential to enhance metabolome coverage, improve analytical efficiency, and reduce costs. Ongoing advancements in instrumentation and computational tools are helping to overcome major challenges in metabolomics, including metabolome complexity, data analysis and integration, and biomarker validation. These developments continue to expand the applications of metabolomics and pharmacometabolomics in both preclinical and clinical research. Ultimately, this review underscores their translational potential in facilitating drug discovery, mitigating risks in clinical trials, and shaping the future of precision medicine.
1. Introduction
More than 30% of compounds entering Phase II clinical trials fail to progress to Phase III, and nearly 60% of drugs entering Phase III do not advance further [1]. Additionally, only 25–60% of patients in clinical trials exhibit the anticipated response to treatment. These challenges are further exacerbated by adverse drug reactions (ADRs), which significantly contribute to drug-related complications, including increased morbidity and mortality [2]. Between 2020 and the third quarter of 2025, the FDA Adverse Event Reporting System (FAERS) Public Dashboard recorded a total of 9,127,716 drug-related adverse events. Of these, 54% were classified as serious adverse events, and 7.84% were associated with ADR-associated deaths (see Figure 1) [3]. Omics and systems biology technologies are emerging as powerful tools enhance the research and development of novel therapeutics. These approaches support the shift toward personalized medicine by enabling more efficient resource management and improved treatment outcomes. Metabolomics plays a critical role throughout the drug development pipeline—from refining candidate molecular targets to conducting safety assessments. It holds promise for both preclinical evaluation and clinical trials [4,5,6]. Key stages such as sample collection and preparation, separation techniques, instrumental analysis, data processing, and visualization remain complex and present numerous challenges. The physicochemical diversity of metabolites necessitates advanced expertise in analytical methods for their isolation, separation, detection, identification and quantification, along with strong capabilities in data interpretation and integration.
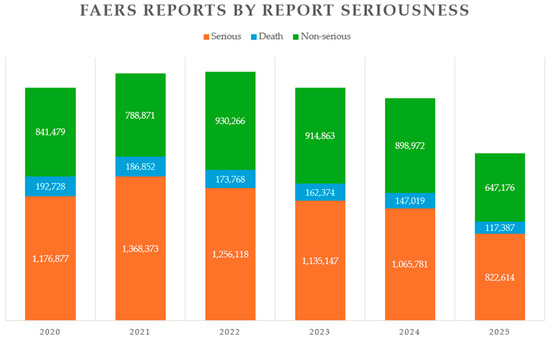
Figure 1.
FDA Adverse Event Reporting System (FAERS) Public Dashboard reports categorized by report seriousness. The statistics cover the period from 2020 through the third quarter of 2025. Serious ADRs are depicted in orange, ADR-related deaths in blue, and non-serious ADRs in green.
1.1. Metabolomics and Pharmacometabolomics in the Era of Precision Medicine
Metabolomics involves the comprehensive profiling and quantification of low-molecular-weight (<1 kDa) molecules in biological fluids, cells, or tissues at a specific time and under defined conditions. A related term, metabonomics, is often described as a subset of metabolomics. It refers to the quantitative measurement of multiparametric, dynamic metabolic responses of a living system to external stimuli [7,8,9,10]. Although the terms metabolomics and metabonomics are often used interchangeably, the literature suggests a subtle distinction between them (Figure 2D). For instance, analyzing the metabolic profiles of two lymphoma subtype cell lines would be considered a metabolomics experiment, whereas examining how these profiles change in response to drug intervention would fall under metabonomics [10,11,12,13,14]. Other related terms—such as metabolic profiling or metabolic phenotyping—are also commonly used in the field.
Metabolomics and proteomics are the omics disciplines most closely linked to the phenotype, as they investigate the products, building blocks, intermediates, and by-products resulting from the regulated expression of genetic information [9,15,16,17]. Metabolome analysis complements other omics analyses by capturing information on metabolite production, fluctuation, modification, translocation, interaction, and turnover. Collectively, all omics layers contribute to a holistic understanding of an individual’s complex phenotype and underlying pathophysiology [7,17,18]. The metabolome also reflects the intricate interplay between genetic factors, nutrition and diet, gut microflora, inheritance, lifestyle, and environmental exposures. The cumulative phenotypic expression of these influences is referred to as the metabotype [19,20,21]. Studying alterations in the metabotype in response to various stimuli, such as diet, gut microbiome changes, pathological states, drug intervention, developmental stages, or environmental factors, can reveal how these influences affect the biochemical processes within the system under investigation.
Metabolomics has grown rapidly over the past 25 years (Figure 3). A PubMed literature search using the Boolean query (metabolomics OR metabonomics OR pharmacometabolomics OR pharmacometabonomics OR “metabolic phenotyping” OR “metabolic profiling”) AND (NMR OR “mass spectrometry”) AND (disease OR health OR “clinical trial” OR “drug discovery”) yields 20,980 results as of 3 October 2025. Figure 3 illustrates the increasing number of articles published per year, reflecting the field’s continued growth and relevance.
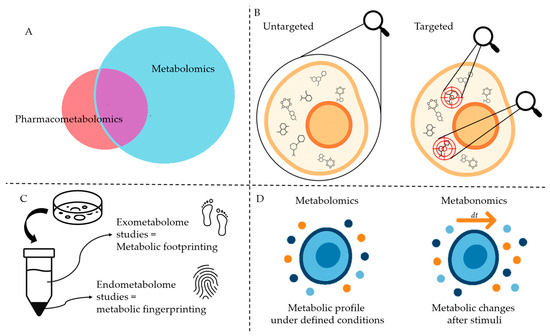
Figure 2.
Schematic representation of terms and definitions presented in this review. (A). Pharmacometabolomics, a subset of Metabolomics (see Figure 4). (B). Targeted and untargeted metabolomics. (C). Metabolic fingerprinting and footprinting. (D). Metabolomics and metabonomics. The image was created using ChatGPT-4.5 as an assisting tool and was further refined by the authors.
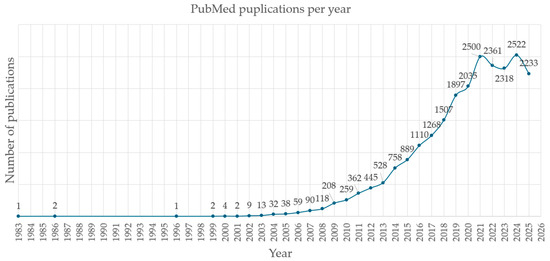
Figure 3.
Yearly number of metabolomics-related publications indexed in PubMed (3 October 2025). Years are plotted on the horizontal axis, and the number of publications on the vertical axis. A steep increase in field-specific publications was observed during the period 2002–2021.
Pharmacometabolomics, an emerging branch of metabolomics (Figure 2A), integrates metabotype data with drug exposure information to better understand and predict treatment outcomes. In other words, pharmacometabolomics leverages the pre-treatment metabolome to interpret post-treatment metabolic changes in response to drug interventions, offering insights into drug efficacy, metabolism, pharmacokinetics, and adverse drug reactions [8,21,22]. This area of research is advancing molecular and clinical pharmacology, drug discovery and development, clinical trial design, and precision medicine. As discussed later in this review, pharmacometabolomics applications underscore its potential as a prognostic tool for optimizing therapeutic strategies [8,21,22] (Figure 4 and Figure 5). Pharmacometabolomics is especially promising in addressing complex, metabolically deregulated diseases, such as cancer, cardiovascular, neuropsychiatric, autoimmune, and neurodegenerative disorders [8,21,22]. These conditions are increasingly understood as overlapping spectra of disease with shared clinical phenotypes, rather than as entirely distinct entities [22,23,24]. Moreover, the growing prevalence of these disorders—combined with the significant proportion of patients who fail to respond to otherwise effective therapies—highlights the urgent need for precise, personalized molecular insights into human diseases, as well as innovative approaches to diagnosis, treatment, and prognosis [21,22].
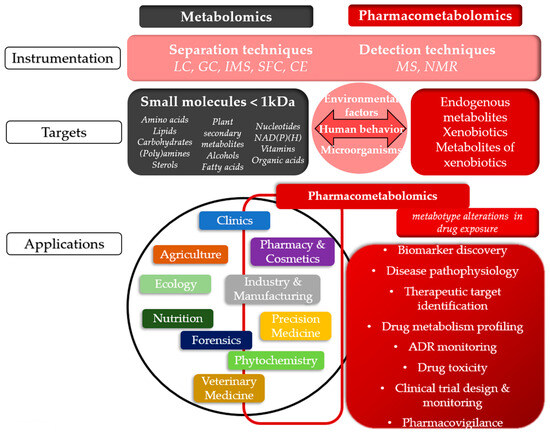
Figure 4.
Comparative overview of Metabolomics and Pharmacometabolomics: Common Techniques, Targets, and Applications. The figure illustrates the shared and specialized aspects of metabolomics and pharmacometabolomics, highlighting their common techniques, chemical targets, and applications across various fields. The concepts shown in black and gray boxes refer to metabolomics, those in red boxes correspond to pharmacometabolomics, while the pink sections represent elements that are shared between both fields. LC: liquid chromatography, GC: gas chromatography, IMS: ion mobility spectrometry, SFC: supercritical fluid chromatography, CE: capillary electrophoresis, MS: mass spectrometry, NMR: nuclear magnetic resonance, ADR: adverse drug reaction.
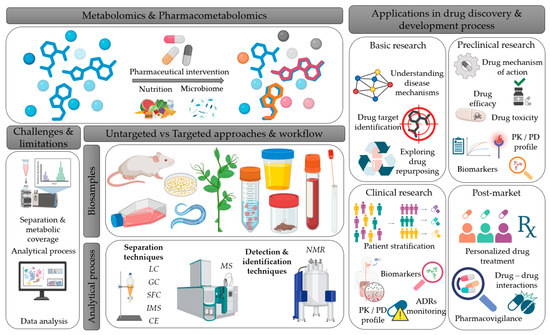
Figure 5.
Schematic overview of Metabolomics and Pharmacometabolomics workflow, linking experimental methods to applications in drug discovery and development. The figure illustrates metabolomics and pharmacometabolomics, along with the factors influencing the metabolic profile (pharmacological treatment, diet, microbiome) and the types of biospecimens that can be analyzed. It also highlights the main analytical techniques, as well as the associated challenges and limitations. Finally, it schematically depicts the applications of metabolomics and pharmacometabolomics across the different stages of drug research and development process. This figure was created with the assistance of BioRender (https://www.biorender.com/?utm_source=moge.ai, accessed on 29 October 2025), an online illustration platform designed for creating scientific figures. LC: liquid chromatography, GC: gas chromatography, IMS: ion mobility spectrometry, SFC: supercritical fluid chromatography, CE: capillary electrophoresis, MS: mass spectrometry, NMR: nuclear magnetic resonance, PK: pharmacokinetics, PD: pharmacodynamics, ADR: adverse drug reaction.
