Untargeted Metabolomics of Dairy Cows as Influenced by the Combinations of Essential Oil Blends and Fumaric Acid as Natural Feed Additives Using RUSITEC
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Approval
2.2. Experimental Design
2.3. Rumen Fluid Sampling and Metabolite Characterization
2.4. Untargeted Metabolome Analysis
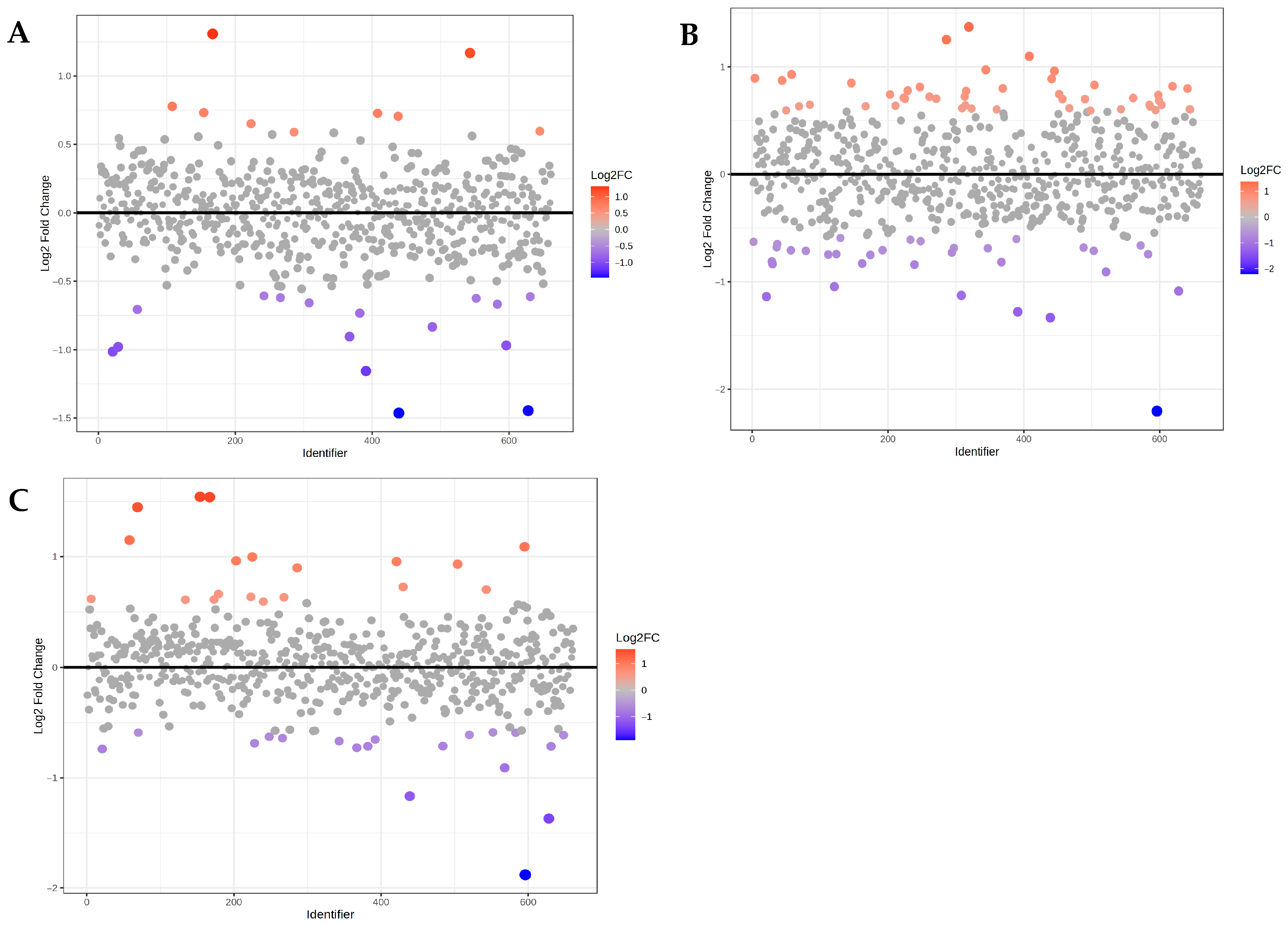
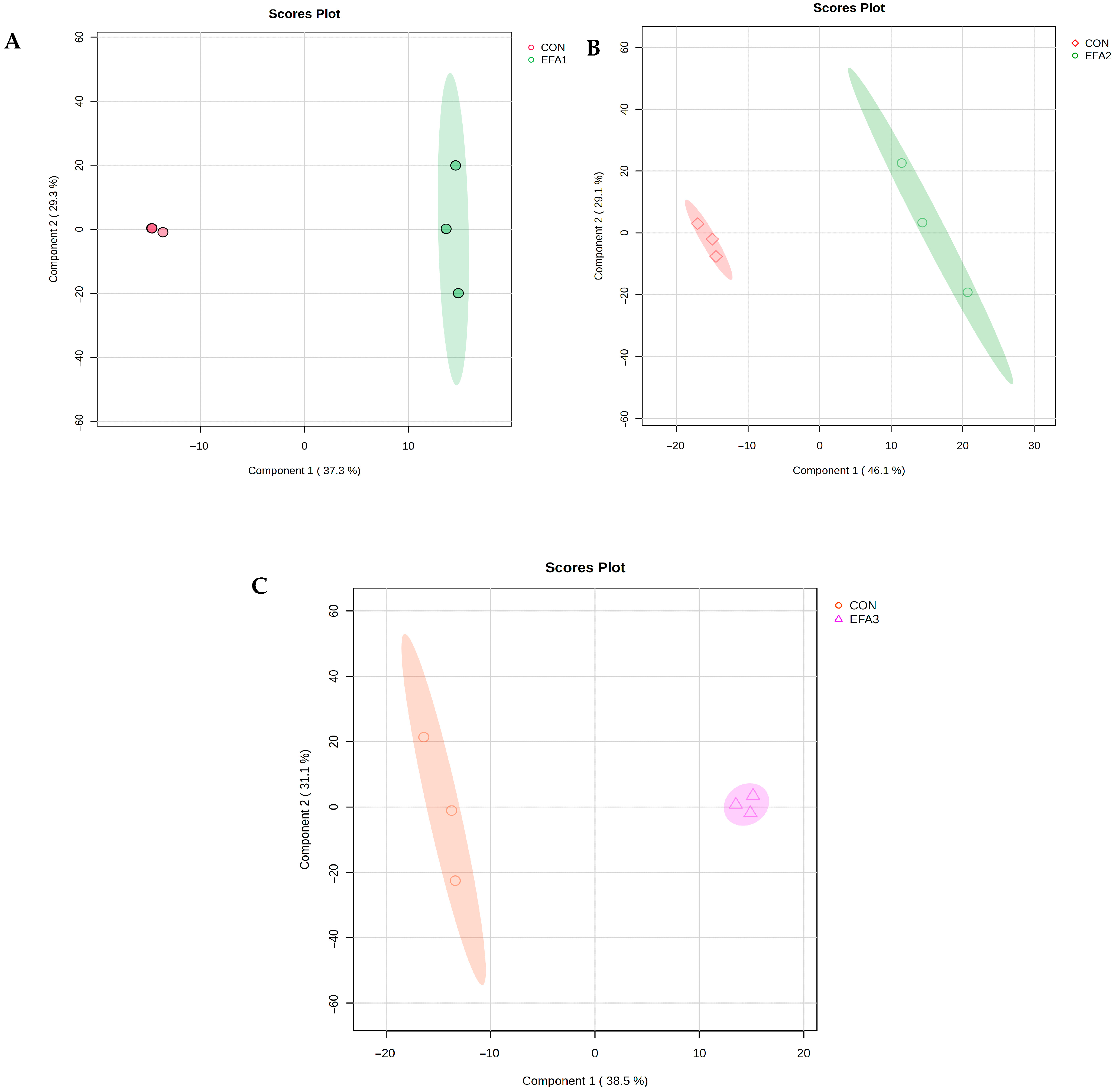
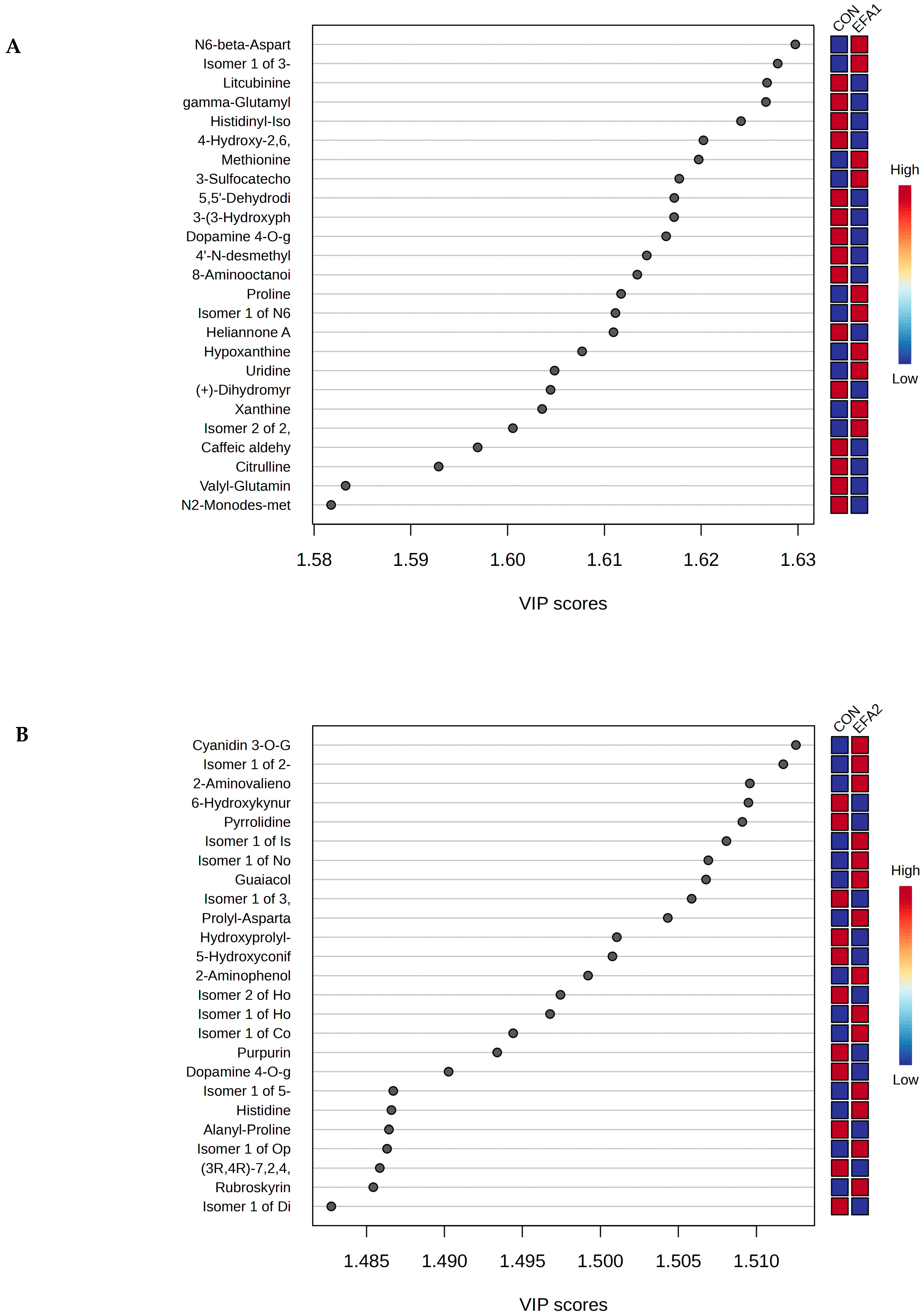
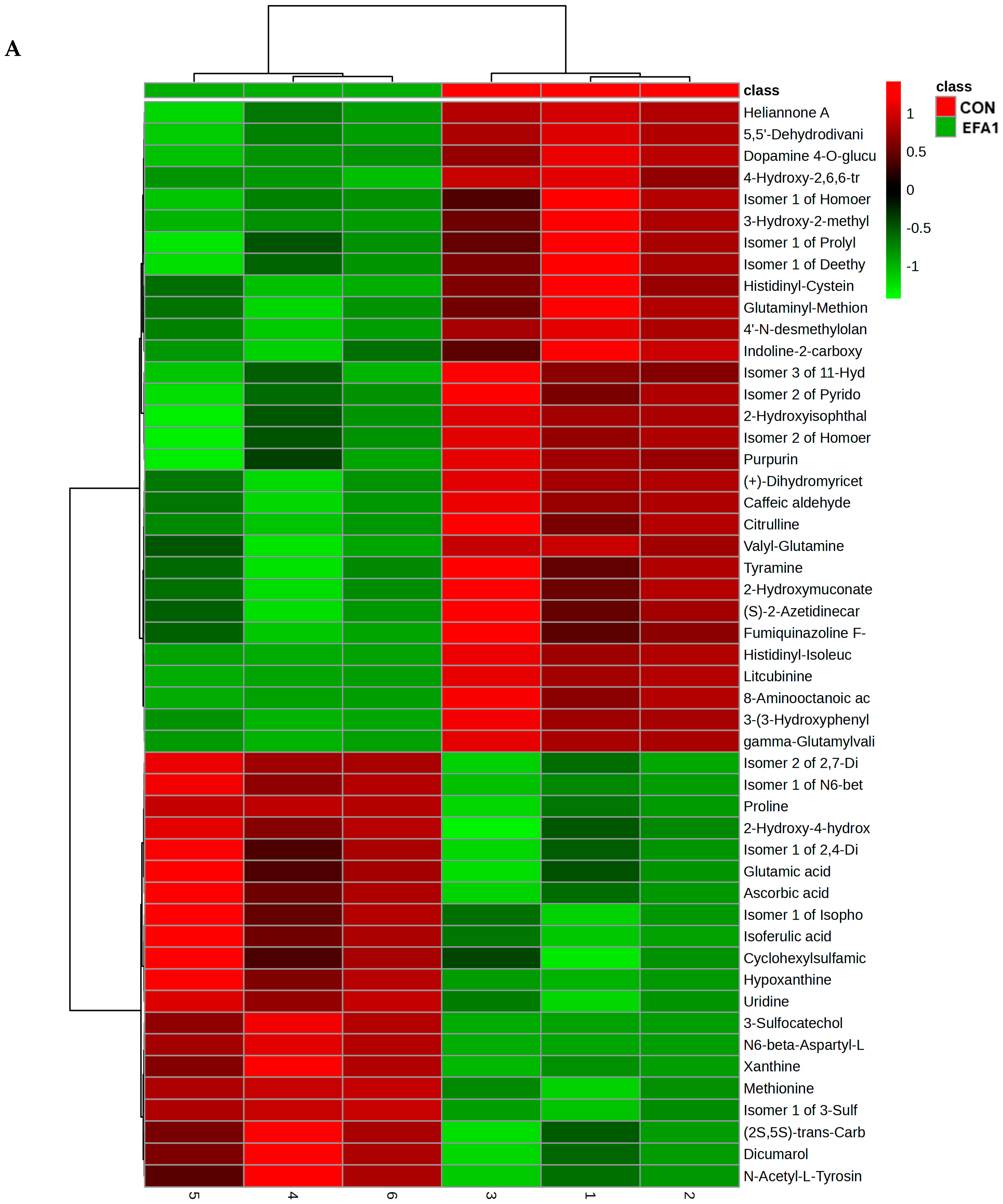
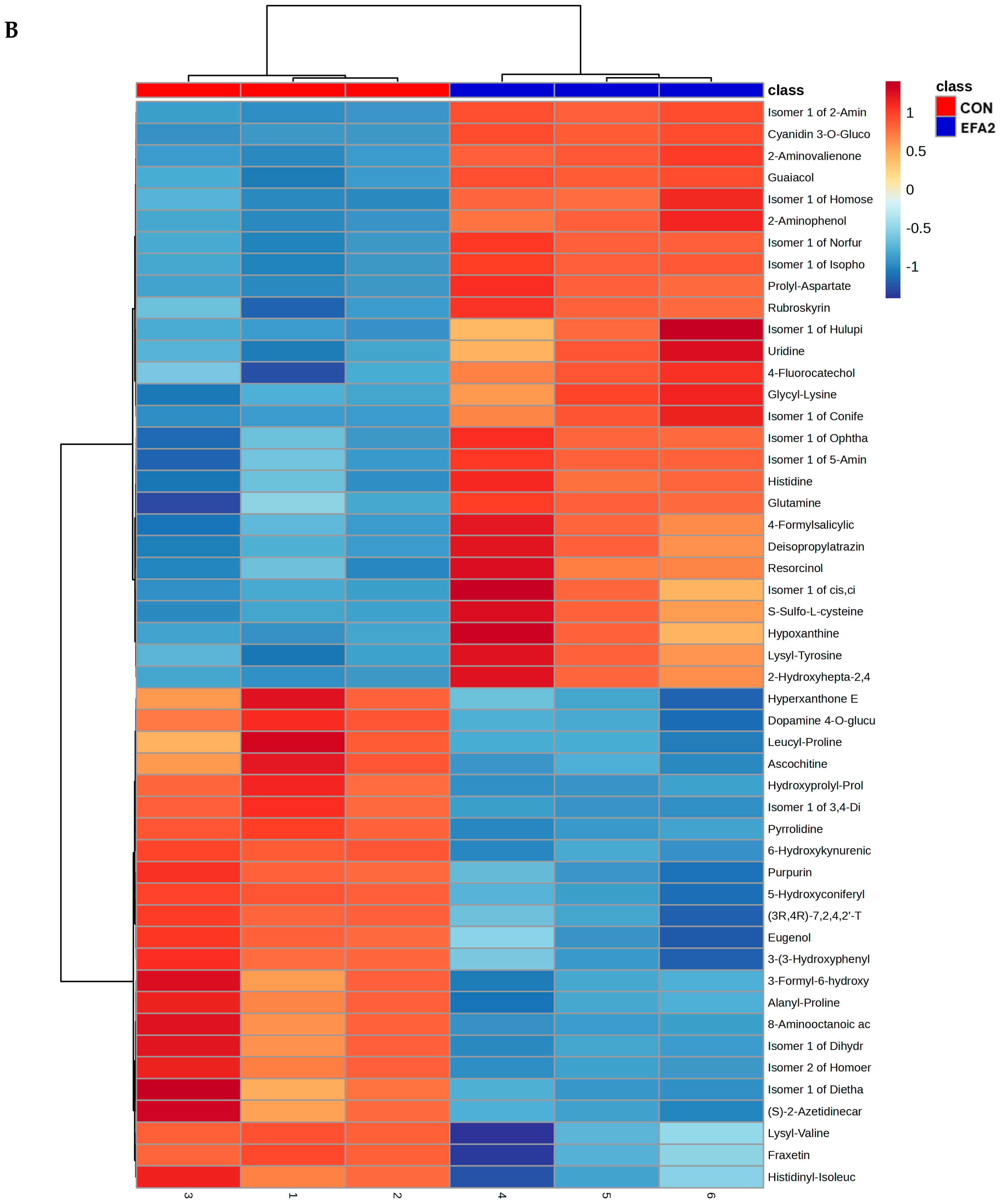
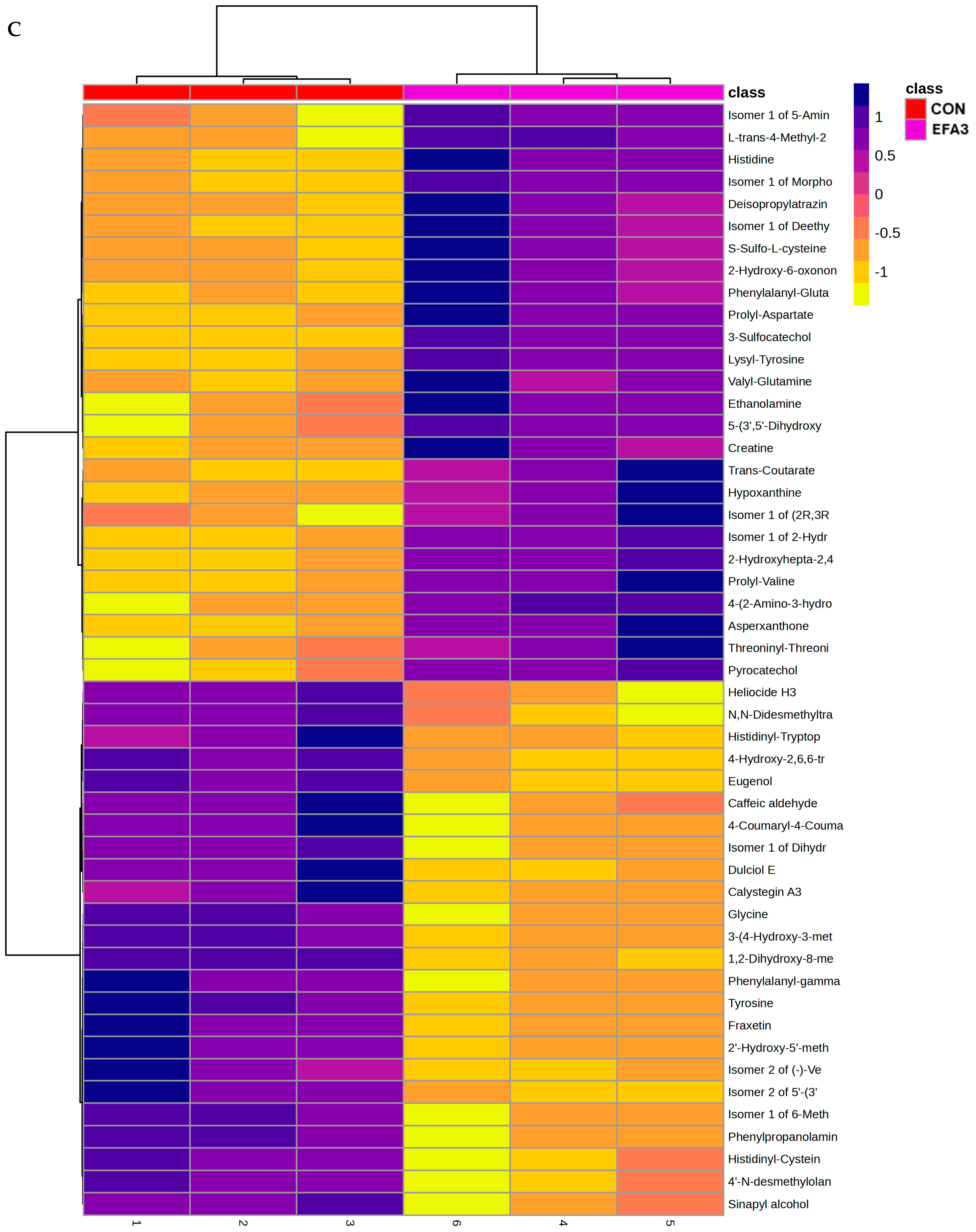
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Knapp, J.R.; Laur, G.L.; Vadas, P.A.; Weiss, W.P.; Tricarico, J.M. Invited Review: Enteric Methane in Dairy Cattle Production: Quantifying the Opportunities and Impact of Reducing Emissions. J. Dairy Sci. 2014, 97, 3231–3261. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. SPECIAL TOPICS—Mitigation of Methane and Nitrous Oxide Emissions from Animal Operations: I. A Review of Enteric Methane Mitigation Options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef]
