Metabolite Genome-Wide Association in Hispanics with Obesity Reveals Genetic Risk and Interactions with Dietary Factors for Type 2 Diabetes
Abstract
1. Introduction
2. Study Population and Methods
Statistical Analysis
3. Results
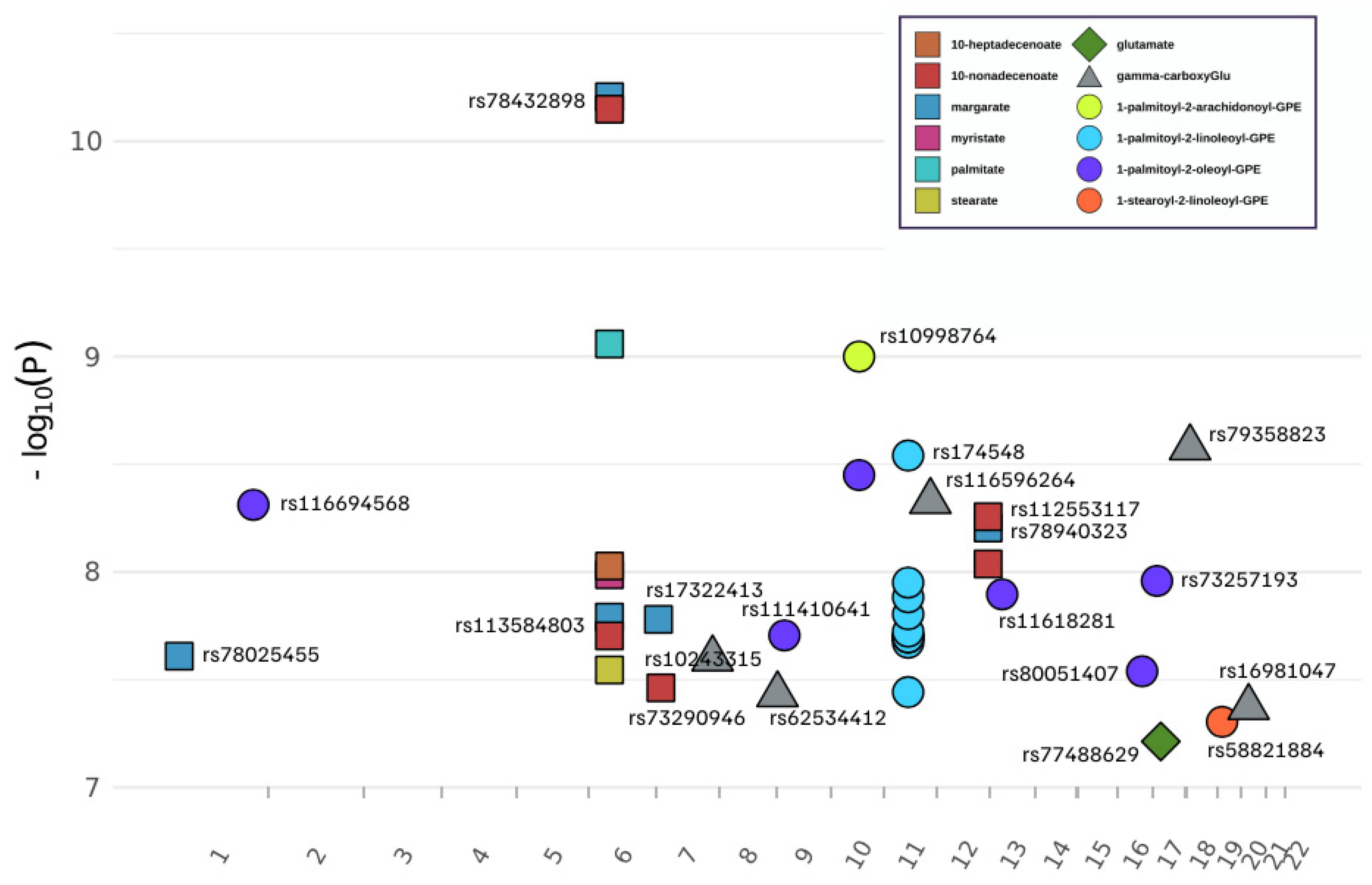
3.1. Genome-Wide Association of 13 Metabolites
3.2. Associations Between mQTLs and T2D Risk
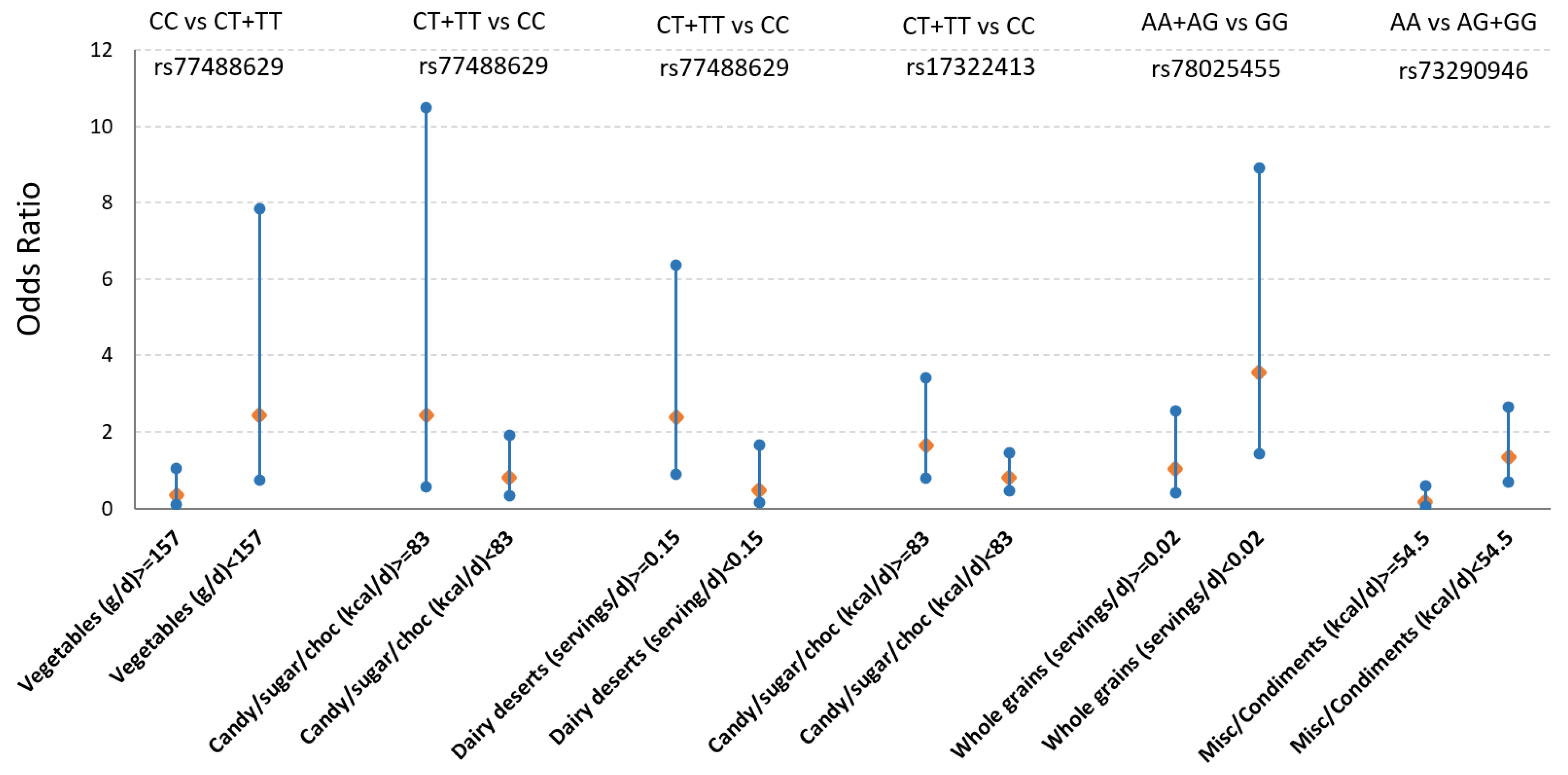
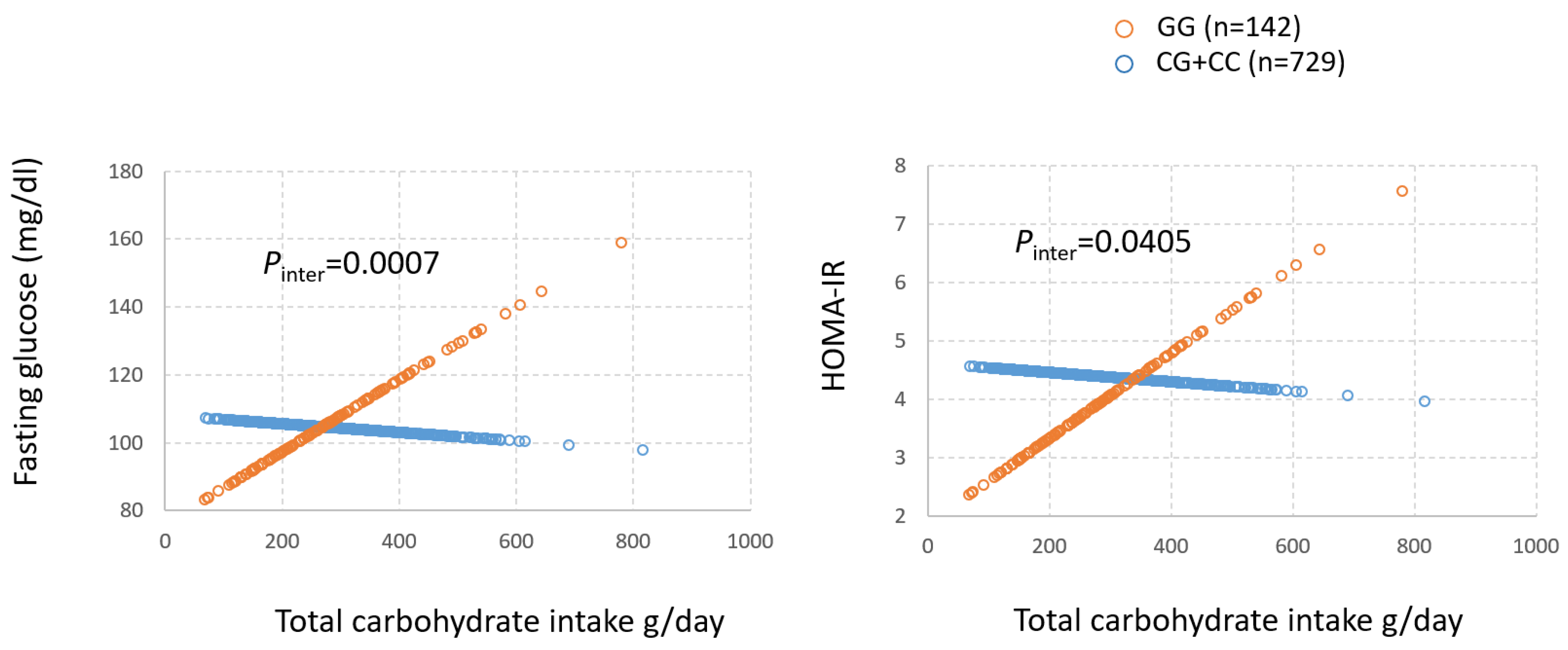
3.3. Gene by Diet (GxD) Interaction on T2D Between mQTLs and Dietary Factors Associated with Obesity Metabolites
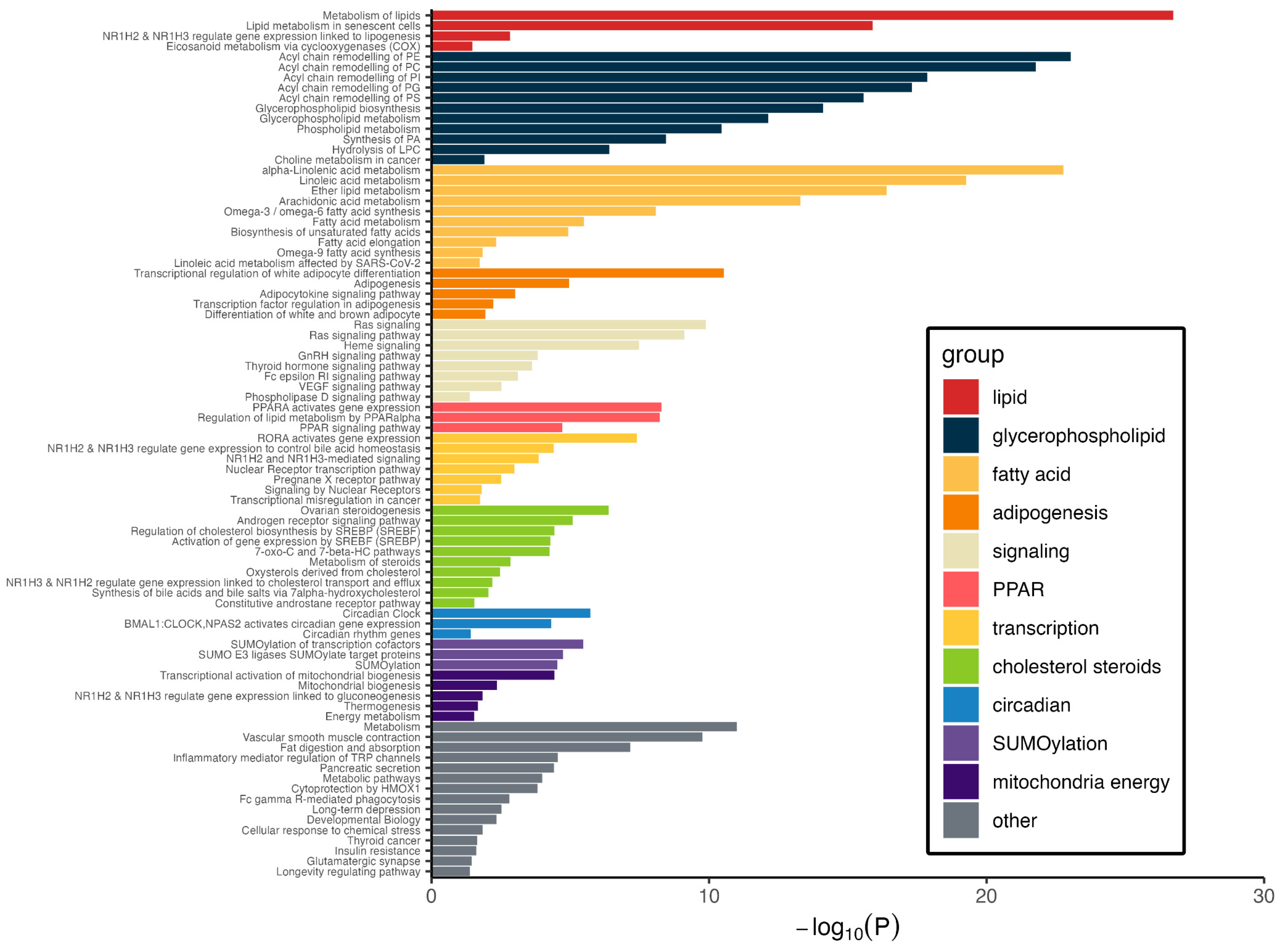
3.4. Pathways Represented by Metabolite–Protein Networks
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | Body mass index. |
| BPRHS | Boston Puerto Rican Health Study. |
| Chr | Chromosome. |
| eQTL | Quantitative trait loci for allele-based expression of genes. |
| FFQ | Food frequency questionnaire. |
| FPG | Fasting plasma glucose. |
| GWAS | Genome-wide association study. |
| GxD | Gene by diet interaction. |
| GTEx | Genotype-Tissue Expression resource. |
| HbA1c | Glycosylated hemoglobin. |
| HOMA-IR | Homeostatic-model-assessment-insulin-resistance. |
| MAF | Minor allele frequency. |
| mQTLs | Metabolite quantitative trait loci. |
| OR | Odds ratio. |
| PC | Phosphatidylcholine. |
| PCA | Principal components analysis. |
