Metabolomic Signatures of MASLD Identified by the Fatty Liver Index Reveal Gamma-Glutamyl Cycle Disruption and Lipid Remodeling
Abstract
1. Introduction
2. Methods
2.1. Data Source and Study Participants
2.2. Metabolomics
2.3. Statistical Analysis
3. Results
3.1. General Characteristics of Participants
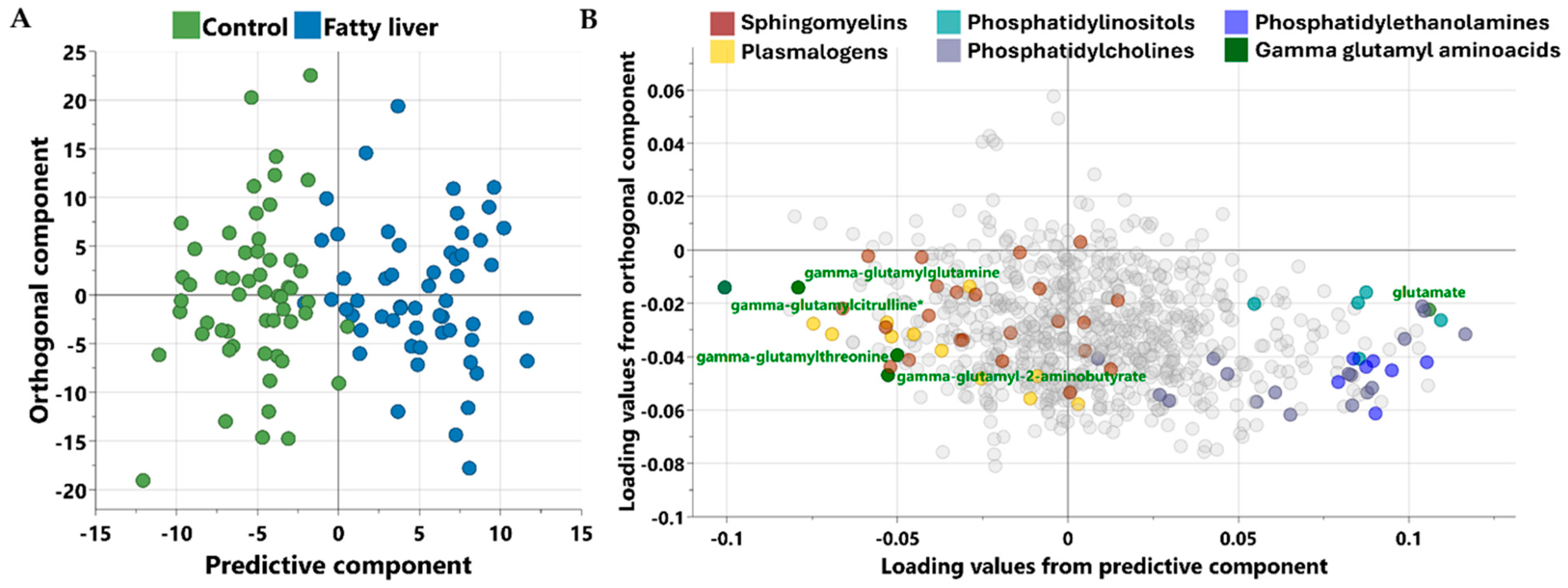
3.2. Multivariate Analysis
3.3. Univariate Analysis
3.4. Functional Enrichment Analysis
3.5. Association Between Metabolites Associated with MASLD and Clinical Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, L. From metabolic dysfunction-associated fatty liver disease to metabolic dysfunction-associated steatotic liver disease: Controversy and consensus. World J. Hepatol. 2023, 15, 1253–1257. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Zelber-Sagi, S.; Lazarus, J.V.; Wong, V.W.; Yilmaz, Y.; Duseja, A.; Eguchi, Y.; Castera, L.; Pessoa, M.G.; Oliveira, C.P.; et al. Global Consensus Recommendations for Metabolic Dysfunction-Associated Steatotic Liver Disease and Steatohepatitis. Gastroenterology 2025, 169, 1017–1032.e1012. [Google Scholar] [CrossRef]
- Diaz, L.A.; Arab, J.P.; Idalsoaga, F.; Perelli, J.; Vega, J.; Dirchwolf, M.; Carreño, J.; Samith, B.; Valério, C.; Moreira, R.O.; et al. Updated recommendations for the management of metabolic dysfunction–associated steatotic liver disease (MASLD) by the Latin American working group. Ann. Hepatol. 2025, 30, 101903. [Google Scholar] [CrossRef]
- Chan, W.K.; Chuah, K.H.; Rajaram, R.B.; Lim, L.L.; Ratnasingam, J.; Vethakkan, S.R. Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD): A State-of-the-Art Review. J. Obes. Metab. Syndr. 2023, 32, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.; Yeo, Y.H.; Li, X.; Li, J.; Zou, B.; Wu, Y.; Ye, Q.; Huang, D.Q.; Zhao, C.; Zhang, J.; et al. 2019 Global NAFLD Prevalence: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2022, 20, 2809–2817.e2828. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, J.; Xie, K.; Tang, C.; Gan, C.; Gao, J. MASLD development: From molecular pathogenesis toward therapeutic strategies. Chin. Med. J. 2025, 138, 1807–1824. [Google Scholar] [CrossRef]
- Bedogni, G.; Bellentani, S.; Miglioli, L.; Masutti, F.; Passalacqua, M.; Castiglione, A.; Tiribelli, C. The Fatty Liver Index: A simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ren, S.; Mi, H.; Wang, M.; He, T.; Zhang, R.; Jiang, W.; Su, C. Fatty liver index as an independent predictor of all-cause and disease-specific mortality. Eur. J. Gastroenterol. Hepatol. 2024, 36, 1453–1463. [Google Scholar] [CrossRef]
- Biciusca, T.; Stan, S.I.; Balteanu, M.A.; Cioboata, R.; Ghenea, A.E.; Danoiu, S.; Bumbea, A.M.; Biciusca, V. The Role of the Fatty Liver Index (FLI) in the Management of Non-Alcoholic Fatty Liver Disease: A Systematic Review. Diagnostics 2023, 13, 3316. [Google Scholar] [CrossRef]