1.2. Currently Applied Metabolomics Strategies and Workflows
In metabolomics, two main strategies are employed: untargeted and targeted (Figure 2B). Untargeted metabolomics, also known as discovery metabolomics, adopts a global-scanning approach aimed at identifying patterns through metabolic fingerprinting and footprinting [7,9,25,26]. This hypothesis-generating strategy distinguishes between the intracellular metabolome (fingerprinting) and the extracellular metabolome (footprinting) (Figure 2C) [7,26,27]. Targeted metabolomics or directed metabolomics, focuses on analyzing a defined set of metabolites. This hypothesis-driven approach generally serves as a confirmatory method that complements untargeted analysis [7,9,25,26].
A general metabolomics workflow consists of data acquisition and data analysis (Figure 6). A typical metabolomics experiment begins with the formulation of a hypothesis, i.e., defining a biological question and designing an appropriate experimental workflow to address it. The next step involves sampling, followed by sample preparation procedures, such as metabolite extraction and preparation for detection [7].
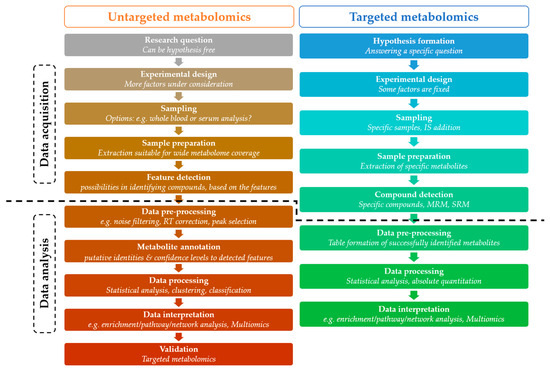
Figure 6.
General metabolomics workflow in targeted and untargeted approaches. The figure illustrates the different modules of targeted and untargeted metabolomics analysis highlighting their basic features and dividing them into data acquisition and data analysis. IS: internal standard, RT: retention time, MRM: multiple reaction monitoring, SRM: single reaction monitoring.
Metabolites are detected using MS and NMR spectroscopy. The acquired raw data undergo preprocessing, which in untargeted MS-based metabolomics includes noise correction, peak detection and alignment, batch effect correction, and compound identification based on spectral databases [7,28]. In NMR-based metabolomics, preprocessing primarily involves spectral alignment, baseline correction, phase adjustment, normalization, and peak deconvolution. Baseline and phase corrections are essential for eliminating instrumental artifacts, while deconvolution algorithms resolve overlapping resonances, particularly in complex biofluids [29,30]. NMR annotation relies on chemical shift reference databases, spin–spin coupling constants, and multidimensional correlation experiments such as COSY (correlation spectroscopy), TOCSY (total correlation spectroscopy), HSQC (heteronuclear single quantum correlation spectroscopy) and HMBC (heteronuclear multiple bond correlation spectroscopy) [30,31]. These 2D NMR techniques enable the elucidation of molecular connectivity and structural relationships (See Table 1).

Table 1.
Two-dimensional NMR techniques commonly used in metabolomics. Comparison of 2D NMR experiments (COSY, TOCSY, HSQC, and HMBC) based on the type of couplings they detect and the structural information they provide. These techniques are essential for metabolite identification and structural elucidation, offering complementary insights into proton–proton and proton–heteronuclear correlations.
Statistical analysis of the data can be univariate or multivariate, revealing significantly altered metabolite levels and uncovering classification or clustering patterns. The significant results obtained from data analysis can be placed in a relevant biological context through enrichment, pathway, and network analyses. Metabolomics data can also be integrated into a multi-omics framework to provide a more holistic metabolic perspective, incorporating potential changes in gene expression, epigenetic regulation or modulation and protein expression, and enzyme activities. The findings can then be validated through follow-up experiments [7,28].
Table 2 summarizes widely used bioinformatics tools, databases, and spectral libraries applied in untargeted MS-based metabolomics workflows. It is important to note that several tools serve multiple functions within the analysis pipeline. For example, MetaboAnalyst supports data preprocessing (e.g., LC-MS spectral processing), peak annotation, statistics analysis and downstream analyses such as enrichment, pathway, and network analysis.

Table 2.
Open access computational tools and databases commonly used in MS-based untargeted metabolomics analyses. The tools are organized in this table based on their data content and the kind of information that they can provide.
2. Analytical Methodologies in Metabolomics and Pharmacometabolomics
Despite significant scientific advances in the field, there is no universally optimal platform for metabotyping. Sensitivity, resolution, throughput, and reproducibility depend on instrumentation and analytical techniques employed in each metabolome study. Selecting an appropriate platform remains a challenging process, demanding knowledge of the potential and applicability of each instrument or method’s strengths and limitations to the specific study goals and biological sample.
MS and NMR spectroscopy are powerful analytical platforms widely used for the detection, identification, and structural elucidation of small molecules in biological samples. Given the vast number of metabolites and their remarkable physiochemical diversity, coupling MS and NMR spectroscopy with separation techniques is essential [7,8,16,25,26,31]. Commonly used separation methods include GC, LC, SFC, IMS, and CE (Table 3). The choice of analytical platform and instrumentation depends on factors, such as sample type, experimental design, cost and available expertise. The following sections discuss the selected separation techniques provide an overview of MS and NMR and explore the prospects of their combination [48,49,50,51,52,53,54].

Table 3.
Comparison of separation techniques based on phase properties and target analytes. The table summarizes key separation techniques (RPLC, HILIC, GC, SFC, IMS, CE) highlighting differences in stationary and mobile phases and compound selectivity based on polarity or ion properties.
2.1. Biosamples and Sample Preparation
Biospecimens are highly complex analytical samples containing compounds that span a wide range of volatility, solubility, hydrophilicity, and concentration levels [55]. They comprise an astonishing diversity of metabolites with varying polarities, and average polarity depending on both the biospecimen type and the sample preparation protocol [19].
Metabolome analysis can be conducted on a plethora of biospecimens, including biofluids (such as plasma, serum, cerebrospinal fluid, urine, saliva, sweat, maternal milk, semen, and bile), as well as stool, breath, tissue, and cell cultures samples [7,9]. Cell cultures are particularly versatile, enabling simultaneous fingerprint (intracellular) and footprint (extracellular medium) profiling [56]. Sample preparation methods, such as homogenization, cell lysis, filtration, extraction, dilution, centrifugation and protein precipitation, or combination thereof, depend on the sample’s nature, complexity, and the objectives of the study [7,50,57]. Protein precipitation is a crucial step prior to analysis, as it enhances chromatographic separation and metabolite detection and identification. Protein precipitation and extraction can also be performed simultaneously by selecting the proper extraction solvent [58,59].
Between sample collection and analysis, metabolites in biospecimens are vulnerable to enzymatic degradation and chemical alterations caused bytotemperature fluctuations, exposure to oxygen, or light-induced structural changes. Quenching is the process of stabilizing and solubilizing metabolites to prevent such degradation [7,52,60]. This step is of great importance in studies of intracellular metabolome or in single-cell metabolomics, but less critical for extracellular material (i.e., plasma or urine) [7,60]. Proper storage conditions, such as keeping the samples in low temperature (freezer at −80 °C), dark, and almost full vials, are essential to avoid degradation and ensure data reliability [50,57].
The choice of extraction methods and solvents depends on the sample properties, the targeted metabolites/metabolite classes, the available or selected separation techniques and detection methods. Sample collection, acquisition, and preparation should be conducted properly to minimize losses of metabolites or potential contamination, thus, enabling metabolomic data integrity [57].
2.2. Separation Techniques
2.2.1. Liquid Chromatography
LC is widely used in metabolomics due to the variety of available columns in length, width, and chemical characteristics of stationary phases, permitting broad metabolic coverage [52,61]. Τhe packing material of the column and the selection of the elution solvents define separation capacity. High-performance LC (HPLC) columns have internal diameters of 2.0–4.6 mm and 5–25 cm length, filled with 3–5 μm particles. Sub-2 μm particles have lately been gaining ground in metabolomics, introducing Ultra HPLC (UHPLC), which improves peak resolution, separation capacity, and metabolic coverage, while reducing the time of the analysis in comparison to HPLC [26,49,50,51,52,62]. HPLC coupled via electrospray ionization with MS (ESI-MS) is a common platform for metabotyping because it allows for the separation of compounds with very different polarities, depending on the system setup and the applied condition, such as the elution system, column selection, and instrument parameters. However, none of the current separation techniques are capable of separating efficiently all the compounds in a sample under investigation. A multidimensional approach, such as combining two (or even three) separation techniques, including Hydrophilic Interaction Liquid Chromatography (HILIC), reversed phase (RP), and/or ion pair chromatography (IPC), provides much wider metabolome coverage. RPLC is particularly effective for separating non-polar, lipophilic metabolites. The most widely used stationary phases in RPLC are non-polar, composed of silica gel modified with C8 or C18 aliphatic chains, while the mobile phase is of a higher polarity. In RPLC, non-polar or less polar compounds are retained longer by the non-polar stationary phase, while more polar compounds are eluted faster by the polar solvent system of the mobile phase [48,51,61,62,63].
On the other hand, polar aqueous compounds, such as those in urine samples, which often exhibit poor peak resolution in RPLC, are usually separated by HILIC [48,49,58]. HILIC utilizes a polar, hydrophilic stationary phase and a polar organic or aprotic mobile phase, such as mixtures of water and methanol or/and acetonitrile. Retention mechanisms involve partitioning, electrostatic interactions, and hydrogen bonding [63]. While RPLC produces narrower and more symmetric chromatograms, HILIC requires longer column equilibration times and has lower loading capacity, which can reduce sensitivity [63]. In cases where HILIC is not efficiently separating targeted metabolites, other options should be tried such as normal phase (U)HPLC, IPC or ion exchange chromatography. Usually, RHLC and HILIC are undertaken separately from one another. During the past decade, attempts were made to mechanically combine the two methodologies, including parallel separation using both columns, column switching, and serial column-coupling [58].
However, LC is not without limitations. Challenges include long runs, high solvent consumption, and the risk of artifact formation, which can lead to metabolite loss or false detection. Ion suppression due to co-eluted metabolites reduces detector response and increases background noise [26,59,64]. In addition, variability in sample preparation, extraction, chromatographic conditions, and instrument settings increases batch effect that makes the comparison of metabolomes across studies difficult [26,64].
2.2.2. Gas Chromatography
In GC, the sample is vaporized upon injection and enters the instrumentation in the gas phase. The mobile phase consists of an inert gas, such as nitrogen or helium, while the stationary phase generally consists of a thin liquid film. Analytical columns in GC are placed inside a heated oven, as the heat aids to elute the compounds of different boiling points. Columns can be either packed columns or capillary columns. The use of the latter has prevailed, as they ensure improved separation performance. The length of capillary columns is noticeably greater than the length of columns used in LC. (e.g., a column in GC can be several meters long, whereas in LC it is usually between 10 and 25 cm). Physicochemical properties of the analytes, such as boiling point and polarity, determine the partitioning between the stationary and the mobile phase. The better a compound is retained in the stationary phase, the longer its elution time will be from the column, thus achieving separation [51,65].