| PE | Phosphatidylethanolamine. |
| PPARA | Peroxisome proliferator-activated receptor alpha. |
| PPI | Protein–protein interaction network. |
| SSB | Sugar-sweetened beverage. |
| SNP | Single-nucleotide polymorphism. |
| SSB | Sugar-sweetened beverages. |
| T2D | Type 2 diabetes. |
References
- American Diabetes Association. Standards of Medical Care in Diabetes-2021 Abridged for Primary Care Providers. Clin. Diabetes 2021, 39, 14–43. [Google Scholar] [CrossRef]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef]
- Klein, S.; Gastaldelli, A.; Yki-Jarvinen, H.; Scherer, P.E. Why does obesity cause diabetes? Cell Metab. 2022, 34, 11–20. [Google Scholar] [CrossRef]
- Florez, J.C. Clinical review: The genetics of type 2 diabetes: A realistic appraisal in 2008. J. Clin. Endocrinol. Metab. 2008, 93, 4633–4642. [Google Scholar] [CrossRef]
- Menke, A.; Casagrande, S.; Geiss, L.; Cowie, C.C. Prevalence of and Trends in Diabetes Among Adults in the United States, 1988–2012. JAMA 2015, 314, 1021–1029. [Google Scholar] [CrossRef]
- Lai, C.Q.; Tucker, K.L.; Choudhry, S.; Parnell, L.D.; Mattei, J.; Garcia-Bailo, B.; Beckman, K.; Burchard, E.G.; Ordovas, J.M. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum. Genet. 2009, 125, 199–209. [Google Scholar] [CrossRef]