- Babu, A.F.; Palomurto, S.; Kärjä, V.; Käkelä, P.; Lehtonen, M.; Hanhineva, K.; Pihlajamäki, J.; Männistö, V. Metabolic signatures of metabolic dysfunction-associated steatotic liver disease in severely obese patients. Dig. Liver Dis. 2024, 56, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Beyoğlu, D.; Popov, Y.V.; Idle, J.R. Metabolomic Hallmarks of Obesity and Metabolic Dysfunction-Associated Steatotic Liver Disease. Int. J. Mol. Sci. 2024, 25, 12809. [Google Scholar] [CrossRef]
- Al Thani, A.; Fthenou, E.; Paparrodopoulos, S.; Al Marri, A.; Shi, Z.; Qafoud, F.; Afifi, N. Qatar Biobank Cohort Study: Study Design and First Results. Am. J. Epidemiol. 2019, 188, 1420–1433. [Google Scholar] [CrossRef]
- Thareja, G.; Al-Sarraj, Y.; Belkadi, A.; Almotawa, M.; Suhre, K.; Albagha, O.M.E. Whole genome sequencing in the Middle Eastern Qatari population identifies genetic associations with 45 clinically relevant traits. Nat. Commun. 2021, 12, 1250. [Google Scholar] [CrossRef]
- Suhre, K.; Stephan, N.; Zaghlool, S.; Triggle, C.R.; Robinson, R.J.; Evans, A.M.; Halama, A. Matching Drug Metabolites from Non-Targeted Metabolomics to Self-Reported Medication in the Qatar Biobank Study. Metabolites 2022, 12, 249. [Google Scholar] [CrossRef]
- Zaghlool, S.B.; Halama, A.; Stephan, N.; Gudmundsdottir, V.; Gudnason, V.; Jennings, L.L.; Thangam, M.; Ahlqvist, E.; Malik, R.A.; Albagha, O.M.E.; et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an Arab population. Nat. Commun. 2022, 13, 7121. [Google Scholar] [CrossRef]
- Amedeo, L.; Stefano, B.; Giorgio, B.; Stefano, B.; Claudio, T. The Fatty liver Index (FLI) 15 years later: A reappraisal. Metab. Target. Organ. Damage 2021, 1, 10. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. Open 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.; Bridgewater, B.; Liu, Q.; Mitchell, M.; Robinson, R.; Dai, H.; Stewart, S.; Dehaven, C.; Miller, L. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 1. [Google Scholar] [CrossRef]
- Rao, G.; Peng, X.; Li, X.; An, K.; He, H.; Fu, X.; Li, S.; An, Z. Unmasking the enigma of lipid metabolism in metabolic dysfunction-associated steatotic liver disease: From mechanism to the clinic. Front. Med. 2023, 10, 1294267. [Google Scholar] [CrossRef]
- Hua, X.; Li, M.; Pan, F.; Xiao, Y.; Cui, W.; Hu, Y. Non-alcoholic fatty liver disease is an influencing factor for the association of SHBG with metabolic syndrome in diabetes patients. Sci. Rep. 2017, 7, 14532. [Google Scholar] [CrossRef]
