Identification of a Novel Lipidomic Biomarker for Hepatocyte Carcinoma Diagnosis: Advanced Boosting Machine Learning Techniques Integrated with Explainable Artificial Intelligence
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Lipidomic Data
2.2. Machine Learning Process and Explainability
2.3. Statistical Analysis
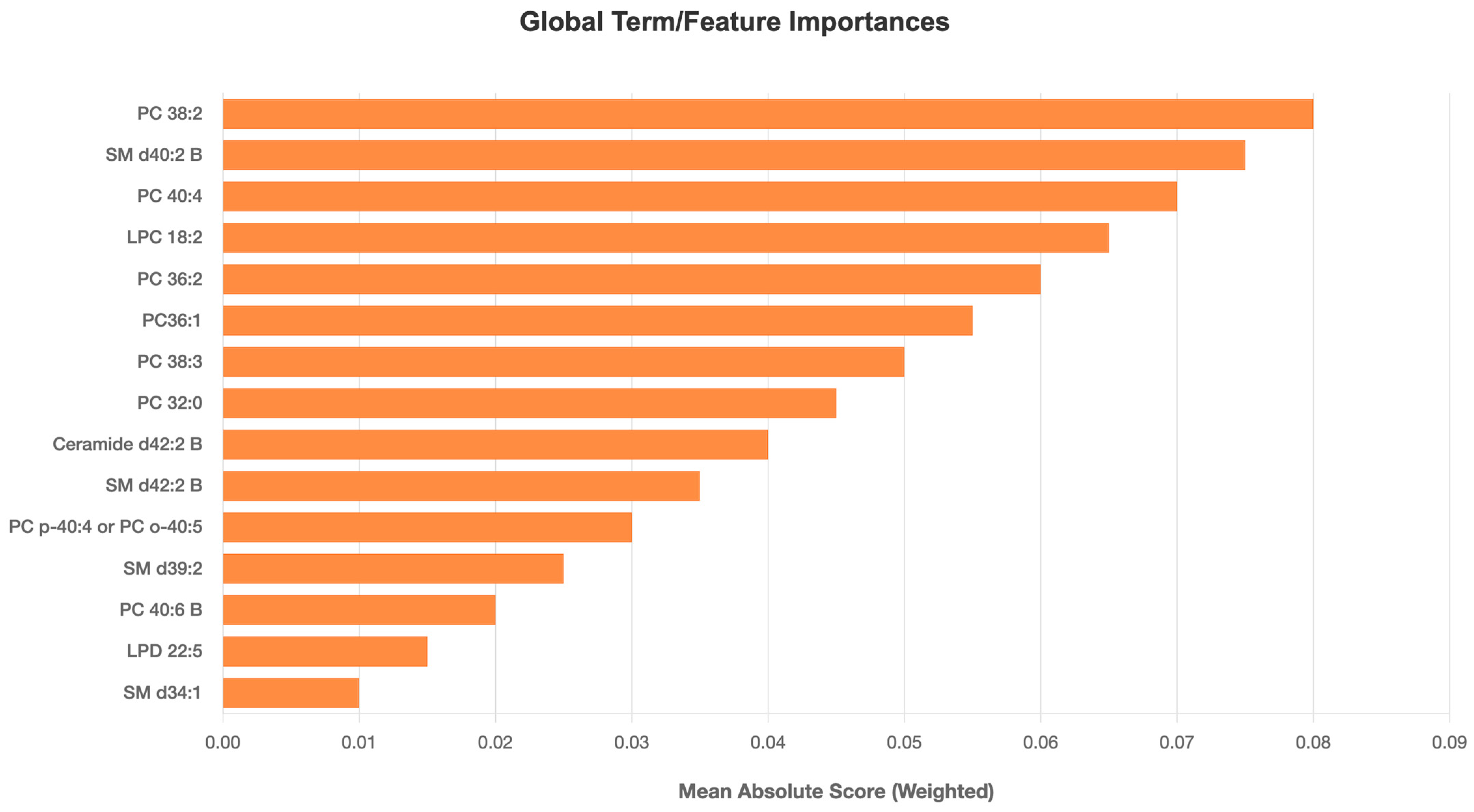
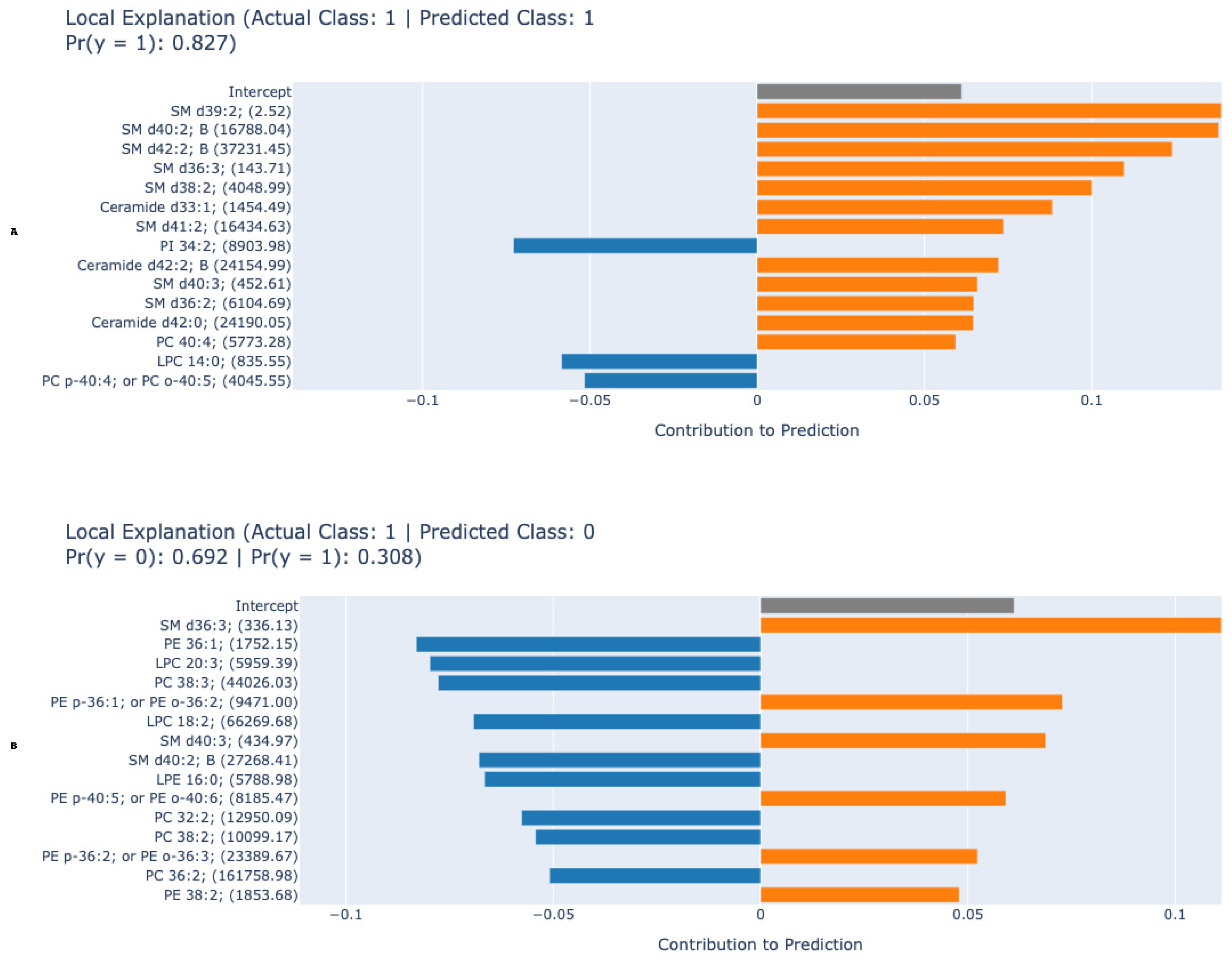
3. Results
4. Discussion
4.1. Machine Learning Model Performance and Interpretability
4.2. Lipid Biomarkers: Pathobiological Implications
4.3. Clinical Translations
4.4. Limitations and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. reviews. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. reviews. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J Clin 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology 2018, 154, 1706–1718.e1701. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Li, Z.; Guan, M.; Lin, Y.; Cui, X.; Zhang, Y.; Zhao, Z.; Zhu, J. Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics. Int. J. Mol. Sci. 2017, 18, 2550. [Google Scholar] [CrossRef]
- Lu, Y.; Li, N.; Gao, L.; Xu, Y.J.; Huang, C.; Yu, K.; Ling, Q.; Cheng, Q.; Chen, S.; Zhu, M.; et al. Acetylcarnitine Is a Candidate Diagnostic and Prognostic Biomarker of Hepatocellular Carcinoma. Cancer Res. 2016, 76, 2912–2920. [Google Scholar] [CrossRef]
- Jia, W.; Yuan, J.; Zhang, J.; Li, S.; Lin, W.; Cheng, B. Bioactive sphingolipids as emerging targets for signal transduction in cancer development. Biochim. Et Biophys. Acta. Rev. Cancer 2024, 1879, 189176. [Google Scholar] [CrossRef]
- DeLeve, L.D. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 2015, 61, 1740–1746. [Google Scholar] [CrossRef]
- Perçın, İ.; Yağin, F.H.; Güldoğan, E.; Yoloğlu, S. ARM: An interactive web software for association rules mining and an application in medicine. In Proceedings of the 2019 International Artificial Intelligence and Data Processing Symposium (IDAP), Malatya, Turkey, 21–22 September 2019; pp. 1–5. [Google Scholar]