- Hu, F.B. Globalization of diabetes: The role of diet, lifestyle, and genes. Diabetes Care 2011, 34, 1249–1257. [Google Scholar] [CrossRef]
- Laakso, M. Biomarkers for type 2 diabetes. Mol. Metab. 2019, 27S, S139–S146. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Rhee, E.P.; Larson, M.G.; Lewis, G.D.; McCabe, E.L.; Shen, D.; Palma, M.J.; Roberts, L.D.; Dejam, A.; Souza, A.L.; et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012, 125, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Gao, H.Y.; Fan, Z.Y.; He, Y.; Yan, Y.X. Metabolomics Signatures in Type 2 Diabetes: A Systematic Review and Integrative Analysis. J. Clin. Endocrinol. Metab. 2020, 105, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Morze, J.; Wittenbecher, C.; Schwingshackl, L.; Danielewicz, A.; Rynkiewicz, A.; Hu, F.B.; Guasch-Ferre, M. Metabolomics and Type 2 Diabetes Risk: An Updated Systematic Review and Meta-analysis of Prospective Cohort Studies. Diabetes Care 2022, 45, 1013–1024. [Google Scholar] [CrossRef]
- Parnell, L.D.; Noel, S.E.; Bhupathiraju, S.N.; Smith, C.E.; Haslam, D.E.; Zhang, X.; Tucker, K.L.; Ordovas, J.M.; Lai, C.Q. Metabolite patterns link diet, obesity, and type 2 diabetes in a Hispanic population. Metabolomics 2021, 17, 88. [Google Scholar] [CrossRef]