- Fodor Duric, L.; Belčić, V.; Oberiter Korbar, A.; Ćurković, S.; Vujicic, B.; Gulin, T.; Muslim, J.; Gulin, M.; Grgurević, M.; Catic Cuti, E. The Role of SHBG as a Marker in Male Patients with Metabolic-Associated Fatty Liver Disease: Insights into Metabolic and Hormonal Status. J. Clin. Med. 2024, 13, 7717. [Google Scholar] [CrossRef]
- Naja, K.; Anwardeen, N.; Albagha, O.; Elrayess, M.A. Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index. Metabolites 2025, 15, 342. [Google Scholar] [CrossRef] [PubMed]
- Bozelli, J.C., Jr.; Azher, S.; Epand, R.M. Plasmalogens and Chronic Inflammatory Diseases. Front. Physiol. 2021, 12, 730829. [Google Scholar] [CrossRef]
- Green, C.D.; Maceyka, M.; Cowart, L.A.; Spiegel, S. Sphingolipids in metabolic disease: The good, the bad, and the unknown. Cell Metab. 2021, 33, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.-K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.; et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef]
- Jang, J.E.; Park, H.S.; Yoo, H.J.; Baek, I.J.; Yoon, J.E.; Ko, M.S.; Kim, A.R.; Kim, H.S.; Park, H.S.; Lee, S.E.; et al. Protective role of endogenous plasmalogens against hepatic steatosis and steatohepatitis in mice. Hepatology 2017, 66, 416–431. [Google Scholar] [CrossRef]
- Liu, Y.; Cong, P.; Zhang, T.; Wang, R.; Wang, X.; Liu, J.; Wang, X.; Xu, J.; Wang, Y.; Wang, J.; et al. Plasmalogen attenuates the development of hepatic steatosis and cognitive deficit through mechanism involving p75NTR inhibition. Redox Biol. 2021, 43, 102002. [Google Scholar] [CrossRef] [PubMed]
- Taiwo, A.; Merrill, R.A.; Wendt, L.; Pape, D.; Thakkar, H.; Maschek, J.A.; Cox, J.; Summers, S.A.; Chaurasia, B.; Pothireddy, N.; et al. Metabolite perturbations in type 1 diabetes associated with metabolic dysfunction-associated steatotic liver disease. Front. Endocrinol. 2025, 16, 1500242. [Google Scholar] [CrossRef]
- Haley, A.P.; Knight-Scott, J.; Caillaud, M.; Gallagher, I.; Park, J.; Li, Y.; Wang, T.; Tanaka, H.; Browning, J.D. Low carbohydrate and low-calorie diets reduce liver fat and lower brain glutamate and myo-inositol levels in patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Metab. Brain Dis. 2025, 40, 199. [Google Scholar] [CrossRef]
- Lee, H.J.; Yeom, J.W.; Yun, J.H.; Jang, H.B.; Yoo, M.-G.; Kim, H.-J.; Koo, S.K.; Lee, H.-J. Increased glutamate in type 2 diabetes in the Korean population is associated with increased plasminogen levels. J. Diabetes 2023, 15, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Sun, Q.; Wang, R.; Wang, Y.; Wang, R. Impacts of glutamate, an exercise-responsive metabolite on insulin signaling. Life Sci. 2024, 341, 122471. [Google Scholar] [CrossRef]