- Gerber, P.J.; Hristov, A.N.; Henderson, B.; Makkar, H.P.S.; Oh, J.; Lee, C.; Meinen, R.; Montes, F.; Ott, T.; Firkins, J.; et al. Technical Options for the Mitigation of Direct Methane and Nitrous Oxide Emissions from Livestock: A Review. Animal 2013, 7, 220–234. [Google Scholar] [CrossRef]
- Liu, Y.; Espinosa, C.D.; Abelilla, J.J.; Casas, G.A.; Lagos, L.V.; Lee, S.A.; Kwon, W.B.; Mathai, J.K.; Navarro, D.M.D.L.; Jaworski, N.W.; et al. Non-Antibiotic Feed Additives in Diets for Pigs: A Review. Anim. Nutr. 2018, 4, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Omonijo, F.A.; Ni, L.; Gong, J.; Wang, Q.; Lahaye, L.; Yang, C. Essential Oils as Alternatives to Antibiotics in Swine Production. Anim. Nutr. 2018, 4, 126–136. [Google Scholar] [CrossRef]
- Zeng, Z.; Xu, X.; Zhang, Q.; Li, P.; Zhao, P.; Li, Q.; Liu, J.; Piao, X. Effects of Essential Oil Supplementation of a Low-Energy Diet on Performance, Intestinal