GC is appropriate for the separation of low molecular weight (semi)volatile chemical entities. Highly polar molecules such as amino acids, sugars, and small organic acids can be analyzed by GC. Derivatization is essential to convert these compounds into their volatile derivatives but also to prevent the conversion of compounds into their stereoisomers due to high temperature (e.g., carbohydrate cyclization). Therefore, chemical processing of molecules via derivatization improves chromatographic performance, as it increases thermal stability and enhances peak shape, resolution, and intensity [51,53,66].
GC presents some advantages over LC, such as efficiency, chromatographic resolution, and high repeatability, both in retention time and response [67]. At the same time, operating and consumable costs are lower compared to LC. As for ionization techniques, GC can employ either hard techniques such as electron impact (EI), or the soft ionization technique of ESI. EI, compared to ESI, produces complex, yet reproducible spectra, allowing for more accurate identification of a compound at the MS/MS level, especially when high-resolution mass analyzers are used, such as time-of-flight (TOF) instrumentation. There are publicly available GC spectral libraries, such as NIST and Fiehnlib, in which experimentally derived spectra can be matched, as well as free and reliable deconvolution software, such as AMDIS. As a result, peak deconvolution of co-eluted compounds in GC is more accurate than in LC [67,68].
However, there are also limitations. Initially, GC-MS is restricted to the analysis of semi-volatile, volatile, and thermally stable compounds, offering limited metabolite coverage compared to LC-MS [53]. Many biological metabolites, such as amino acids, carbohydrates, organic acids, nucleotides, and lipids are not naturally volatile. Moreover, living organisms, and consequently the biomolecules that compose them, have evolved to remain stable only within a defined physiological temperature range. Furthermore, more extensive sample processing is required prior to analysis due to derivatization. The sample preparation time and the conditions required for derivatization may lead to the destabilization of metabolites and the creation of artifacts in biological samples [53,69]. In recent years, multidimensional GC×GC approaches are being explored to achieve better separation and reduce co-eluted compounds in highly complex mixtures like biosamples [53,69].
Volatile compounds are now considered a separate field, volatolomics, in which GC is the dominant separation technique before applying MS. Volatolomics finds applications in life sciences and beyond, as volatile components may originate from organisms (mammals, plants, insects), or from human or natural activity (industry, natural environment). Certain biofluids are indicative candidates for such analyses, including urine, sweat, and exhaled breath [54,65].
2.2.3. Supercritical Fluid Chromatography
SFC was first reported by Klesper and colleagues in 1962, but it gained traction in the late 1980s, with the introduction of Ultra High Performance SFC, which employs sub-2 μm columns [70]. The main solvent in SFC is supercritical CO2 under high pressure, though combinations with co-solvents, such as ethanol and acetonitrile, enable adjustments of retention times. The main advantage of SFC is its ability to simultaneously separate polar and non-polar metabolites, thus offering wide metabolome coverage in a single run [20,48,70]. SFC is compatible with MS and can be directly coupled with a supercritical fluid extraction, providing efficient, automated extraction and separation from unprocessed biosamples, such as dried blood or serum spots, due to the high diffusivity and low viscosity of supercritical CO2. Moreover, SFC is ecofriendly and cost-effective, due to the use of recyclable CO2 instead of organic and chlorinated solvents. SFC is also applied in chiral separation, particularly in pharmaceutical studies of metabolism and toxicity [20,48,70]. While SFC shows promise in lipidomics and urine analysis [59], its application in metabolomics is limited compared to LC, due to the lack of comprehensive data libraries, including retention times and spectra, restricted stationary phase options, and challenges in quantitation [59].
2.2.4. Ion Mobility Spectrometry
IMS separates ions based on their mobility in buffer gas, under an electric field, whereas the ion mobilities depend on their size, charge, and shape. Smaller and more compact ions exhibit higher mobility and thus attain higher drift velocity under the same electric field. Common types of IMS, based on ion separation, include Drift Tube IMS (DTIMS), which uses a constant low electric field, applied in a drift tube filled with a high-pressure buffer gas, such as nitrogen or helium, whilst traveling wave IMS (TWIMS) uses an oscillating electric field. Ion separation in IMS is governed by the probability of collisions with buffer gas molecules, quantified as the collisional cross section (CCS, Å2), which reflects the ion’s size, shape, and charge distribution [49,71,72]. IMS can be hyphenated to LC-MS, enabling separation of isobaric compounds, and decreasing noise interference, thus improving specificity and accuracy of the measurement. The LC-IMS-MS approach enables the detection of low-abundant small molecules, improving library matching and reducing false discovery rates. This occurs because the combination of two different separation techniques enhances the probability of accurate metabolite annotation, as more physicochemical properties and molecular characteristics of each compound are taken into account.
IMS is a rapid technique, permitting high-throughput separation in milliseconds, compatible with TOF and Orbitrap analyzers. Although SFC-IMS-MS has been used in a few studies until now, its potential for wider coverage of metabolome is promising. IMS coupled with LC-MS enhances metabolic tissue imaging, unveiling cellular metabolic heterogeneity and complexity. However, advanced bioinformatics tools are needed to exploit the wealth of such data [71,72,73]. Moreover, LC-IMS-MS can differentiate structural isomers, such as phase Ι drug metabolites that have undergone hydroxylation at different sites and distinguish drugs and their metabolic derivatives from endogenous isobaric metabolites [49,72]. IMS coupled to MS and Infrared Spectroscopy (IR) further enables gas-phase structural characterization of metabolites and peptides [73], while IMS alone can also shed light onto protein–drug interactions [74].
2.2.5. Capillary Electrophoresis
In CE, the analyte is injected into a narrow, fused silica capillary filled with a background electrolyte in the aqueous mobile phase. Separation occurs through the application of an electric field, and analytes migrate through the capillary, in free solution, at a rate that depends on their charge, size, and shape [48,51,75,76,77]. Polar ionogenic metabolites are ideal for CE-MS analysis [77].
It should be noted that separation in both IMS and CE depends on charge, size, and shape. The main differences between the two techniques lie in the fact that in CE, the separation occurs in a liquid phase, whereas in IMS it takes place in the gas phase. The migration time of an ion through the capillary depends on its electrophoretic mobility, the length of the capillary, the applied voltage, and the electro-osmotic mobility generated by the charge on the capillary wall [48,75]. That electro-osmotic flow is responsible for the migration of neutral analytes [51]. On the other hand, in IMS, ion separation is based on their mobility in the gas phase, through collisions with the buffer gas.
CE can be coupled with MS, although it is more technically challenging to set up than LC, and is not suitable for separating hydrophobic compounds. The separation efficiency and sensitivity of CE are very high compared to chromatographic-based methods, as there is no mass transfer (partitioning) between phases [77,78]. Additionally, injection volumes in CE are in the nanoliter range, meaning that only microliter-level volumes of biosamples are required [51,77,78]. The use of aqueous solvents makes the technique more environmentally friendly compared to LC, where costly organic solvents are often used. CE analysis is enantioselective, which is important for the separation of stereoisomers [52,79]. CE is performed across different levels of biochemical analysis, such as genomics and proteomics, which may facilitate multiomics data integration in the future.
2.3. Instrumental Analysis for Detection and Identification of Metabolites
2.3.1. Mass Spectrometry
MS is the preferred method for qualitative and quantitative metabolome profiling [61]. The molecules in the sample under investigation are converted into ionized particles (cations or anions). The ionization source can vary. The generated ions are accelerated and separated based on their mass-to-charge ratio (m/z), using electric and/or magnetic fields. The method of separation depends on the type of mass analyzer used. The separated ions reach a detector, which records their intensities and m/z values, generating the mass spectrum [16,28,51]. It is a common practice in metabolomics to acquire MS spectra of the same sample in both negative and positive ionization mode, thus enhancing metabolome coverage.
ESI, being a soft ionization method, is most widely employed in metabolomics, showing little or no in-source fragmentation. Other ionization techniques include atmospheric pressure photoionization (APPI), atmospheric-pressure chemical ionization (APCI) and matrix-assisted laser desorption ionization (MALDI). The combination of different ionization methods can expand metabolic coverage [26,61,62]. High mass accuracy is important in metabolomics, as many different metabolites with similar molecular weights may co-elute [62]. Common analyzers include Orbitrap, single or triple quadrupole and quadrupole ion trap in targeted metabolomics, and Orbitrap, Fourier Transform Ion Cyclotron Resonance (FT-ICR), TOF, and quadrupole TOF (QTOF) in untargeted metabotyping. QTRAP analyzers, which resemble a triple quadrupole, except that the third quadrupole has been replaced by an ion trap, are also utilized [16,61,64]. Samples are typically introduced into the mass spectrometer after chromatographic or electrophoretic separation [63], as direct infusion analysis often causes ion suppression, causing significant loss of signal [61]. Despite its high cost of the analysis and limitations in distinguishing isobaric compounds, MS remains a cornerstone of metabolomics [62].
MS spectra are not always reproducible among different instrumental platforms and also not between the same instruments with different instrumental parameters setup. Typical MS-based approaches in metabolomics apply statistical methods to identify metabolites whose concentrations are significantly different between control and studied groups. LC is typically coupled to tandem MS, where the instrument has two mass analyzers, the first measures m/z of ionized metabolites (MS1 scan), then these ions are entering a collision cell to collide with gas molecules producing ion fragments, which are then analyzed in the second mass analyzer (m/z of fragments, MS/MS) [80]. The combined MS/MS data, retention time, and precursor ion m/z enable more accurate compound identification. The precursor ion is selected either in a dependent or independent manner. In DDA approaches (data-dependent acquisition), precursor ions are automatically selected based on intensity thresholds after a full MS1 scan, whereas in data-independent acquisition (DIA) approach, fragmentation occurs to all precursor ions within a specified m/z range, regardless of intensity (Figure 7). DIA MS/MS spectra are highly complex because they contain fragment ion from multiple precursor ions, making it challenging to match precursor and fragment ions and therefore identify the metabolite [9,56,80,81]. The current application of MS/MS in state-of-the-art MS-based untargeted metabolomics approaches provides an affordable and reliable tool for metabolite identification, reducing significantly the required time and effort.
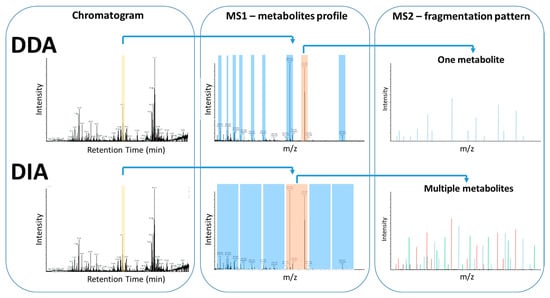
Figure 7.