- Lehn-Stefan, A.; Peter, A.; Machann, J.; Schick, F.; Randrianarisoa, E.; Heni, M.; Wagner, R.; Birkenfeld, A.L.; Fritsche, A.; Häring, H.U.; et al. Elevated Circulating Glutamate Is Associated With Subclinical Atherosclerosis Independently of Established Risk Markers: A Cross-Sectional Study. J. Clin. Endocrinol. Metab. 2021, 106, e982–e989. [Google Scholar] [CrossRef] [PubMed]
- Berkeley, L.I.; Cohen, J.F.; Crankshaw, D.L.; Shirota, F.N.; Nagasawa, H.T. Hepatoprotection by L-cysteine-glutathione mixed disulfide, a sulfhydryl-modified prodrug of glutathione. J. Biochem. Mol. Toxicol. 2003, 17, 95–97. [Google Scholar] [CrossRef] [PubMed]

| Test | Variable | Control Group (FLI < 30) n = 55 | MASLD Group (FLI ≥ 60) n = 55 | p Value |
|---|---|---|---|---|
| FLI | 22.2 (18.8–25.7) | 69.3 (62.4–84.1) | <0.001 | |
| General characteristics | Sex (M/F) | 23/32 | 23/32 | 0.99 |
| Age | 42.9 (11.8) | 44.2 (10.3) | 0.55 | |
| BMI (kg/m2) | 28.9 (26.2–30.9) | 28.7 (27.6–30.4) | 0.781 | |
| Waist to hip ratio | 0.81 (0.08) | 0.89 (0.09) | <0.001 | |
| Systolic blood pressure (mmHg) | 109 (101–116) | 120 (112–132) | <0.001 | |
| Diastolic blood pressure (mmHg) | 73 (67–78) | 78 (73–86.5) | 0.001 | |
| Handgrip left | 28.5 (22–37.7) | 28 (20–39) | 0.599 | |
| Handgrip right | 30.5 (22–42) | 32 (24–40) | 0.972 | |
| Blood sugar | Fasting blood glucose (mmol/L) | 4.9 (4.74–5.3) | 5.4 (4.8–6.2) | 0.016 |
| HbA1C (%) | 5.45 (5.3–5.7) | 5.7 (5.4–6.32) | 0.016 | |
| C-peptide (ng/mL) | 1.9 (1.51–2.53) | 2.74 (2.1–4.03) | <0.001 | |
| Insulin (uU/mL) | 8 (5–9) | 14.2 (9–20) | <0.001 | |
| Lipid profile | Total cholesterol (mmol/L) | 4.68 (1.06) | 5.4 (0.97) | <0.001 |
| HDL-cholesterol (mmol/L) | 1.49 (0.42) | 1.15 (0.27) | <0.001 | |
| LDL-cholesterol (mmol/L) | 2.71 (0.94) | 3.2 (1.03) | 0.011 | |
| Triglyceride (mmol/L) | 0.83 (0.64–1.25) | 2.18 (1.51–2.86) | <0.001 | |
| Cardiac function | NT-proBNP (pg/mL) | 33 (18.5–57.5) | 22.5 (11.8–43.7) | 0.097 |
| Homocysteine (µmol/L) | 8 (6.95–10.1) | 8.1 (6.9–9.4) | 0.477 | |
| Kidney function | Creatinine (µmol/L) | 66.16 (16.21) | 65.44 (12.94) | 0.910 |
| Urea (mmol/L) | 4.4 (3.4–5.3) | 4.3 (3.5–5) | 0.560 | |
| Bicarbonate (mmol/L) | 27 (26–29) | 27 (25–28) | 0.051 | |
| Total protein (g/L) | 71.25 (4.18) | 74.06 (3.37) | <0.001 | |
| Liver function | Alkaline phosphatase (U/L) | 59 (50.5–72) | 74 (63.5–85) | <0.001 |
| ALT (U/L) | 16 (11.5–21) | 32 (21–40.5) | <0.001 | |
| AST (U/L) | 16 (13–19) | 23 (18–31) | <0.001 | |
| GGT (U/L) | 12 (8.5–15) | 44 (24.5–84) | <0.001 | |
| Albumin (g/L) | 44.96 (2.45) | 45.31 (3.14) | 0.521 | |
| Bilirubin (µmol/L) | 6.9 (4.5–8) | 6 (4–7) | 0.209 | |
| Hormones | SHBG (nmol/L) | 54 (35.5–78.75) | 31 (21–42) | <0.001 |
| T4 (pmol/L) | 13.4 (12.35–14.1) | 13.2 (12.25–14.47) | 0.901 | |
| T3 (pmol/L) | 4.33 (0.53) | 4.52 (0.59) | 0.097 | |