Morphology and Microflora, Immune Properties and Antioxidant Activities in Weaned Pigs. Anim. Sci. J. 2015, 86, 279–285. [Google Scholar] [CrossRef]
- Blanch, M.; Carro, M.D.; Ranilla, M.J.; Viso, A.; Vázquez-Añón, M.; Bach, A. Influence of a Mixture of Cinnamaldehyde and Garlic Oil on Rumen Fermentation, Feeding Behavior and Performance of Lactating Dairy Cows. Anim. Feed. Sci. Technol. 2016, 219, 313–323. [Google Scholar] [CrossRef]
- Kholif, A.E.; Olafadehan, O.A. Essential Oils and Phytogenic Feed Additives in Ruminant Diet: Chemistry, Ruminal Microbiota and Fermentation, Feed Utilization and Productive Performance. Phytochem. Rev. 2021, 20, 1087–1108. [Google Scholar] [CrossRef]
- Tian, Q.; Piao, X. Essential Oil Blend Could Decrease Diarrhea Prevalence by Improving Antioxidative Capability for Weaned Pigs. Animals 2019, 9, 847. [Google Scholar] [CrossRef]
- Cardozo, P.W.; Calsamiglia, S.; Ferret, A.; Kamel, C. Screening for the Effects of Natural Plant Extracts at Different pH on in Vitro Rumen Microbial Fermentation of a High-Concentrate Diet for Beef Cattle. J. Anim. Sci. 2005, 83, 2572–2579. [Google Scholar] [CrossRef]
- Kholif, A.E.; Kassab, A.Y.; Azzaz, H.H.; Matloup, O.H.; Hamdon, H.A.; Olafadehan, O.A.; Morsy, T.A. Essential Oils Blend with a Newly Developed Enzyme Cocktail Works Synergistically to Enhance Feed Utilization and Milk Production of Farafra Ewes in the Subtropics. Small Rumin. Res. 2018, 161, 43–50. [Google Scholar] [CrossRef]
- Kholif, A.E.; Olafadehan, O.L.; Kholif, A.M.M.; Ghavipanje, N.; Vargas-Bello-Pérez, E.; Anele, U.Y. The Role of Encapsulated Essential Oils in Reducing Methane Production from Ruminant Animals—A Review. Ann. Anim. Sci. 2025; in press. [Google Scholar] [CrossRef]
- Gray, D.; Dele, P.A.; Alabi, J.O.; Adelusi, O.O.; Wuaku, M.; Okedoyin, D.O.; Anotaenwere, C.C.; Ike, K.A.; Oderinwale, O.A.; Kholif, A.E.; et al. Comparative Properties of Anise, Clove, Oregano and Peppermint Essential Oils Used Individually or Combined on Nutrient Digestibility and Greenhouse Gas Emissions in Concentrate-and Fiber-Based Diets. Agric. Conspec. Sci. 2025, 90, 71–81. [Google Scholar]