Comparison of DDA and DIA data acquisition methods. Both approaches, in most cases, acquire a MS1 scan of a mixture of molecules injected into MS, following LC separation. In the case of DDA, individual precursor ions are selected, isolated, and fragmented to acquire a corresponding MS/MS. In the case of DIA, instead of individual precursor ions, a pool of precursor ions is selected for a specific m/z window and all of them are fragmented simultaneously to acquire a consensus MS/MS.
Metabolite identification framework involves the following steps: metabolite annotation, mass-based search, spectral interpretation, and spectral matching [79]. Ion annotation is the process of clustering compounds by their elution profiles. Extracted ion chromatograms (XICs) group peaks likely originating from the same compound, aiding in resolving overlapping peaks, or identifying potential isomers [79,81]. Mass-based searches deploy m/z values to create a list of putative identifications, considering adduct ions, neutral losses, and isotopes. Spectral interpretation entails in silico fragmentation predictions to match experimental spectra with a hypothetical MS/MS spectra “collection”. Findings are verified via spectral matching, where the experimental MS/MS spectra are aligned with those of reference compounds in spectral databases [55,79].
MS-based metabolomics faces several challenges, including calculating false discovery rates (FDR) unpredictable fragmentation patterns, rather low reproducibility in LC-MS/MS [55,81]. Low-abundance metabolites are often disregarded due to software limitations. An integrated feature extraction strategy in LC-MS/MS, based on classifying detected features by confidence levels, can improve metabolic coverage of low-abundance compounds. Such an approach could potentially reveal significant early-stage disease metabolic biomarkers [6,82].
2.3.2. Nuclear Magnetic Resonance Spectroscopy
As noted in G. Watson’s book, NMR uses radiofrequency waves to stimulate atomic nuclei (e.g., 1H, 13C, 15N, 19F, 31P), for their parallel spin to alternate in antiparallel, when magnetic field is applied, providing detailed structural information. Multidimensional NMR analysis combines spectra from different nuclei, permitting the discovery of novel compounds. NMR is extensively used in pharmaceutical science and industry to quantify active drugs and their metabolites in complex mixtures like biospecimens, natural product extracts and drug formulations [51].
NMR is among the most used analytical techniques in metabolomics, due to several advantages. In NMR, sample preparation is rather simple, it is a non-destructive analysis that enables quantitation of metabolites, as signal intensity is directly correlated to metabolite concentration. As for the equipment, the system is highly automated, offering stable performance and requiring less frequent maintenance compared to mass spectrometry instruments. Considering rapid performance, automation and precise quantitation, NMR is suitable for large-scale population metabolomics studies. Generally used in untargeted analysis, NMR, unlike MS, provides high reproducibility, which is maintained even between different instruments [19]. However, NMR’s sensitivity is relatively low, typically characterizing up to 200 metabolites per scan at concentrations >1 μM, highly polar compounds and inorganic, metal ions included [31]. The latest technological achievements in NMR equipment, such as high-frequency magnets (>600 MHz) and the development of cryoprobes have increased sensitivity and resolution and decreased limit of detection [83]. The technique is applicable to semi-solid and solid samples (e.g., cells, tissues, organs, biopsies), through magic-angle spinning (MAS) NMR [84]. Moreover, NMR enables in vitro and in vivo fluxomics analyses, permitting real-time evaluation of drug response at the molecular level, considering its capacity for precise quantitation [31,83,85].
2.4. Quantitation in LC-MS/MS and NMR
Quantitation in metabolomics depends on the orthogonal approach used in each experiment. In targeted metabolomics, commercially available isotope-labeled internal standards (e.g., 2H, 13C, 15N and 18O) are used for validation and absolute quantitation. The limited availability and high cost of these standards confine the practicality of this approach. However, isotope-labeled internal standards serve in method development and validation, such as selectivity, limits of detection and quantitation, reproducibility, and accuracy [16,60,64]. Untargeted metabolomics relies on relative quantitation comparing spectral patterns and intensities across samples to detect differential features or distinctive marks [60,64].
NMR spectroscopy is inherently quantitative, with signal intensity proportional to the number of nuclei in each molecule. That is a salient advantage of NMR, factoring in the rapidness of the technique. qNMR (quantitative) methods include relative and absolute quantitation. Relative qNMR is widely used, enabling differential comparisons in human metabolome studies. Observed changes in metabolite levels are not assessed individually, i.e., metabolite-by-metabolite, but each metabolite is studied in correlation with others. Another qNMR application in drug discovery is detecting metabolome changes before and after treatment, thus defining pharmacological response in early stages of treatment, providing assessments of drug toxicity. Absolute qNMR is independent of the molecules in question and employs internal standards for precise concentration measurements [86,87].
2.5. MS and NMR Combination
While most metabolomic studies employ either MS or NMR, combining the two techniques holds great promise, offering complementary strengths. MS and NMR synergy widens metabolome coverage, improves measurement accuracy, enabling monitoring of low-abundant metabolites, all with time- and cost-effectiveness. MS provides compound identification based on accurate mass determination and molecular fragmentation pattern, while NMR enables precise quantitation. Moreover, the latter enables structural elucidation of unidentified metabolites, using chemical shifts and coupling constants, and information on molecular dynamics. This combination is popular in natural product-based drug discovery studies, allowing the characterization of unknown metabolites. These metabolites are found either in the extracts of the primary material, or as xenobiotic metabolites within the biological system in which their effects are being investigated.
However, combining MS and NMR data can be challenging because of the fundamental differences in data acquisition that are based on different physicochemical properties of the compounds. Chemical derivatization with agents, such as 15N-cholamine to tag specific functional groups has been used to bridge this gap [88,89]. 15N-cholaminecarries both an NMR-detectable isotope and permanent charge that increases sensitivity of its MS detection. This approach could find applications in biomarker discovery and identification of unknown endogenous or xenobiotic metabolites [90]. Despite these efforts, the unification of MS and NMR spectral databases and the development of analytical and computational tools, such as NMR/MS translator and SUMMIT MS/NMR, are needed to allow and simplify data integration and application [89] (Table 4). Another factor that limits the widespread application of combined MS and NMR approaches is the high cost of acquisition and maintenance of each instrumentation and their consumables, as well as the fact that it is uncommon for a single laboratory to specialize in both techniques simultaneously.

Table 4.
NMR and MS comparison. The table summarizes the basic differences between the two common techniques used in metabolomics data acquisition.
3. Metabolomic Applications in Drug Discovery and Development
In the era of precision, phenomic medicine, systems biology, and systems pharmacology, omics technologies are pivotal for deciphering the molecular basis of diseases. Among them, metabolomics holds particular for diagnosing and monitoring diseases, assessing disease progression, predicting onset or outcomes, and guiding the design of therapeutic strategies and interventions. By comparing metabolites levels and identifying metabolic signatures between diseased and healthy individuals, metabolomics can reveal potential disease biomarkers in various biospecimens. These biomarkers offer diagnostic, prognostic, monitoring, and mechanistic insights. Metabolomics approaches have been applied across a wide range of diseases, including diabetes, obesity, cardiovascular diseases, neurodegenerative disorders, and cancer [91,92,93,94,95,96,97,98]. Moreover, emerging areas, such as exposome research, longevity studies, extracellular vehicle research, and pharmacometabolomics are gaining increasing attention. The exposome encompasses all environmental exposures an individual experiences throughout their life; consequently, pharmaceutical interventions are also considered forms of external chemical exposure [5,6,13,17,18,71,88,99,100]. For example, metabolic reprogramming, a hallmark of cancer, differs between cancer types and among individuals. Metabolomics can guide the development of more effective metabolism-based anticancer therapies. In addition, metabolic reprogramming plays a role in modulating tumor growth [101,102], immune evasion, and metastasis [103,104], further highlighting its therapeutic relevance [17,18,105].
Metabotype information, when integrated with other omics data (genomics, transcriptomics, proteomics) and clinical data, offers a comprehensive view of drug exposure effects. This integration helps clarify mechanisms underlying disease pathophysiology, heterogeneity, and interindividual variability in pharmacotherapy outcomes, as well as pharmacokinetics and pharmacodynamic (PK/PD) properties [2,4,6,9,15,16,18,22,24,106]. Importantly, metabolomics reflects an individual’s physiology, lifestyle, and environmental exposures, making it a valuable tool in data interpretation [4,15]. Comparative analyses of pre- and post- treatment metabotypes provide molecular-level insights into drug mechanisms of action, impacted metabolic pathways, and efficacy networks [6,15,22,24,106,107]. In addition, off-target drug identification through multi-omics data integration (e.g., structural proteomics, transcriptomics) enhances our understanding of drug mechanisms and potential adverse reactions [108].
Pharmacometabolomics can support toxicology by elucidating toxicity mechanisms within complex metabolic networks and identifying the metabolic pathways affected by drug exposure. Endogenous metabolites provide baseline information about an individual’s physiological state, serving as a foundation for predicting drug response and treatment outcomes. On the other hand, exogenous metabolomics-which includes xenobiotics, such as environmental pollutants (air, water), nutrients, and pharmateuticals—helps map the metabolic fate of a drug molecule. This can reveal the formation of toxic metabolites and chemically define the causes of toxicity or interindividual variability in treatment response. This approach is particularly valuable for clarifying chronic, acute, and organ-specific toxicities, especially when integrated with pharmacogenomic data [2,5,15,106,107,109,110].
The microbiome can also have an impact on disease and drug response, affecting PK/PD properties of several drugs, such as metformin, antibiotics, antihypertensives and anticancer immunotherapeutics [17,100]. Metabolomics, by incorporating microbiome data, can assess its impact on drug treatment.
Pharmacometabolomics application is vital in preclinical and clinical phase of drug discovery and development. Pre- and post-dose comparative analyses provide drug response prediction information and drug effect assessment. For example, pre-dose biofluid samples in clinical trials, compared with post-dose samples, and combined with clinical data, may reveal predictive biomarkers for toxicity or efficacy. Short term post-dose biofluid samples are used for pharmacokinetic studies, whereas long term post-dose biofluids can be exploited in pharmacovigilance studies [2].
In pre-clinical stages of drug discovery, pharmacometabolomic studies provide information concerning drug efficacy, safety, metabolism and pharmacokinetics [2,8,79]. The overlap of metabolites across species can be both a challenge and an advantage. For example, while it complicates the identification of specific metabolite origins in microbiome studies, it increases the reliability of animal models used in the early stages of drug research and development, for pharmacodynamic and pharmacokinetic properties, ADME (absorption, distribution, metabolism, excretion) assessments and toxicological and teratogenic evaluations. Moreover, putative biomarkers detected in animal models, can predict treatment efficacy, toxicity, disease progression, or prognosis after the intervention, contributing to prioritization of drug candidates and the exclusion of likely failures in later-phase development [2,18,55,100,110,111].
Drug resistance, particularly in cancer, is another significant area. Intratumor cell heterogeneity may result in different treatment responses, and single-cell metabolomics is a promising approach in discriminating treatment-responsive from non-responsive cells enabling personalized therapy [6,111,112,113].