| Metabolites | Super-Pathway | Sub-Pathway | Estimate | SE | p-Value | FDR |
|---|---|---|---|---|---|---|
| Glutamate | Amino Acid | Glutamate Metabolism | 0.537 | 0.069 | 4.4 × 10−12 | 3.6 × 10−9 |
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1) * | Lipid | Plasmalogen | −0.379 | 0.056 | 1.1 × 10−9 | 4.4 × 10−7 |
| 1-palmitoyl-2-palmitoleoyl-GPC (16:0/16:1) * | Lipid | Phosphatidylcholine | 0.496 | 0.083 | 2.9 × 10−8 | 6.1 × 10−6 |
| Gamma-glutamylcitrulline * | Peptide | Gamma-glutamyl Amino Acid | −0.524 | 0.089 | 5.8 × 10−8 | 8.0 × 10−6 |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPC (P-16:0/18:2) * | Lipid | Plasmalogen | −0.369 | 0.064 | 9.8 × 10−8 | 1.2 × 10−5 |
| Sphingomyelin (d18:2/24:1, d18:1/24:2) * | Lipid | Sphingomyelins | −0.240 | 0.045 | 6.6 × 10−7 | 6.8 × 10−5 |
| 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0) * | Lipid | Plasmalogen | −0.243 | 0.047 | 1.2 × 10−6 | 1.1 × 10−4 |
| 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | Lipid | Phosphatidylethanolamine | 0.511 | 0.100 | 1.4 × 10−6 | 1.2 × 10−4 |
| Sphingomyelin (d18:1/24:1, d18:2/24:0) * | Lipid | Sphingomyelins | −0.200 | 0.042 | 6.5 × 10−6 | 3.8 × 10−4 |
| Cysteine-glutathione disulfide | Amino Acid | Glutathione Metabolism | −0.760 | 0.172 | 9.1 × 10−6 | 4.4 × 10−4 |
| 1-palmitoyl-2-arachidonoyl-GPI (16:0/20:4) * | Lipid | Phosphatidylinositol | 0.370 | 0.080 | 1.3 × 10−5 | 5.9 × 10−4 |
| 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | Lipid | Phosphatidylethanolamine | 0.424 | 0.093 | 1.5 × 10−5 | 6.6 × 10−4 |
| 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) * | Lipid | Phosphatidylethanolamine | 0.404 | 0.091 | 2.2 × 10−5 | 8.9 × 10−4 |
| 1-(1-enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1) * | Lipid | Plasmalogen | −0.330 | 0.075 | 2.8 × 10−5 | 1.0 × 10−3 |
| 1-myristoyl-2-arachidonoyl-GPC (14:0/20:4) * | Lipid | Phosphatidylcholine | 0.473 | 0.109 | 3.4 × 10−5 | 1.1 × 10−3 |
| 1-myristoyl-2-palmitoyl-GPC (14:0/16:0) | Lipid | Phosphatidylcholine | 0.451 | 0.104 | 3.5 × 10−5 | 1.1 × 10−3 |
| Gamma-glutamylglutamine | Peptide | Gamma-glutamyl Amino Acid | −0.287 | 0.067 | 4.3 × 10−5 | 1.2 × 10−3 |
| Palmitoyl sphingomyelin (d18:1/16:0) | Lipid | Sphingomyelins | −0.143 | 0.035 | 8.4 × 10−5 | 2.1 × 10−3 |
| Serine | Amino Acid | Glycine, Serine and Threonine Metabolism | −0.150 | 0.038 | 1.2 × 10−4 | 2.9 × 10−3 |
| 1-palmitoyl-2-docosahexaenoyl-GPE (16:0/22:6) * | Lipid | Phosphatidylethanolamine | 0.472 | 0.119 | 1.3 × 10−4 | 3.1 × 10−3 |
| 1-palmitoyl-2-linoleoyl-GPI (16:0/18:2) | Lipid | Phosphatidylinositol | 0.311 | 0.081 | 2.0 × 10−4 | 4.6 × 10−3 |
| 1-palmitoyl-2-dihomo-linolenoyl-GPC (16:0/20:3n3 or 6) * | Lipid | Phosphatidylcholine | 0.222 | 0.058 | 2.3 × 10−4 | 4.9 × 10−3 |