- Alabi, J.O.; Dele, P.A.; Okedoyin, D.O.; Wuaku, M.; Anotaenwere, C.C.; Adelusi, O.O.; Gray, D.; Ike, K.A.; Oderinwale, O.A.; Subedi, K.; et al. Synergistic Effects of Essential Oil Blends and Fumaric Acid on Ruminal Fermentation, Volatile Fatty Acid Production and Greenhouse Gas Emissions Using the Rumen Simulation Technique (RUSITEC). Fermentation 2024, 10, 114. [Google Scholar] [CrossRef]
- Alabi, J.O.; Okedoyin, D.O.; Anotaenwere, C.C.; Wuaku, M.; Gray, D.; Adelusi, O.O.; Ike, K.A.; Olagunju, L.K.; Dele, P.A.; Anele, U.Y. Essential Oil Blends with or without Fumaric Acid Influenced In Vitro Rumen Fermentation, Greenhouse Gas Emission, and Volatile Fatty Acids Production of a Total Mixed Ration. Ruminants 2023, 3, 373–384. [Google Scholar] [CrossRef]
- Remling, N.; Riede, S.; Meyer, U.; Beineke, A.; Breves, G.; Flachowsky, G.; Dänicke, S. Influence of Fumaric Acid on Ruminal Parameters and Organ Weights of Growing Bulls Fed with Grass or Maize Silage. Animal 2017, 11, 1754–1761. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, X.; Ma, X.; Geng, S.; Jiang, X.; Huang, Q.; Hu, C.; Han, X. Intestinal Microbiome-Metabolome Responses to Essential Oils in Piglets. Front. Microbiol. 2018, 9, 385734. [Google Scholar] [CrossRef]
- Lin, B.; Lu, Y.; Wang, J.; Liang, Q.; Liu, J. The Effects of Combined Essential Oils along with Fumarate on Rumen Fermentation and Methane Production in Vitro. J. Anim. Feed Sci. 2012, 21, 198–210. [Google Scholar] [CrossRef]
- Monteiro, M.S.; Carvalho, M.; Bastos, M.L.; Guedes de Pinho, P. Metabolomics Analysis for Biomarker Discovery: Advances and Challenges. Curr. Med. Chem. 2013, 20, 257–271. [Google Scholar] [CrossRef]
- Foroutan, A.; Goldansaz, S.A.; Lipfert, M.; Wishart, D.S. Protocols for NMR Analysis in Livestock Metabolomics. Methods Mol. Biol. 2019, 1996, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; He, D.; Yang, Z.; Fu, Y.; Zheng, H.; Xing, Z.; Shen, W.; Shi, A.; Tao, J. Metabolomic