Biomarkers are essential throughout drug research and development, predicting drug efficacy, toxicity, and response [99]. Preclinically detected putative predictive biomarkers allow early observation of drug properties, while their validation improves the efficacy of later-phase clinical trials [4,23,110,114]. Mass spectrometry allows high-throughput analysis of endogenous and exogenous metabolites, facilitating simultaneous assessment of toxicity, efficacy, and drug metabolites. This approach also identifies molecular targets and biochemical pathways affected by drugs, even not directly linked to them [6,101,102]. Integrating pharmacometabolomics and pharmacogenomics findings can prevent drug–drug interactions, define therapeutic windows and enable precise dose selection, based on patients’ metabolic profiles or disease stages [4,6,99,114].
Metabolomics and pharmacometabolomics are important tools in clinical trials, by directing participant selection in clinical trials, stratifying patients by disease stage and patients’ treatment evaluation status [4,99,100,114]. In addition, these approaches could mitigate bioethical concerns about clinical trials, by restraining drug testing and exposure duration to patients likely to benefit and identifying toxicity potential early. Pharmacometabolomics approaches, by reducing clinical trial duration, risk and cost of drug development, accelerate the development of safer, more effective new therapies [4,114]. Pharmacometabolomics can also be applied in drug rescue and drug repurposing studies [4,6,114]. Continued advancements in these fields will further refine their applications, benefiting both research and patient care.
As the global population ages, polypharmacy, and ADRs are responsible for approximately 5% of urgent hospital admissions [115,116]. Moreover, the study of Bouvy et al., 2015 indicated that 5% of patients experience an ADR during hospitalization [116]. Beeler et al., 2023, indicate that approximately 2.3% of hospital admissions per year are caused by ADRs [117]. According to FAERS, approximately 172,000 deaths per year were associated with ADRs between 2020 and 2024 (Figure 1). Pharmacometabolomics can intensify pharmacovigilance (Phase IV drug development), by analyzing non-invasive samples in large-scale studies, identifying patients at risk for ADRs early, and improving drug safety.
The following table (Table 5) summarizes selected studies retrieved from PubMed, demonstrating the application of metabolomics in various stages of drug discovery and development. The table reports the analytical platform used (MS or NMR), the sample type, and the specific objective of the metabolomics approach in each case, such as biomarker discovery, toxicity assessment, or drug mechanism elucidation.

Table 5.
Selected publications from PubMed concerning metabolomics applications in drug discovery and development. The table lists the applications, whereas it specifies the disease or condition to which they are applied. The table also indicates whether MS or NMR was employed as the analytical platform and provides corresponding literature references.
4. Current Limitations and Challenges
A major challenge in metabolomics and pharmacometabolomics lies in the chemical complexity of metabolomes. Small molecules differ in carbon chain length, functional groups, and ligands, resulting in a wide range of physicochemical properties, such as lipophilicity and chirality [17]. These variations, combined with the large number of metabolites present in biological samples, complicate their separation, detection, and analysis.
Limitations in current separation and detection techniques include the need for specialized methods tailored to different metabolite classes (e.g., hydrophobic, hydrophilic), as well as the high costs associated instrumentation, solvents, and internal standards [26,59,61,62,63,64]. Another significant challenge in MS-based metabolomics is ionization efficiency, which depends on both the analyte’s properties and the composition of the mobile phase [140]. Internal standards are typically used to mitigate this issue.
While MS offers high sensitivity and a broad analytical range-enabling the quantification of both low- and high-abundance metabolites-low-abundance signals can be masked by more dominant ones. Errors can occur at any stage of a metabolomics experiment, including study design, sample collection and storage, sample preparation, data acquisition, analysis, and interpretation. External factors, such as compound volatility and environmental conditions (e.g., temperature, humidity) may also impact metabolite detection [7,99]. In pharmacometabolomics, physiological variables, such as gender, age, diet, gut microbiota, and lifestyle further complicate data analysis [17,18]. Additionally, temporal and tissue-specific variations, as well as intracellular compartmentalization, must be considered for accurate biological interpretation [99,141].
Batch effects represent another major limitation, especially in cross-study integration and comparison. These refer to systematic variations introduced during sample collection, storage, preparation, analysis, and are unrelated to biological variability. If not properly addressed, batch effects can distort statistical analyses and lead to misleading conclusions-a common issue in all high-throughput omics studies.
Metabolomics and pharmacometabolomics are inherently multidisciplinary fields, requiring expertise in analytical chemistry, statistics, computational biology, and biological interpretation. Metabolite annotation is challenging because many compounds are involved in multiple metabolic pathways [141]. The high throughput and heterogenous nature of the resulting data demands advanced computational tools for processing, statistical analysis, compound annotation, and biochemical interpretation (Table 2). Platforms like MetaboAnalyst and MZMine offer ‘’end-to-end” solutions, while custom programming remain essential for tailored workflows. Nonetheless, multi-omics data visualization and integration continue to present major challenges [99,100,141,142].
Compound identification remains a major challenge in untargeted metabolomics. Despite the availability of spectral databases (e.g., HMDB, MetaboLights, BMRB, Bermingham Metabolite Library) and computational tools (see Table 2), many metabolites remain unannotated after data analysis [18,99,100,141]. This highlights the ongoing need to expand publicly available datasets and to develop more advanced computational tools to improve metabolite annotation and identification.
In recent years, artificial intelligence (AI) has significantly advanced metabolite annotation by enabling automated spectral interpretation and structure prediction through machine learning (ML) and deep learning (DL) approaches. AI models trained on thousands of reference MS/MS spectra can recognize characteristic fragmentation patterns, predict in silico spectra from molecular structures, and match them against experimental data. These models also perform feature prioritization and confidence scoring, enabling researchers to focus on the most reliable annotations [143].
Tools such as SIRIUS, via its CSI:FingerID module, employ ML-based fragmentation tree computation and molecular fingerprint prediction. Additionally, the extension CANOPUS uses DLalgorithms for chemical class prediction [34]. In parallel, NetID applies network inference and probabilistic scoring to link experimental features with plausible biochemical transformations [144].
MS enables and facilitates the detection of drug products, substrates, and endogenous metabolites that are not directly linked to specific targets. However, when accounting for interindividual heterogeneity, the resulting data can be highly variable, making it challenging to distinguish specific biomarkers from non-specific ones. Furthermore, the complexity of human diseases—along with phenotypic and molecular variation between individuals—suggest that identifying metabolic panels rather than single biomarkers may yield more robust and reliable predictions [99].
Validating potential biomarkers identified in pharmacometabolomics studies on a large scale and translating them into clinical practice remains a significant challenge [2,100]. Large-scale studies require standardized protocols for biospecimen and biofluid collection, proper storage, and the establishment of biobanks, as well as a skilled workforce to conduct and interpret the data [99]. In addition, most pharmacometabolomics research is conducted in small populations, which limits its generalizability and broader clinical applicability [2,100].
5. Conclusions
Metabolomics provides a holistic view of biological systems, spanning from genotype to phenotype, and incorporating environmental, lifestyle, and drug-related factors. This comprehensive insight makes it a powerful tool in drug discovery and development, enabling precise interventions and advancing translational research.
Instrumental analysis is central to metabolomics, but the process is complex and requires a thorough understanding of the strengths and limitations of available techniques for metabolite separation, detection, and identification. LC-MS is widely used due to its broad metabolic coverage but is time-consuming, expensive and suffers from issues with reproducibility. Emerging techniques, such as IMS and SFC, are gaining ground due to their time- and cost-efficient nature, with added environmental benefits. MS and NMR, used either separately or in combination, are essential for metabolome analysis across all stages of drug discovery and development.
In the era of Precision Medicine, metabolomics and pharmacometabolomics can guide every step of drug discovery and development—from revealing disease mechanisms and identifying candidate therapeutic targets, to deciphering the mechanism of action of a drug, to assessing drug metabolism, and evaluating toxicity and efficacy. These strategies also enhance clinical trial design and monitoring, facilitating the development of personalized treatments. While challenges remain, ongoing advancements in the field continue to address these limitations, further expanding the potential applications of metabolomics in research and clinical practice.
Author Contributions
Conceptualization, E.V.S. and M.A.; investigation, E.V.S.; writing—original draft preparation, E.V.S.; writing—review and editing, E.V.S., K.P. and M.A.; visualization, E.V.S. and M.A.; supervision, K.P. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Norman, G.A. Phase II Trials in Drug Development and Adaptive Trial Design. JACC Basic Transl. Sci. 2019, 4, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Phapale, P. Pharmaco-metabolomics opportunities in drug development and clinical research. Anal. Sci. Adv. 2021, 2, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Qlik Sense. Available online: https://fis.fda.gov/sense/app/95239e26-e0be-42d9-a960-9a5f7f1c25ee/sheet/7a47a261-d58b-4203-a8aa-6d3021737452/state/analysis (accessed on 1 October 2025).
- Burt, T.; Nandal, S. Pharmacometabolomics in Early-Phase Clinical Development. Clin Transl Sci. 2016, 9, 128–138. [Google Scholar] [CrossRef]
- Neavin, D.; Kaddurah-Daouk, R.; Weinshilboum, R. Pharmacometabolomics informs pharmacogenomics. Metabolomics 2016, 12, 121. Available online: https://link.springer.com/article/10.1007/s11306-016-1066-x (accessed on 29 October 2025). [CrossRef]
- Pang, H.; Hu, Z. Metabolomics in drug research and development: The recent advances in technologies and applications. Acta Pharm. Sin. B 2023, 13, 3238–3251. [Google Scholar] [CrossRef]
- Nalbantoglu, S. Metabolomics: Basic Principles and Strategies. In Molecular Medicine; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Everett, J.R. NMR-based pharmacometabonomics: A new paradigm for personalised or precision medicine. Prog. Nucl. Magn. Reason. Spectrosc. 2017, 102–103, 1–14. [Google Scholar] [CrossRef]
- Alves, S.; Paris, A.; Rathahao-Paris, E. Mass spectrometry-based metabolomics for an in-depth questioning of human health. Adv. Clin. Chem. 2020, 99, 147–191. [Google Scholar] [CrossRef]
- Ramsden, J. Metabolomics and Metabonomics. In Bioinformatics; Springer: London, UK, 2015; pp. 265–270. [Google Scholar] [CrossRef]
- Kaur, R.; Gupta, S.; Kulshrestha, S.; Khandelwal, V.; Pandey, S.; Kumar, A.; Sharma, G.; Kumar, U.; Parashar, D.; Das, K. Metabolomics-Driven Biomarker Discovery for Breast Cancer Prognosis and Diagnosis. Cells 2024, 14, 5. [Google Scholar] [CrossRef]
- Yang, Q.J.; Zhao, J.R.; Hao, J.; Li, B.; Huo, Y.; Han, Y.L.; Wan, L.L.; Li, J.; Huang, J.; Lu, J.; et al. Serum and urine metabolomics study reveals a distinct diagnostic model for cancer cachexia. J. Cachexia Sarcopenia Muscle 2018, 9, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Luo, Y.; Shen, Y.; Hu, Y.; Zhu, F.; Wu, J.; Liu, Y. Integrated Metabonomics and Network Pharmacology to Reveal the Action Mechanism Effect of Shaoyao Decoction on Ulcerative Colitis. Drug Des. Dev. Ther. 2022, 16, 3739–3776. [Google Scholar] [CrossRef]
- Hang, J.; Chen, Y.; Liu, L.; Chen, L.; Fang, J.; Wang, F.; Wang, M. Antitumor effect and metabonomics of niclosamide micelles. J. Cell Mol. Med. 2022, 26, 4814–4824. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Song, H.E.; Lee, H.Y.; Yoo, H.J. Mass Spectrometry-based Metabolomics in Translational Research. Adv. Exp. Med. Biol. 2021, 1310, 509–531. Available online: https://pubmed.ncbi.nlm.nih.gov/33834448/ (accessed on 29 October 2025).
- Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; del Bosque-Plata, L. The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Han, Y.; Yan, G.L.; Wu, F.F.; Wang, X.J. Metabolomics biotechnology, applications, and future trends: A systematic review. RSC Adv. 2019, 9, 37245–37257. [Google Scholar] [CrossRef]
- Iruzubieta, P.; Arias-Loste, M.T.; Barbier-Torres, L.; Martinez-Chantar, M.L.; Crespo, J. The need for biomarkers in diagnosis and prognosis of drug-induced liver disease: Does metabolomics have any role? Biomed Res. Int. 2015, 2015, 386186. [Google Scholar] [CrossRef]
- van de Velde, B.; Guillarme, D.; Kohler, I. Supercritical fluid chromatography—Mass spectrometry in metabolomics: Past, present, and future perspectives. J. Chromatogr. B 2020, 1161, 122444. [Google Scholar] [CrossRef]
- Kantae, V.; Krekels, E.H.J.; Esdonk, M.J.V.; Lindenburg, P.; Harms, A.C.; Knibbe, A.J.C.; Van der Graaf, P.H.; Hankemeier, T. Integration of pharmacometabolomics with pharmacokinetics and pharmacodynamics: Towards personalized drug therapy. Metabolomics 2017, 13, 9. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Weinshilboum, R.M. Pharmacometabolomics: Implications for Clinical Pharmacology and Systems Pharmacology. Clin. Pharmacol. Ther. 2014, 95, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Katsila, T.; Konstantinou, E.; Lavda, I.; Malakis, H.; Papantoni, I.; Skondra, L.; Patrinos, G.P. Pharmacometabolomics-aided Pharmacogenomics in Autoimmune Disease. EBioMedicine 2016, 5, 40–45. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Weinshilboum, R. Metabolomic signatures for drug response phenotypes: Pharmacometabolomics enables precision medicine. Clin. Pharmacol. Ther. 2015, 98, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Begou, O.; Gika, H.G.; Wilson, I.D.; Theodoridis, G. Hyphenated MS-based targeted approaches in metabolomics. Analyst. R. Soc. Chem. 2017, 142, 3079–3100. [Google Scholar] [CrossRef]
- Aszyk, J.; Byliński, H.; Namieśnik, J.; Kot-Wasik, A. Main strategies, analytical trends and challenges in LC-MS and ambient mass spectrometry–based metabolomics. TrAC—Trends Anal. Chem. 2018, 108, 278–295. [Google Scholar] [CrossRef]
- Barderas, M.G.; Laborde, C.M.; Posada, M.; de la Cuesta, F.; Zubiri, I.; Vivanco, F.; Alvarez-Llamas, G. Metabolomic Profiling for Identification of Novel Potential Biomarkers in Cardiovascular Diseases. J Biomed. Biotechnol. 2011, 2011, 790132. [Google Scholar] [CrossRef]
- Chen, Y.; Li, E.M.; Xu, L.Y. Guide to Metabolomics Analysis: A Bioinformatics Workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef]
- Emwas, A.H.; Saccenti, E.; Gao, X.; McKay, R.T.; Martins Dos Santos, V.A.P.; Roy, R.; Wishart, D.S. Recommended strategies for spectral processing and post-processing of 1D 1H-NMR data of biofluids with a particular focus on urine. Metabolomics 2018, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Moco, S. Studying Metabolism by NMR-Based Metabolomics. Front. Mol. Biosci. 2022, 9, 882487. Available online: https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2022.882487/full (accessed on 29 October 2025). [CrossRef] [PubMed]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Nagana Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. Nmr spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, L.B.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]
- Heuckeroth, S.; Damiani, T.; Smirnov, A.; Mokshyna, O.; Brungs, C.; Korf, A.; Smith, J.A.; Stincone, P.; Dreolin, N.; Nothias, L.F.; et al. Reproducible mass spectrometry data processing and compound annotation in MZmine 3. Nat. Protoc. 2024, 19, 2597–2641. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Ji, F.; Chen, Y.; Cai, Z. statTarget: A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data. Anal. Chim. Acta 2018, 1036, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2012, 41, D781–D786. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523. [Google Scholar] [CrossRef]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. Available online: https://jcheminf.biomedcentral.com/articles/10.1186/s13321-016-0115-9 (accessed on 29 October 2025). [CrossRef]
- Broadhurst, D.I. QC:MXP Repeat Injection Based Quality Control, Batch Correction, Exploration & Data Cleaning (Version 2) Zendono. Available online: https://github.com/broadhurstdavid/QC-MXP (accessed on 29 October 2025).
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.C.; Gutenbrunner, P.; Kenar, E.; et al. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Salguero, P.; Petek, M.; Martinez-Mira, C.; Balzano-Nogueira, L.; Ramšak, Ž.; McIntyre, L.; Gruden, K.; Tarazona, S.; Conesa, A. PaintOmics 4: New tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. 2022, 50, W551–W559. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035. [Google Scholar] [CrossRef]
- Conroy, M.J.; Andrews, R.M.; Andrews, S.; Cockayne, L.; Dennis, E.A.; Fahy, E.; Gaud, C.; Griffiths, W.J.; Jukes, G.; Kolchin, M.; et al. LIPID MAPS: Update to databases and tools for the lipidomics community. Nucleic Acids Res. 2023, 52, D1677–D1682. [Google Scholar] [CrossRef]
- Tobolkina, E.; González-Rui, V.; Meister, I.; De Figueiredo, M.; Guillarme, D.; Boccard, J.; Rudaz, S. Challenges in ESI-MS-based Untargeted Metabolomics. Chimia 2022, 76, 90–100. [Google Scholar] [CrossRef]
- Moses, T.; Burgess, K. Right in two: Capabilities of ion mobility spectrometry for untargeted metabolomics. Front. Mol. Biosci. 2023, 10, 1230282. [Google Scholar] [CrossRef]
- Anh, N.K.; Thu, N.Q.; Tien, N.T.N.; Long, N.P.; Nguyen, H.T. Advancements in Mass Spectrometry-Based Targeted Metabolomics and Lipidomics: Implications for Clinical Research. Molecules 2024, 29, 5934. [Google Scholar] [CrossRef]
- Watson, D.G. Pharmaceutical Analysis: A Textbook for Pharmacy Students and Pharmaceutical Chemists, 3nd Greek ed.; ΕΠΙΣΤHΜOΝΙΚΕΣ ΕΚΔOΣΕΙΣ ΠAΡΙAΣΙAΝOΥ A.Ε.: Attica, Greece, 2015. [Google Scholar]
- Ovbude, S.T.; Sharmeen, S.; Kyei, I.; Olupathage, H.; Jones, J.; Bell, R.J.; Powers, R.; Hage, D.S. Applications of chromatographic methods in metabolomics: A review. J. Chromatogr. B 2024, 1239, 124124. [Google Scholar] [CrossRef]
- Zaid, A.; Hassan, N.H.; Marriott, P.J.; Wong, Y.F. Comprehensive Two-Dimensional Gas Chromatography as a Bioanalytical Platform for Drug Discovery and Analysis. Pharmaceutics 2023, 15, 1121. [Google Scholar] [CrossRef] [PubMed]
- Myridakis, A.; Wen, Q.; Boshier, P.R.; Parker, A.G.; Belluomo, I.; Handakas, E.; Hanna, G.B. Global Urinary Volatolomics with (GC×)GC-TOF-MS. Anal. Chem. 2023, 95, 17170. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Hoerr, V.; Duggan, G.E.; Zbytnuik, L.; Poon, K.K.H.; Große, C.; Neugebauer, U.; Methling, K.; Löffler, B.; Vogel, H.J. Characterization and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 2016, 16, 82. [Google Scholar] [CrossRef]
- Lepoittevin, M.; Blancart-Remaury, Q.; Kerforne, T.; Pellerin, L.; Hauet, T.; Thuillier, R. Comparison between 5 extractions methods in either plasma or serum to determine the optimal extraction and matrix combination for human metabolomics. Cell. Mol. Biol. Lett. 2023, 28, 43. Available online: https://pubmed.ncbi.nlm.nih.gov/37210499/ (accessed on 29 October 2025). [CrossRef]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): The state of the art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Plumb, R.S.; Wilson, I.D. Current practice of liquid chromatography-mass spectrometry in metabolomics and metabonomics. J. Pharm. Biomed. Anal. 2014, 87, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef]
- Lopes, A.S.; Cruz, E.C.S.; Sussulini, A.; Klassen, A. Metabolomic strategies involving mass spectrometry combined with liquid and gas chromatography. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; Volume 965, pp. 77–98. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef] [PubMed]
- Rainville, P.D.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Advances in liquid chromatography coupled to mass spectrometry for metabolic phenotyping. TrAC Trends Anal. Chem. 2014, 61, 181–191. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Vaidyanathan, S. Towards quantitative mass spectrometry-based metabolomics in microbial and mammalian systems. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150363. [Google Scholar] [CrossRef]
- Giannoukos, S.; Agapiou, A.; Brkić, B.; Taylor, S. Volatolomics: A broad area of experimentation. J. Chromatogr. B 2019, 1105, 136–147. [Google Scholar] [CrossRef]