| Gamma-glutamyl-2-aminobutyrate | Peptide | Gamma-glutamyl Amino Acid | −0.307 | 0.081 | 2.7 × 10−4 | 5.7 × 10−3 |
| 1-stearoyl-2-docosahexaenoyl-GPC (18:0/22:6) | Lipid | Phosphatidylcholine | 0.301 | 0.081 | 3.1 × 10−4 | 6.0 × 10−3 |
| Sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1) * | Lipid | Sphingomyelins | −0.133 | 0.036 | 3.3 × 10−4 | 6.3 × 10−3 |
| 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | Lipid | Phosphatidylethanolamine | 0.368 | 0.099 | 3.5 × 10−4 | 6.3 × 10−3 |
| Sphingomyelin (d18:2/18:1) * | Lipid | Sphingomyelins | −0.194 | 0.053 | 3.6 × 10−4 | 6.4 × 10−3 |
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) * | Lipid | Phosphatidylethanolamine | 0.352 | 0.097 | 4.5 × 10−4 | 6.8 × 10−3 |
| Sphingomyelin (d18:2/24:2) * | Lipid | Sphingomyelins | −0.215 | 0.060 | 4.9 × 10−4 | 7.3 × 10−3 |
| Gamma-glutamylthreonine | Peptide | Gamma-glutamyl Amino Acid | −0.213 | 0.061 | 7.8 × 10−4 | 1.1 × 10−2 |
| Sphingomyelin (d18:2/16:0, d18:1/16:1) * | Lipid | Sphingomyelins | −0.126 | 0.036 | 8.9 × 10−4 | 1.2 × 10−2 |
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (P-16:0/18:1) * | Lipid | Plasmalogen | −0.193 | 0.057 | 1.1 × 10−3 | 1.4 × 10−2 |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (P-16:0/18:2) * | Lipid | Plasmalogen | −0.275 | 0.082 | 1.2 × 10−3 | 1.4 × 10−2 |
| Enriched Pathways | p-Value | FDR |
|---|---|---|
| Phosphatidylethanolamine | 5.1 × 10−7 | 4.7 × 10−5 |
| Sphingomyelins | 5.0 × 10−5 | 2.3 × 10−3 |
| Plasmalogen | 1.0 × 10−4 | 3.1 × 10−3 |
| Gamma-glutamyl amino acids | 3.1 × 10−4 | 3.4 × 10−3 |
| Phosphatidylcholine | 5.4 × 10−4 | 1.2 × 10−2 |
| Phosphatidylinositol | 1.1 × 10−3 | 2.0 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naja, K.; Anwardeen, N.; Elrayess, M.A. Metabolomic Signatures of MASLD Identified by the Fatty Liver Index Reveal Gamma-Glutamyl Cycle Disruption and Lipid Remodeling. Metabolites 2025, 15, 687. https://doi.org/10.3390/metabo15110687
Naja K, Anwardeen N, Elrayess MA. Metabolomic Signatures of MASLD Identified by the Fatty Liver Index Reveal Gamma-Glutamyl Cycle Disruption and Lipid Remodeling. Metabolites. 2025; 15(11):687. https://doi.org/10.3390/metabo15110687
Chicago/Turabian StyleNaja, Khaled, Najeha Anwardeen, and Mohamed A. Elrayess. 2025. "Metabolomic Signatures of MASLD Identified by the Fatty Liver Index Reveal Gamma-Glutamyl Cycle Disruption and Lipid Remodeling" Metabolites 15, no. 11: 687. https://doi.org/10.3390/metabo15110687
APA StyleNaja, K., Anwardeen, N., & Elrayess, M. A. (2025). Metabolomic Signatures of MASLD Identified by the Fatty Liver Index Reveal Gamma-Glutamyl Cycle Disruption and Lipid Remodeling. Metabolites, 15(11), 687. https://doi.org/10.3390/metabo15110687