- Tetik, B.; Mert Doğan, G.; Paşahan, R.; Durak, M.A.; Güldoğan, E.; Saraç, K.; Önal, Ç.; Yıldırım, İ.O. Multi-parameter-based radiological diagnosis of Chiari Malformation using Machine Learning Technology. Int. J. Clin. Pract. 2021, 75, e14746. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Southam, A.D. Monitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomics. Metabolomics Off. J. Metabolomic Soc. 2016, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Poirion, O.; Lu, L.; Garmire, L. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2017, 24, 1248–1259. [Google Scholar] [CrossRef]
- Yagin, F.H.; Colak, C.; Algarni, A.; Gormez, Y.; Guldogan, E.; Ardigò, L.P. Hybrid explainable artificial intelligence models for targeted metabolomics analysis of diabetic retinopathy. Diagnostics 2024, 14, 1364. [Google Scholar] [CrossRef]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2018, 1, 206–215. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Caruana, R.; Gehrke, J. Intelligible models for classification and regression. In Proceedings of the Knowledge Discovery and Data Mining, Beijing, China, 12–16 August 2012. [Google Scholar]
- Hajduch, E.; Lachkar, F.; Ferré, P.; Foufelle, F. Roles of Ceramides in Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2021, 10, 792. [Google Scholar] [CrossRef]
- Barupal, D.K.; Ramos, M.L.; Florio, A.A.; Wheeler, W.A.; Weinstein, S.J.; Albanes, D.; Fiehn, O.; Graubard, B.I.; Petrick, J.L.; McGlynn, K.A. Identification of pre-diagnostic lipid sets associated with liver cancer risk using untargeted lipidomics and chemical set analysis: A nested case-control study within the ATBC cohort. Int. J. Cancer 2024, 154, 454–464. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Ileri, K. Comparative analysis of CatBoost, LightGBM, XGBoost, RF, and DT methods optimised with PSO to estimate the number of k-barriers for intrusion detection in wireless sensor networks. Int. J. Mach. Learn. Cybern. 2025, 16, 6937–6956. [Google Scholar] [CrossRef]