- Fritsche-Guenther, R.; Gloaguen, Y.; Bauer, A.; Tobias, O.; Kempa, S.; Fleming, C.A.; Redmond, H.P.; Kirwan, J.A. Optimized workflow for on-line derivatization for targeted metabolomics approach by gas chromatography-mass spectrometry. Metabolites 2021, 11, 888. Available online: https://pubmed.ncbi.nlm.nih.gov/34940646/ (accessed on 29 October 2025). [CrossRef] [PubMed]
- Rey-Stolle, F.; Dudzik, D.; Gonzalez-Riano, C.; Fernández-García, M.; Alonso-Herranz, V.; Rojo, D.; Barbas, C.; García, A. Low and high resolution gas chromatography-mass spectrometry for untargeted metabolomics: A tutorial. Anal. Chim. Acta 2022, 1210, 339043. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: The combination of targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Frolova, N.; Orlova, A.; Popova, V.; Bilova, T.; Frolov, A.; Timiryazev, K. GC-MS-based metabolomics-Part 1 Gas chromatography-mass spectrometry (GC-MS) and its place in the plant metabolomics toolbox. ChemRxiv 2025. [Google Scholar] [CrossRef]
- Shulaev, V.; Isaac, G. Supercritical fluid chromatography coupled to mass spectrometry—A metabolomics perspective. J Chromatogr B Anal. Technol Biomed. Life Sci. 2018, 1092, 499–505. [Google Scholar] [CrossRef]
- Dodds, J.N.; Baker, E.S. Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead. J. Chromatogr. B 2019, 30, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Sans, M.; Feider, C.L.; Eberlin, L.S. Advances in mass spectrometry imaging coupled to ion mobility spectrometry for enhanced imaging of biological tissues. Curr. Opin. Chem. Biol. 2018, 42, 138–146. [Google Scholar] [CrossRef]
- Campuzano, I.D.G.; Lippens, J.L. Ion mobility in the pharmaceutical industry: An established biophysical technique or still niche? Curr. Opin. Chem. Biol. 2018, 42, 147–159. [Google Scholar] [CrossRef]
- Eyers, C.E.; Vonderach, M.; Ferries, S.; Jeakock, K.; Eyers, P.A. Understanding protein-drug interactions using ion mobility-mass spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, G.; Yan, X.; Zhang, Z.; Wojcik, R.; Champion, M.M.; Dovichi, N.J. Capillary zone electrophoresis for bottom-up analysis of complex proteomes. Proteomics 2015, 16, 188. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, C.M.; D’amico, C.I.; Kennedy, R.T. Advances in Capillary Electrophoresis and the Implications for Drug Discovery. Expert Opin. Drug Discov. 2016, 12, 213. [Google Scholar] [CrossRef]
- Zhang, W.; Hankemeier, T.; Ramautar, R. Next-generation capillary electrophoresis–mass spectrometry approaches in metabolomics. Curr Opin Biotechnol. 2017, 43, 1–7. [Google Scholar] [CrossRef]
- Gomes, F.P.; Yates, J.R. Recent Trends of Capillary Electrophoresis-Mass Spectrometry in Proteomics Research. Mass Spectrom. Rev. 2019, 38, 445. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef]
- Wang, R.; Yin, Y.; Zhu, Z.J. Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology. Anal. Bioanal. Chem. 2019, 411, 4349–4357. [Google Scholar] [CrossRef]
- Jiang, F.; Liu, Q.; Li, Q.; Zhang, S.; Qu, X.; Zhu, J.; Zhong, G.; Huang, M. Signal Drift in Liquid Chromatography Tandem Mass Spectrometry and Its Internal Standard Calibration Strategy for Quantitative Analysis. Anal. Chem. 2020, 92, 7690–7698. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Cai, B.; Huan, T. Enhancing Metabolome Coverage in Data-Dependent LC-MS/MS Analysis through an Integrated Feature Extraction Strategy. Anal Chem. 2019, 91, 14433–14441. [Google Scholar] [CrossRef]
- Leenders, J.; Frédérich, M.; De Tullio, P. Nuclear magnetic resonance: A key metabolomics platform in the drug discovery process. Drug Discov. Today Technol. 2015, 13, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Torres, C.; Wong, A. Current Developments in µMAS NMR Analysis for Metabolomics. Metabolites 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Würtz, P.; Kangas, A.J.; Soininen, P.; Lawlor, D.A.; Smith, G.D.; Ala-Korpela, M. Quantitative Serum Nuclear Magnetic Resonance Metabolomics in Large-Scale Epidemiology: A Primer on -Omic Technologies. Am. J. Epidemiol. 2017, 186, 1084–1096. [Google Scholar] [CrossRef]
- Singh, S.; Roy, R. The application of absolute quantitative 1H NMR spectroscopy in drug discovery and development. Expert Opin. Drug Discov. 2016, 11, 695–706. [Google Scholar] [CrossRef]
- Crook, A.A.; Powers, R. Quantitative NMR-Based Biomedical Metabolomics: Current Status and Applications. Molecules 2020, 25, 5128. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Pereira, D.M.; Valentão, P.; Andrade, P.B. Hybrid MS/NMR methods on the prioritization of natural products: Applications in drug discovery. J. Pharm. Biomed. Anal. 2018, 147, 234–249. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef]
- Tayyari, F.; Gowda, G.A.N.; Gu, H.; Raftery, D. 15N-Cholamine—A smart isotope tag for combining NMR- and MS-based metabolite profiling. Anal. Chem. 2013, 85, 8715–8721. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Geng, W.; Sun, H.; Liu, C.; Huang, F.; Cao, J.; Xia, L.; Zhao, H.; Zhai, J.; Li, Q.; et al. Integrative metabolomic characterisation identifies altered portal vein serum metabolome contributing to human hepatocellular carcinoma. Gut 2022, 71, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.; Tan, S.; Zhu, Z.; Bulloch, G.; Long, E.; Huang, W.; He, M.; Wang, W. Metabolomic phenotyping of obesity for profiling cardiovascular and ocular diseases. J. Transl. Med. 2023, 21, 384. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Gu, J.; Zhang, M.; Su, F.; Su, W.; Xie, Y. NMR-Based Metabolomics to Analyze the Effects of a Series of Monoamine Oxidases-B Inhibitors on U251 Cells. Biomolecules 2023, 13, 600. [Google Scholar] [CrossRef]
- Cacciatore, S.; Zadra, G.; Bango, C.; Penney, K.L.; Tyekucheva, S.; Yanes, O.; Loda, M. Metabolic Profiling in Formalin-Fixed and Paraffin-Embedded Prostate Cancer Tissues. Mol. Cancer Res. 2017, 15, 439. [Google Scholar] [CrossRef]
- Hundertmark, M.J.; Agbaje, O.F.; Coleman, R.; George, J.T.; Grempler, R.; Holman, R.R.; Lamlum, H.; Lee, J.; Milton, J.E.; Niessen, H.G.; et al. Design and rationale of the EMPA-VISION trial: Investigating the metabolic effects of empagliflozin in patients with heart failure. ESC Heart Fail. 2021, 8, 2580–2590. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, X.; Bu, X.; Li, Y.; Wang, M.; Zhang, B.; Sun, W.; Li, C. Application of plasma metabolome for monitoring the effect of rivaroxaban in patients with nonvalvular atrial fibrillation. PeerJ 2022, 10, e13853. [Google Scholar] [CrossRef]
- Shao, Y.; Li, T.; Liu, Z.; Wang, X.; Xu, X.; Li, S.; Xu, G.; Le, W. Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography-mass spectrometry. Mol. Neurodegener. 2021, 16, 4. [Google Scholar] [CrossRef]
- Sánchez-Castillo, A.; Heylen, E.; Hounjet, J.; Savelkouls, K.G.; Lieuwes, N.G.; Biemans, R.; Dubois, L.J.; Reynders, K.; Rouschop, K.M.; Vaes, R.D.W.; et al. Targeting serine/glycine metabolism improves radiotherapy response in non-small cell lung cancer. Br. J. Cancer 2023, 130, 568. [Google Scholar] [CrossRef]
- Tolstikov, V. Metabolomics: Bridging the Gap between Pharmaceutical Development and Population Health. Metabolites 2016, 6, 20. [Google Scholar] [CrossRef]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef]
- Yin, K.; Lee, J.; Liu, Z.; Kim, H.; Martin, D.R.; Wu, D.; Liu, M.; Xue, X. Mitophagy protein PINK1 suppresses colon tumor growth by metabolic reprogramming via p53 activation and reducing acetyl-CoA production. Cell Death Differ. 2021, 28, 2421. [Google Scholar] [CrossRef]
- Tan, B.; Jaulin, A.; Bund, C.; Outilaft, H.; Wendling, C.; Chenard, M.P.; Alpy, F.; Cicek, A.E.; Namer, I.J.; Tomasetto, C.; et al. Matrix Metalloproteinase-11 Promotes Early Mouse Mammary Gland Tumor Growth through Metabolic Reprogramming and Increased IGF1/AKT/FoxO1 Signaling Pathway, Enhanced ER Stress and Alteration in Mitochondrial UPR. Cancers 2020, 12, 2357. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chen, H.; Yin, D.; Wang, J.; Wang, S.; Yang, F.; Li, J.; Mu, T.; Li, J.; Zhao, J.; et al. Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma. NPJ Precis. Oncol. 2024, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Liu, W.; Xia, S.; Zhou, Y.; Tang, M.; Xu, M.; Lin, M.; Li, X.; Wang, Q. Warburg effect enhanced by AKR1B10 promotes acquired resistance to pemetrexed in lung cancer-derived brain metastasis. J. Transl. Med. 2023, 21, 547. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016. [Google Scholar] [PubMed]
- Tran, D.T.; Dahlin, A. Pharmacometabolomics: General Applications of Metabolomics in Drug Development and Personalized Medicine. In Metabolomics; Springer: Cham, Switzerland, 2023; pp. 127–164. Available online: https://link.springer.com/chapter/10.1007/978-3-031-39094-4_5 (accessed on 29 October 2025).