- Haslam, D.E.; Liang, L.; Wang, D.D.; Kelly, R.S.; Wittenbecher, C.; Perez, C.M.; Martinez, M.; Lee, C.H.; Clish, C.B.; Wong, D.T.W.; et al. Associations of network-derived metabolite clusters with prevalent type 2 diabetes among adults of Puerto Rican descent. BMJ Open Diabetes Res. Care 2021, 9, e002298. [Google Scholar] [CrossRef]
- Tucker, K.L.; Mattei, J.; Noel, S.E.; Collado, B.M.; Mendez, J.; Nelson, J.; Griffith, J.; Ordovas, J.M.; Falcon, L.M. The Boston Puerto Rican Health Study, a longitudinal cohort study on health disparities in Puerto Rican adults: Challenges and opportunities. BMC Public Health 2010, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Noel, S.E.; Newby, P.K.; Ordovas, J.M.; Tucker, K.L. A traditional rice and beans pattern is associated with metabolic syndrome in Puerto Rican older adults. J. Nutr. 2009, 139, 1360–1367. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.L.; Bianchi, L.A.; Maras, J.; Bermudez, O.I. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am. J. Epidemiol. 1998, 148, 507–518. [Google Scholar] [CrossRef]
- Smith, U. Abdominal obesity: A marker of ectopic fat accumulation. J. Clin. Investig. 2015, 125, 1790–1792. [Google Scholar] [CrossRef]
- Jacob, E.; Wulff, M.W.M. A Comparison of Various Normalization Methods for LC/MS Metabolomics Data. Adv. Biosci. Biotechnol. 2018, 9, 339–351. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 October 2025).