- Mishra, A.; Jatti, V.S.; Sefene, E.M.; Paliwal, S. Explainable artificial intelligence (XAI) and supervised machine learning-based algorithms for prediction of surface roughness of additively manufactured polylactic acid (PLA) specimens. Appl. Mech. 2023, 4, 668–698. [Google Scholar] [CrossRef]
- Arslan, A.K.; Yagin, F.H.; Algarni, A.; Al-Hashem, F.; Ardigò, L.P. Combining the Strengths of the Explainable Boosting Machine and Metabolomics Approaches for Biomarker Discovery in Acute Myocardial Infarction. Diagnostics 2024, 14, 1353. [Google Scholar] [CrossRef] [PubMed]
- Guldogan, E.; Yagin, F.H.; Pinar, A.; Colak, C.; Kadry, S.; Kim, J. A proposed tree-based explainable artificial intelligence approach for the prediction of angina pectoris. Sci. Rep. 2023, 13, 22189. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Trovato, F.M.; Zia, R.; Artru, F.; Mujib, S.; Jerome, E.; Cavazza, A.; Coen, M.; Wilson, I.; Holmes, E.; Morgan, P. Lysophosphatidylcholines modulate immunoregulatory checkpoints in peripheral monocytes and are associated with mortality in people with acute liver failure. J. Hepatol. 2023, 78, 558–573. [Google Scholar] [CrossRef]
- Yin, Y.; Feng, W.; Chen, J.; Chen, X.; Wang, G.; Wang, S.; Xu, X.; Nie, Y.; Fan, D.; Wu, K. Immunosuppressive tumor microenvironment in the progression, metastasis, and therapy of hepatocellular carcinoma: From bench to bedside. Exp. Hematol. Oncol. 2024, 13, 72. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Armistead, J.; Höpfl, S.; Goldhausen, P.; Müller-Hartmann, A.; Fahle, E.; Hatzold, J.; Franzen, R.; Brodesser, S.; Radde, N.E.; Hammerschmidt, M. A sphingolipid rheostat controls apoptosis versus apical cell extrusion as alternative tumour-suppressive mechanisms. Cell Death Dis. 2024, 15, 746. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, L.; Sun, J.; Zheng, L.; Tang, Y.; Tang, H. Hepatocellular carcinoma: Pathogenesis, molecular mechanisms, and treatment advances. Front. Oncol. 2025, 15, 1526206. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, T.; Su, Y.; Lin, Z.; Liu, X.; Jiao, Y.; Liu, J.; Chen, E. High expression of SMPD4 promotes liver cancer and is associated with poor prognosis. BMC Res. Notes 2025, 18, 159. [Google Scholar] [CrossRef]