- Fillet, M.; Frédérich, M. The emergence of metabolomics as a key discipline in the drug discovery process. Drug Discov. Today Technol. 2015, 13, 19–24. [Google Scholar] [CrossRef]
- Chowdhury, S.; Zielinski, D.C.; Dalldorf, C.; Rodrigues, J.V.; Palsson, B.O.; Shakhnovich, E.I. Empowering drug off-target discovery with metabolic and structural analysis. Nat. Commun. 2023, 14, 3390. [Google Scholar] [CrossRef]
- Coen, M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. NMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chem. Res. Toxicol. 2008, 21, 9–27. [Google Scholar] [CrossRef]
- Ramirez, T.; Daneshian, M.; Kamp, H.; Bois, F.Y.; Clench, M.R.; Coen, M.; Donley, B.; Fischer, S.M.; Ekman, D.R.; Fabian, E.; et al. T4 Report* Metabolomics in toxicology and preclinical research. Altex 2013, 30, 209–225. [Google Scholar] [CrossRef]
- Abouleila, Y.; Ali, A.; Masuda, K.; Mashaghi, A.; Shimizu, Y. Capillary microsampling-based single-cell metabolomics by mass spectrometry and its applications in medicine and drug discovery. Cancer Biomark. 2022, 33, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Buglakova, E.; Ekelöf, M.; Schwaiger-Haber, M.; Schlicker, L.; Molenaar, M.R.; Shahraz, M.; Stuart, L.; Eisenbarth, A.; Hilsenstein, V.; Patti, G.J.; et al. Spatial single-cell isotope tracing reveals heterogeneity of de novo fatty acid synthesis in cancer. Nat. Metab. 2024, 6, 1695–1711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, M.; Li, M.; Qin, S.; Miao, D.; Bai, Y. Dynamic single-cell metabolomics reveals cell-cell interaction between tumor cells and macrophages. Nat. Commun. 2025, 16, 4582. [Google Scholar] [CrossRef]
- Beyoǧlu, D.; Idle, J.R. Metabolomics and its potential in drug development. Biochem Pharmacol. 2013, 85, 12–20. [Google Scholar] [CrossRef]
- Komagamine, J. Prevalence of urgent hospitalizations caused by adverse drug reactions: A cross-sectional study. Sci. Rep. 2024, 14, 6058. [Google Scholar] [CrossRef]
- Bouvy, J.C.; De Bruin, M.L.; Koopmanschap, M.A. Epidemiology of Adverse Drug Reactions in Europe: A Review of Recent Observational Studies. Drug Saf. 2015, 38, 437. [Google Scholar] [CrossRef] [PubMed]
- Beeler, P.E.; Stammschulte, T.; Dressel, H. Hospitalisations Related to Adverse Drug Reactions in Switzerland in 2012–2019: Characteristics, In-Hospital Mortality, and Spontaneous Reporting Rate. Drug Saf. 2023, 46, 753–763. Available online: https://pubmed.ncbi.nlm.nih.gov/37335465/ (accessed on 29 October 2025). [CrossRef]
- Crestani, E.; Harb, H.; Charbonnier, L.M.; Leirer, J.; Motsinger-Reif, A.; Rachid, R.; Phipatanakul, W.; Kaddurah-Daouk, R.; Chatila, T.A. Untargeted Metabolomic Profiling Identifies Disease-specific Signatures in Food Allergy and Asthma. J. Allergy Clin Immunol. 2019, 145, 897. [Google Scholar] [CrossRef]
- Yue, L.; Zeng, P.; Li, Y.; Chai, Y.; Wu, C.; Gao, B. Nontargeted and targeted metabolomics approaches reveal the key amino acid alterations involved in multiple myeloma. PeerJ 2022, 10, e12918. [Google Scholar] [CrossRef]
- Prince, N.; Chu, S.H.; Chen, Y.; Mendez, K.M.; Hanson, E.; Green-Snyder, L.; Brooks, E.; Korrick, S.; Lasky-Su, J.A.; Kelly, R.S. Phenotypically driven subgroups of ASD display distinct metabolomic profiles. Brain Behav. Immun. 2023, 111, 21. [Google Scholar] [CrossRef]
- Reher, R.; Aron, A.T.; Fajtová, P.; Stincone, P.; Wagner, B.; Pérez-Lorente, A.I.; Liu, C.; Ben Shalom, I.Y.; Bittremieux, W.; Wang, M.; et al. Native metabolomics identifies the rivulariapeptolide family of protease inhibitors. Nat Commun. 2022, 13, 4619. [Google Scholar] [CrossRef] [PubMed]
- Rattner, J.I.; Kopciuk, K.A.; Vogel, H.J.; Tang, P.A.; Shapiro, J.D.; Tu, D.; Jonker, D.J.; Siu, L.L.; O’Callaghan, C.J.; Bathe, O.F. Clinical and metabolomic characterization of Brivanib-Induced hypertension in metastatic colorectal cancer. Cancer Med. 2023, 12, 16019. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, K.; Zhao, H.; Hao, Z.; Yang, Y.; Gao, M.; Zhao, D. Integrated lipidomics and network pharmacology analysis to reveal the mechanisms of berberine in the treatment of hyperlipidemia. J. Transl. Med. 2022, 20, 412. [Google Scholar] [CrossRef]
- Xie, C.; Gao, X.; Sun, D.; Zhang, Y.; Krausz, K.W.; Qin, X.; Gonzalez, F.J. Metabolic profiling of the novel hypoxia-inducible factor 2α inhibitor PT2385 in vivo and in vitro. Drug Metab. Dispos. 2018, 46, 336–345. Available online: https://pubmed.ncbi.nlm.nih.gov/29363499/ (accessed on 29 October 2025). [CrossRef] [PubMed]
- Li, Y.; Girgis, M.; Jayatilake, M.; Serebrenik, A.A.; Cheema, A.K.; Kaytor, M.D.; Singh, V.K. Pharmacokinetic and metabolomic studies with a BIO 300 Oral Powder formulation in nonhuman primates. Sci. Rep. 2022, 12, 13475. [Google Scholar] [CrossRef]
- Jieu, B.; Sykorova, E.B.; Rohleder, C.; Marcolini, E.; Hoffmann, A.E.; Koethe, D.; Leweke, F.M.; Couttas, T.A. Alterations to sphingolipid metabolism from antipsychotic administration in healthy volunteers are restored following the use of cannabidiol. Psychiatry Res. 2024, 339, 116005. [Google Scholar] [CrossRef]
- Jangsiripornpakorn, J.; Srisuk, S.; Chailurkit, L.; Nimitphong, H.; Saetung, S.; Ongphiphadhanakul, B. The glucose-lowering effect of low-dose diacerein and its responsiveness metabolic markers in uncontrolled diabetes. BMC Res. Notes 2022, 15, 91. [Google Scholar] [CrossRef]
- Zhao, Q.; Lu, Y.; Duan, J.; Du, D.; Pu, Q.; Li, F. Gut microbiota depletion and FXR inhibition exacerbates zonal hepatotoxicity of sunitinib. Theranostics 2024, 14, 7219. [Google Scholar] [CrossRef]
- Boguszewicz, Ł.; Jamroz, E.; Ciszek, M.; Emich-Widera, E.; Kijonka, M.; Banasik, T.; Skorupa, A.; Sokół, M. NMR-based metabolomics in pediatric drug resistant epilepsy—Preliminary results. Sci. Rep. 2019, 9, 15035. [Google Scholar] [CrossRef]
- Moreno-Gordaliza, E.; González-Nicolás, M.Á.; Lázaro, A.; Barbas, C.; Gómez-Gómez, M.M.; López-Gonzálvez, Á. Untargeted metabolomics analysis of serum and urine unveils the protective effect of cilastatin on altered metabolic pathways during cisplatin-induced acute kidney injury. Biochem. Pharmacol. 2024, 227, 116435. [Google Scholar] [CrossRef] [PubMed]
- Latham, B.D.; Oskin, D.S.; Crouch, R.D.; Vergne, M.J.; Jackson, K.D. Cytochromes P450 2C8 and 3A Catalyze the Metabolic Activation of the Tyrosine Kinase Inhibitor Masitinib. Chem. Res. Toxicol. 2022, 35, 1467–1481. [Google Scholar] [CrossRef]
- Sonn, B.J.; Heard, K.J.; Heard, S.M.; D’Alessandro, A.; Reynolds, K.M.; Dart, R.C.; Rumack, B.H.; Monte, A.A. Metabolomic markers predictive of hepatic adaptation to therapeutic dosing of acetaminophen. Clin. Toxicol. 2021, 60, 221. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Chen, B.; Yan, Z.; Gao, J.; Tang, J.; Zhou, C. Metabolites profiling and pharmacokinetics of troxipide and its pharmacodynamics in rats with gastric ulcer. Sci. Rep. 2020, 10, 13619. [Google Scholar] [CrossRef]
- Lesche, D.; Sigurdardottir, V.; Leichtle, A.B.; Nakas, C.T.; Christians, U.; Englberger, L.; Fiedler, M.; Largiadèr, C.R.; Mohacsi, P.; Sistonen, J. Targeted and global pharmacometabolomics in everolimus-based immunosuppression: Association of co-medication and lysophosphatidylcholines with dose requirement. Metabolomics 2017, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Fokina, V.M.; Xu, M.; Rytting, E.; Abdel-Rahman, S.Z.; West, H.; Oncken, C.; Clark, S.M.; Ahmed, M.S.; Hankins, G.D.V.; Nanovskaya, T.N. Pharmacokinetics of bupropion and its pharmacologically active metabolites in pregnancy. Drug Metab. Dispos. 2016, 44, 1832–1838. [Google Scholar] [CrossRef]
- Lin, L.H.; Ghasemi, M.; Burke, S.M.; Mavis, C.K.; Nichols, J.R.; Torka, P.; Mager, D.E.; Hernandez-Ilizaliturri, F.J.; Goey, A.K.L. Population Pharmacokinetics and Pharmacodynamics of Carfilzomib in Combination with Rituximab, Ifosfamide, Carboplatin, and Etoposide in Adult Patients with Relapsed/Refractory Diffuse Large B Cell Lymphoma. Target. Oncol. 2023, 18, 685. [Google Scholar] [CrossRef]
- de Almeida, V.; Alexandrino, G.L.; Aquino, A.; Gomes, A.F.; Murgu, M.; Dobrowolny, H.; Guest, P.C.; Steiner, J.; Martins-de-Souza, D. Changes in the blood plasma lipidome associated with effective or poor response to atypical antipsychotic treatments in schizophrenia patients. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 101, 109945. [Google Scholar] [CrossRef]
- Guan, X.; Xu, J.; Xiu, M.; Li, X.; Liu, H.; Wu, F. Kynurenine pathway metabolites and therapeutic response to olanzapine in female patients with schizophrenia: A longitudinal study. CNS Neurosci. Ther. 2022, 28, 1539. [Google Scholar] [CrossRef]
- Wang, H.; Fang, Z.Z.; Zheng, Y.; Zhou, K.; Hu, C.; Krausz, K.W.; Sun, D.; Idle, J.R.; Gonzalez, F.J. Metabolic profiling of praziquantel enantiomers. Biochem. Pharmacol. 2014, 90, 166–178. [Google Scholar] [CrossRef]
- Bieber, S.; Letzel, T.; Kruve, A. Electrospray Ionization Efficiency Predictions and Analytical Standard Free Quantification for SFC/ESI/HRMS. J. Am. Soc. Mass Spectrom. 2023, 34, 1511–1518. Available online: https://pubmed.ncbi.nlm.nih.gov/37358930/ (accessed on 29 October 2025). [CrossRef] [PubMed]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef] [PubMed]
- Mildau, K.; Ehlers, H.; Meisenburg, M.; Del Pup, E.; Koetsier, R.A.; Torres Ortega, L.R.; de Jonge, N.F.; Singh, K.S.; Ferreira, D.; Othibeng, K.; et al. Effective data visualization strategies in untargeted metabolomics. Nat. Prod. Rep. 2024, 42, 982–1019. [Google Scholar] [CrossRef]
- Coler, E.A.; Chen, W.; Melnik, A.V.; Morton, J.T.; Aksenov, A.A. Metabolomics in the era of artificial intelligence. Microbiota Host 2024, 2, e230017. [Google Scholar] [CrossRef]
- Chen, L.; Lu, W.; Wang, L.; Xing, X.; Chen, Z.; Teng, X.; Zeng, X.; Muscarella, A.D.; Shen, Y.; Cowan, A.; et al. Metabolite discovery through global annotation of untargeted metabolomics data. Nat. Methods 2021, 18, 1377–1385. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).