- Lopez-Ibanez, J.; Pazos, F.; Chagoyen, M. MBROLE 2.0-functional enrichment of chemical compounds. Nucleic Acids Res. 2016, 44, W201–W204. [Google Scholar] [CrossRef]
- Smith, C.E.; Parnell, L.D.; Lai, C.Q.; Rush, J.E.; Adin, D.B.; Ordovas, J.M.; Freeman, L.M. Metabolomic profiling in dogs with dilated cardiomyopathy eating non-traditional or traditional diets and in healthy controls. Sci. Rep. 2022, 12, 22585. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Parnell, L.D.; Blokker, B.A.; Dashti, H.S.; Nesbeth, P.D.; Cooper, B.E.; Ma, Y.; Lee, Y.C.; Hou, R.; Lai, C.Q.; Richardson, K.; et al. CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData Min. 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005, 95, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Parnell, L.D.; McCaffrey, K.S.; Brooks, A.W.; Smith, C.E.; Lai, C.Q.; Christensen, J.J.; Wiley, C.D.; Ordovas, J. Rate-limiting enzymes in cardiometabolic health and aging in humans. Lifestyle Genom. 2023, 16, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Chande, A.T.; Rishishwar, L.; Conley, A.B.; Valderrama-Aguirre, A.; Medina-Rivas, M.A.; Jordan, I.K. Ancestry effects on type 2 diabetes genetic risk inference in Hispanic/Latino populations. BMC Med. Genet. 2020, 21, 132. [Google Scholar] [CrossRef]
- Polfus, L.M.; Darst, B.F.; Highland, H.; Sheng, X.; Ng, M.C.Y.; Below, J.E.; Petty, L.; Bien, S.; Sim, X.; Wang, W.; et al. Genetic discovery and risk characterization in type 2 diabetes across diverse populations. HGG Adv. 2021, 2, 100029. [Google Scholar] [CrossRef]
- Qi, Q.; Stilp, A.M.; Sofer, T.; Moon, J.Y.; Hidalgo, B.; Szpiro, A.A.; Wang, T.; Ng, M.C.Y.; Guo, X.; MEta-Analysis of Type 2 Diabetes in African Americans (MEDIA) Consortium; et al. Genetics of Type 2 Diabetes in U.S. Hispanic/Latino Individuals: Results From the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes 2017, 66, 1419–1425. [Google Scholar] [CrossRef]
- Malik, V.S.; Schulze, M.B.; Hu, F.B. Intake of sugar-sweetened beverages and weight gain: A systematic review. Am. J. Clin. Nutr. 2006, 84, 274–288. [Google Scholar] [CrossRef]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Despres, J.P.; Willett, W.C.; Hu, F.B. Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: A meta-analysis. Diabetes Care 2010, 33, 2477–2483. [Google Scholar] [CrossRef]
- Zhou, B.; Ichikawa, R.; Parnell, L.D.; Noel, S.E.; Zhang, X.; Bhupathiraju, S.N.; Smith, C.E.; Tucker, K.L.; Ordovas, J.M.; Lai, C.Q. Metabolomic Links between Sugar-Sweetened Beverage Intake and Obesity. J. Obes. 2020, 2020, 7154738. [Google Scholar] [CrossRef]
- Shetty, S.S.; Suchetha, K.N.; Harshini, D.; Sharmila, K.P.; Rai, S. Association of FADS2 rs174575 gene polymorphism and insulin resistance in type 2 diabetes mellitus. Afr. Health Sci. 2020, 20, 1770–1776. [Google Scholar] [CrossRef]
- Bronnikov, G.E.; Aboulaich, N.; Vener, A.V.; Stralfors, P. Acute effects of insulin on the activity of mitochondrial GPAT1 in primary adipocytes. Biochem. Biophys. Res. Commun. 2008, 367, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, G. PPARs Integrate the Mammalian Clock and Energy Metabolism. PPAR Res. 2014, 2014, 653017. [Google Scholar] [CrossRef] [PubMed]
- Tsekmekidou, X.; Tsetsos, F.; Koufakis, T.; Georgitsi, M.; Papanas, N.; Papazoglou, D.; Roumeliotis, A.; Panagoutsos, S.; Thodis, E.; Theodoridis, M.; et al. Variants in clock genes could be associated with lower risk of type 2 diabetes in an elderly Greek population. Maturitas 2021, 152, 20–25. [Google Scholar] [CrossRef]
- Resseguie, M.; Song, J.; Niculescu, M.D.; da Costa, K.A.; Randall, T.A.; Zeisel, S.H. Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J. 2007, 21, 2622–2632. [Google Scholar] [CrossRef]
- Bhaswant, M.; Poudyal, H.; Brown, L. Mechanisms of enhanced insulin secretion and sensitivity with n-3 unsaturated fatty acids. J. Nutr. Biochem. 2015, 26, 571–584. [Google Scholar] [CrossRef]
- Liu, Y.; Dou, X.; Zhou, W.Y.; Ding, M.; Liu, L.; Du, R.Q.; Guo, L.; Qian, S.W.; Tang, Y.; Yang, Q.Q.; et al. Hepatic Small Ubiquitin-Related Modifier (SUMO)-Specific Protease 2 Controls Systemic Metabolism Through SUMOylation-Dependent Regulation of Liver-Adipose Tissue Crosstalk. Hepatology 2021, 74, 1864–1883. [Google Scholar] [CrossRef]