- Zeng, J.; Zhao, D.; Tai, Y.-L.; Jiang, X.; Su, L.; Wang, X.; Gurley, E.; Hylemon, P.B.; Fan, J.; Ase, S.O. Dysregulated sphingolipid metabolism contributes to NASH-HCC disease progression. Physiology 2023, 38, 5734514. [Google Scholar] [CrossRef]
- Mass-Sanchez, P.B.; Krizanac, M.; Štancl, P.; Leopold, M.; Engel, K.M.; Buhl, E.M.; van Helden, J.; Gassler, N.; Schiller, J.; Karlić, R. Perilipin 5 deletion protects against nonalcoholic fatty liver disease and hepatocellular carcinoma by modulating lipid metabolism and inflammatory responses. Cell Death Discov. 2024, 10, 94. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]

| Variable | HCC | Control |
|---|---|---|
| Age (years), mean ± SD | 57.8 ± 5.6 | 56.9 ± 5.2 |
| AGE DISTRIBUTION | ||
| ≤54 years, n (%) | 61 (27.9) | 65 (29.7) |
| 55–59 years, n (%) | 90 (41.1) | 99 (45.2) |
| 60–64 years, n (%) | 48 (21.9) | 42 (19.1) |
| ≥65 years, n (%) | 20 (9.1) | 13 (5.9) |
| Body Mass Index (kg/m2), mean ± SD | 27.2 ± 4.1 | 25.8 ± 3.6 |
| BMI CATEGORIES | ||
| <25 kg/m2, n (%) | 61 (27.8) | 84 (38.3) |
| 25 to <30 kg/m2, n (%) | 108 (49.3) | 106 (48.4) |
| ≥30 kg/m2, n (%) | 50 (22.8) | 29 (13.2) |
| EDUCATIONAL LEVEL | ||
| Elementary/no vocational, n (%) | 45 (20.5) | 69 (31.5) |
| Elementary/vocational, n (%) | 112 (51.1) | 104 (47.9) |
| Beyond elementary, n (%) | 62 (28.3) | 45 (20.5) |
| SMOKING INTENSITY CATEGORIES | ||
| <25 pack-years, n (%) | 52 (23.7) | 70 (31.9) |
| 25–34 pack-years, n (%) | 44 (20.0) | 51 (23.2) |
| 35–44 pack-years, n (%) | 49 (22.3) | 52 (23.7) |
| ≥45 pack-years, n (%) | 74 (33.7) | 46 (21.0) |
| ALCOHOL CONSUMPTION CATEGORIES | ||
| Non-drinker, n (%) | 14 (6.3) | 14 (6.3) |
| >0 to <1 drink/day, n (%) | 68 (31.0) | 105 (47.9) |
| 1 to <2 drinks/day, n (%) | 60 (27.4) | 54 (24.7) |
| ≥2 drinks/day, n (%) | 62 (28.3) | 35 (16.4) |
| DIABETES STATUS | ||
| No diabetes, n (%) | 198 (90.4) | 214 (97.7) |
| Diabetes present, n (%) | 21 (9.6) | 5 (2.3) |
| Metrics/Model | SVM | CatBoost | Random Forest | XGBoost | EBM |