- Zhao, J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007, 64, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Cobb, M.H. Sumoylation regulates the transcriptional activity of MafA in pancreatic beta cells. J. Biol. Chem. 2009, 284, 3117–3124. [Google Scholar] [CrossRef]
- He, X.; Lai, Q.; Chen, C.; Li, N.; Sun, F.; Huang, W.; Zhang, S.; Yu, Q.; Yang, P.; Xiong, F.; et al. Both conditional ablation and overexpression of E2 SUMO-conjugating enzyme (UBC9) in mouse pancreatic beta cells result in impaired beta cell function. Diabetologia 2018, 61, 881–895. [Google Scholar] [CrossRef]
- Dai, X.Q.; Plummer, G.; Casimir, M.; Kang, Y.; Hajmrle, C.; Gaisano, H.Y.; Manning Fox, J.E.; MacDonald, P.E. SUMOylation regulates insulin exocytosis downstream of secretory granule docking in rodents and humans. Diabetes 2011, 60, 838–847. [Google Scholar] [CrossRef]
- Ferdaoussi, M.; Dai, X.; Jensen, M.V.; Wang, R.; Peterson, B.S.; Huang, C.; Ilkayeva, O.; Smith, N.; Miller, N.; Hajmrle, C.; et al. Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional beta cells. J. Clin. Investig. 2015, 125, 3847–3860. [Google Scholar] [CrossRef]
- Abe, J.I.; Imanishi, M.; Li, S.; Zhang, A.; Ko, K.A.; Samanthapudi, V.S.K.; Lee, L.L.; Bojorges, A.P.; Gi, Y.J.; Hobbs, B.P.; et al. An ERK5-NRF2 Axis Mediates Senescence-Associated Stemness and Atherosclerosis. Circ. Res. 2023, 133, 25–44. [Google Scholar] [CrossRef] [PubMed]
- Shitashige, M.; Satow, R.; Honda, K.; Ono, M.; Hirohashi, S.; Yamada, T. Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology 2008, 134, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Garaulet, M.; Corbalan-Tutau, M.D.; Madrid, J.A.; Baraza, J.C.; Parnell, L.D.; Lee, Y.C.; Ordovas, J.M. PERIOD2 variants are associated with abdominal obesity, psycho-behavioral factors, and attrition in the dietary treatment of obesity. J. Am. Diet Assoc. 2010, 110, 917–921. [Google Scholar] [CrossRef]
- Garaulet, M.; Esteban Tardido, A.; Lee, Y.C.; Smith, C.E.; Parnell, L.D.; Ordovas, J.M. SIRT1 and CLOCK 3111T> C combined genotype is associated with evening preference and weight loss resistance in a behavioral therapy treatment for obesity. Int. J. Obes. 2012, 36, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Dashti, H.S.; Vetter, C.; Lane, J.M.; Smith, M.C.; Wood, A.R.; Weedon, M.N.; Rutter, M.K.; Garaulet, M.; Scheer, F.; Saxena, R. Assessment of MTNR1B Type 2 Diabetes Genetic Risk Modification by Shift Work and Morningness-Eveningness Preference in the UK Biobank. Diabetes 2020, 69, 259–266. [Google Scholar] [CrossRef]
- Gutierrez-Cuevas, J.; Galicia-Moreno, M.; Monroy-Ramirez, H.C.; Sandoval-Rodriguez, A.; Garcia-Banuelos, J.; Santos, A.; Armendariz-Borunda, J. The Role of NRF2 in Obesity-Associated Cardiovascular Risk Factors. Antioxidants 2022, 11, 235. [Google Scholar] [CrossRef]
- Strom, J.; Xu, B.; Tian, X.; Chen, Q.M. Nrf2 protects mitochondrial decay by oxidative stress. FASEB J. 2016, 30, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Pallauf, K.; Duckstein, N.; Hasler, M.; Klotz, L.O.; Rimbach, G. Flavonoids as Putative Inducers of the Transcription Factors Nrf2, FoxO, and PPARgamma. Oxidative Med. Cell. Longev. 2017, 2017, 4397340. [Google Scholar] [CrossRef] [PubMed]
- Bhori, M.; Rastogi, V.; Tungare, K.; Marar, T. A review on interplay between obesity, lipoprotein profile and nutrigenetics with selected candidate marker genes of type 2 diabetes mellitus. Mol. Biol. Rep. 2022, 49, 687–703. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, G.; Simcox, J.; Seldin, M.M.; Parnell, T.J.; Stubben, C.; Just, S.; Begaye, L.; Lusis, A.J.; Villanueva, C.J. Targeted deletion of Tcf7l2 in adipocytes promotes adipocyte hypertrophy and impaired glucose metabolism. Mol. Metab. 2019, 24, 44–63. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Schramm, K.; Adamski, J.; Gieger, C.; Herder, C.; Carstensen, M.; Peters, A.; Rathmann, W.; Roden, M.; et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015, 11, e1005274. [Google Scholar] [CrossRef]