|---|---|---|---|---|---|
| Accuracy | 0.765 (0.725–0.805) | 0.785 (0.747–0.824) | 0.797 (0.759–0.834) | 0.806 (0.769–0.843) | 0.870 (0.838–0.901) |
| F1-Score | 0.775 (0.735–0.814) | 0.793 (0.755–0.831) | 0.804 (0.766–0.841) | 0.812 (0.776–0.849) | 0.871 (0.839–0.902) |
| Sensitivity | 0.808 (0.75–0.858) | 0.822 (0.765–0.87) | 0.831 (0.775–0.878) | 0.840 (0.785–0.886) | 0.877 (0.826–0.917) |
| Specificity | 0.721 (0.657–0.78) | 0.749 (0.686–0.805) | 0.763 (0.701–0.817) | 0.772 (0.71–0.826) | 0.863 (0.81–0.906) |
| AUC | 0.828 (0.764–0.893) | 0.848 (0.785–0.910) | 0.858 (0.797–0.919) | 0.866 (0.806–0.927) | 0.918 (0.870–0.966) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yagin, F.H.; Colak, C.; Al-Hashem, F.; Alzakari, S.A.; Alhussan, A.A.; Aghaei, M. Identification of a Novel Lipidomic Biomarker for Hepatocyte Carcinoma Diagnosis: Advanced Boosting Machine Learning Techniques Integrated with Explainable Artificial Intelligence. Metabolites 2025, 15, 716. https://doi.org/10.3390/metabo15110716
Yagin FH, Colak C, Al-Hashem F, Alzakari SA, Alhussan AA, Aghaei M. Identification of a Novel Lipidomic Biomarker for Hepatocyte Carcinoma Diagnosis: Advanced Boosting Machine Learning Techniques Integrated with Explainable Artificial Intelligence. Metabolites. 2025; 15(11):716. https://doi.org/10.3390/metabo15110716
Chicago/Turabian StyleYagin, Fatma Hilal, Cemil Colak, Fahaid Al-Hashem, Sarah A. Alzakari, Amel Ali Alhussan, and Mohammadreza Aghaei. 2025. "Identification of a Novel Lipidomic Biomarker for Hepatocyte Carcinoma Diagnosis: Advanced Boosting Machine Learning Techniques Integrated with Explainable Artificial Intelligence" Metabolites 15, no. 11: 716. https://doi.org/10.3390/metabo15110716
APA StyleYagin, F. H., Colak, C., Al-Hashem, F., Alzakari, S. A., Alhussan, A. A., & Aghaei, M. (2025). Identification of a Novel Lipidomic Biomarker for Hepatocyte Carcinoma Diagnosis: Advanced Boosting Machine Learning Techniques Integrated with Explainable Artificial Intelligence. Metabolites, 15(11), 716. https://doi.org/10.3390/metabo15110716