- Cuevas, A.G.; Wang, K.; Williams, D.R.; Mattei, J.; Tucker, K.L.; Falcon, L.M. The Association Between Perceived Discrimination and Allostatic Load in the Boston Puerto Rican Health Study. Psychosom. Med. 2019, 81, 659–667. [Google Scholar] [CrossRef]
| Type 2 Diabetes (T2D, n = 520) | No Diabetes (Non-T2D, n = 783) | Metabolomics (n = 806) | Full Cohort (n = 1303) | ||
|---|---|---|---|---|---|
| Age (SD) | 58.8 (7.2) * | 56.2 (7.7) | 57.2 (7.4) | 57.2 (7.6) | |
| Female (%) | 365 (70.2%) | 563 (71.9%) | 570 (70.7%) | 928 (71.2%) | |
| Body mass index (BMI, SD) | 33.6 (6.8) * | 30.9 (6.2) | 32.1 (6.7) | 31.9 (6.6) | |
| Waist (cm) | 106.3 (15.0) * | 98.9 (13.9) | 102.2 (14.8) | 101.8 (14.8) | |
| Obese # (n, %) | 414 (79.6%) * | 519 (66.2%) | 584 (72.5%) | 933 (71.6%) | |
| Fasting glucose (mg/dL) | 155.9 (69.6) * | 97.1 (11.2) | 119.9 (49.9) | 120.4 (53.2) | |
| Fasting insulin (mg/dL) | 24.0 (35.7) * | 14.1 (9.3) | 18.5 (24.6) | 18.0 (24.2) | |
| Glycosylated hemoglobin (HbA1c, %) | 8.3 (2.0) * | 6.1 (0.8) | 7.0 (1.7) | 7.0 (1.8) | |
| HOMA-IR | 9.9 (24.3) * | 3.4 (2.5) | 5.9 (9.3) | 6.0 (15.8) | |
| T2D medication | 431 (82.9%) * | 0 | 254 (31.5%) | 431 (33.1%) | |
| Hypertension (n, %) | 439 (84.4%) * | 459 (58.6%) | 448 (52.1%) | 719 (55.2%) | |
| Smoking (n, %) | Non-smoker | 241 (46.3%) | 347 (44.3%) | 373 | 588 |
| Past smoker | 167 (32.1%) | 231 (29.5%) | 248 | 398 | |
| Current smoker | 112 (21.5%) * | 203 (25.9%) | 183 | 315 | |
| Alcohol use (n, %) | Non-drinker | 161 (31.0%) | 219 (28.0%) | 226 | 380 |
| Past-drinker | 185 (35.6%) | 209 (26.7%) | 243 | 394 | |
| Current drinker | 172 (33.1%) * | 351 (44.8%) | 334 | 523 | |
| Education | 2.4 (1.0) * | 2.6 (1.0) | 2.5 (1.0) | 2.5 (1.0) | |
| Physical activity score | 30.8 (4.3) * | 31.8 (4.9) | 31.5 (4.7) | 31.4 (4.7) | |
| Total energy intake (kcal, SD) | 2076 (887) | 2153 (897) | 2174 (975) | 2122 (894) | |
| Metabolite Dass | COmpID | Metabolite | SNP | Chr | Position * | Associated Gene | p-Value | SNP Beta | SNP Beta SE | Minor/Major | MAF # |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate Metabolism | 57 | glutamate | rs77488629 | 17 | 19448308 | SLC47Al | 6.12 × 10−8 | 0.164 | 0.030 | T/C | 0.015 |
| 38754 | gamma-carboxyglutamate | rs10243315 | 7 | 142900460 | TAS2R40 | 2.53 × 10−8 | 0.621 | 0.110 | C/T | 0.012 | |
| 38754 | gamma-carboxyglutamate | rs62534412 | 9 | 2869593 | KIAA0020 | 3.71 × 10−8 | 0.634 | 0.114 | C/T | 0.011 | |
| 38754 | gamma-carboxyglutamate | rs116596264 | 11 | 118043675 | SCN2B | 4.70 × 10−9 | 0.623 | 0.105 | C/T | 0.013 | |
| 38754 | gamma-carboxyglutamate | rs79358823 | 18 | 10407315 | LOC105371988 | 2.65 × 10−9 | 0.652 | 0.108 | C/T | 0.010 | |
| 38754 | gamma-carboxyglutamate | rs16981047 | 20 | 19708935 | SLC24A3 | 4.25 × 10−8 | 0.512 | 0.092 | C/G | 0.014 | |
| Long Chain Fatty Acid | 1121 | margarate (17:0) | rs78025455 | 1 | 23890507 | ID3 | 2.46 × 10−8 | 0.458 | 0.081 | A/G | 0.014 |
| 1121 | margarate (17:0) | rs113584803 | 6 | 52435180 | TRAM2 | 1.62 × 10−8 | 0.407 | 0.071 | G/A | 0.019 | |
| 1121 | margarate (17:0) | rs78432898 | 6 | 52574289 | ? | 6.22 × 10−11 | 0.441 | 0.066 | C/T | 0.021 | |
| 1121 | margarate (17:0) | rs17322413 | 7 | 5879004 | ZNF815P | 1.66 × 10−8 | 0.334 | 0.058 | T/C | 0.029 | |
| 1121 | margarate (17:0) | rs78940323 | 12 | 129114888 | TMEM132C | 6.25 × 10−9 | 0.407 | 0.069 | A/G | 0.018 | |
| 1365 | myristate (14:0) | rs78432898 | 6 | 52574289 | ? | 1.03 × 10−8 | 0.484 | 0.083 | C/T | 0.021 | |
| 1336 | palmitate (16:0) | rs78432898 | 6 | 52574289 | ? | 8.74 × 10−10 | 0.342 | 0.055 | C/T | 0.021 | |
| 1358 | stearate (18:0) | rs78432898 | 6 | 52574289 | ? | 2.85 × 10−8 | 0.203 | 0.036 | C/T | 0.021 | |
| 33971 | 10-heptadecenoate (17:1n7) | rs78432898 | 6 | 52574289 | ? | 9.38 × 10−9 | 0.641 | 0.110 | C/T | 0.021 | |
| 33972 | 10-nonadecenoate (19:1n9) | rs113584803 | 6 | 52435180 | TRAM2 | 1.98 × 10−8 | 0.625 | 0.110 | G/A | 0.019 | |
| 33972 | 10-nonadecenoate (19:1n9) | rs78432898 | 6 | 52574289 | ? | 7.11 × 10−11 | 0.679 | 0.102 | C/T | 0.021 | |
| 33972 | 10-nonadecenoate (19:1n9) | rs73290946 | 7 | 12023844 | TMEM106B | 3.45 × 10−8 | 0.541 | 0.097 | G/A | 0.022 | |
| 33972 | 10-nonadecenoate (19:1n9) | rs78940323 | 12 | 129114888 | TMEM132C | 9.18 × 10−9 | 0.622 | 0.107 | A/G | 0.018 | |
| 33972 | 10-nonadecenoate (19:1n9) | rs112553117 | 12 | 129117140 | TMEM132C | 5.54 × 10−9 | 0.555 | 0.094 | T/C | 0.024 | |
| Phosphatidyle thanolamine (PE) | 52464 | 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) | rs10998764 | 10 | 71170572 | TACR2 | 1.00 × 10−9 | 1.332 | 0.214 | A/G | 0.010 |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174533 | 11 | 61549025 | MYRF | 2.13 × 10−8 | 0.388 | 0.068 | A/G | 0.341 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs102274 | 11 | 61557826 | TMEM258 | 1.12 × 10−8 | 0.398 | 0.069 | C/T | 0.341 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174546 | 11 | 61569830 | FADS1 | 2.01 × 10−8 | 0.390 | 0.068 | T/C | 0.341 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174547 | 11 | 61570783 | FADS1 | 2.01 × 10−8 | 0.390 | 0.068 | C/T | 0.342 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174548 | 11 | 61571348 | FADS1 | 2.88 × 10−9 | 0.402 | 0.067 | G/C | 0.403 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174549 | 11 | 61571382 | FADS1 | 1.57 × 10−8 | 0.396 | 0.069 | A/G | 0.333 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174550 | 11 | 61571478 | FADS1 | 2.01 × 10−8 | 0.390 | 0.068 | C/T | 0.341 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174555 | 11 | 61579760 | FADS1 | 1.89 × 10−8 | 0.392 | 0.069 | C/T | 0.335 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174566 | 11 | 61592362 | FADS2 | 3.61 × 10−8 | 0.369 | 0.066 | G/A | 0.421 | |
| 42449 | 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | rs174570 | 11 | 61597212 | FADS2 | 1.31 × 10−8 | 0.429 | 0.074 | T/C | 0.239 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs116694568 | 1 | 211536212 | TRAF5 | 4.88 × 10−9 | 1.765 | 0.297 | G/A | 0.016 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs111410641 | 9 | 20368408 | MLLT3 | 1.97 × 10−8 | 0.837 | 0.147 | T/C | 0.061 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs10998764 | 10 | 71170572 | TACR2 | 3.55 × 10−9 | 2.080 | 0.347 | A/G | 0.010 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs11618281 | 13 | 31488958 | MEDAG | 1.27 × 10−8 | 1.373 | 0.238 | A/G | 0.026 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs80051407 | 16 | 62592922 | ? | 2.89 × 10−8 | 1.890 | 0.336 | G/T | 0.013 | |
| 19263 | 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | rs73257193 | 17 | 9779676 | GLP2R | 1.10 × 10−8 | 2.045 | 0.353 | A/G | 0.012 | |
| 52446 | 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) | rs58821884 | 19 | 11159230 | SMARCA4 | 4.96 × 10−8 | 1.065 | 0.193 | A/G | 0.021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, C.-Q.; Parnell, L.D.; Li, Z.; Noel, S.E.; Bhupathiraju, S.N.; Tucker, K.L.; Ordovás, J.M. Metabolite Genome-Wide Association in Hispanics with Obesity Reveals Genetic Risk and Interactions with Dietary Factors for Type 2 Diabetes. Metabolites 2025, 15, 697. https://doi.org/10.3390/metabo15110697
Lai C-Q, Parnell LD, Li Z, Noel SE, Bhupathiraju SN, Tucker KL, Ordovás JM. Metabolite Genome-Wide Association in Hispanics with Obesity Reveals Genetic Risk and Interactions with Dietary Factors for Type 2 Diabetes. Metabolites. 2025; 15(11):697. https://doi.org/10.3390/metabo15110697
Chicago/Turabian StyleLai, Chao-Qiang, Laurence D. Parnell, Zhuoheng Li, Sabrina E. Noel, Shilpa N. Bhupathiraju, Katherine L. Tucker, and José M. Ordovás. 2025. "Metabolite Genome-Wide Association in Hispanics with Obesity Reveals Genetic Risk and Interactions with Dietary Factors for Type 2 Diabetes" Metabolites 15, no. 11: 697. https://doi.org/10.3390/metabo15110697
APA StyleLai, C.-Q., Parnell, L. D., Li, Z., Noel, S. E., Bhupathiraju, S. N., Tucker, K. L., & Ordovás, J. M. (2025). Metabolite Genome-Wide Association in Hispanics with Obesity Reveals Genetic Risk and Interactions with Dietary Factors for Type 2 Diabetes. Metabolites, 15(11), 697. https://doi.org/10.3390/metabo15110697