Analysis of Rumen Fluid in Tan Sheep Reveals Sex-Specific Key Metabolites and Pathways Associated with Residual Feed Intake. Sci. Rep. 2025, 15, 22115. [Google Scholar] [CrossRef]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A Comparison of the Microbiome and the Metabolome of Different Regions of the Equine Hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef]
- O’Callaghan, T.F.; Vázquez-Fresno, R.; Serra-Cayuela, A.; Dong, E.; Mandal, R.; Hennessy, D.; McAuliffe, S.; Dillon, P.; Wishart, D.S.; Stanton, C.; et al. Pasture Feeding Changes the Bovine Rumen and Milk Metabolome. Metabolites 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, J.; Lei, Y.; Bai, Y.; Jia, L.; Li, Z.; Liu, T.; Xu, Y.; Sun, J.; Wang, Y.; et al. Oregano Essential Oils Promote Rumen Digestive Ability by Modulating Epithelial Development and Microbiota Composition in Beef Cattle. Front. Nutr. 2021, 8, 722557. [Google Scholar] [CrossRef] [PubMed]
- Okedoyin, D.O.; Alabi, J.O.; Anotaenwere, C.C.; Wuaku, M.; Gray, D.; Adelusi, O.O.; Ike, K.A.; Dele, P.A.; Oderinwale, O.A.; Idowu, M.D.; et al. Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends. Ruminants 2024, 4, 329–351. [Google Scholar] [CrossRef]
- Huang, Y.; Cheng, S.; Shi, J.; He, P.; Ma, Y.; Yang, R.; Zhang, X.; Cao, Y.; Lei, Z. Enhancing Holstein Steers Growth Performance: Oregano Essential Oil’s Impact on Rumen Development, Functionality and Microorganism. Anim. Microbiome 2025, 7, 44. [Google Scholar] [CrossRef]
- Czerkawski, J.W.; Breckenridge, G. Design and Development of a Long-Term Rumen Simulation Technique (Rusitec). Br. J. Nutr. 1977, 38, 371–384. [Google Scholar] [CrossRef]
- Idowu, M.; Taiwo, G.; Sidney, T.; Morenikeji, O.B.; Pech Cervantes, A.; Estrada-Reyes, Z.M.; Wilson, M.; Ogunade, I.M. The Differential Plasma and Ruminal Metabolic Pathways and Ruminal Bacterial Taxa Associated with Divergent Residual Body Weight Gain Phenotype in Crossbred Beef Steers. Transl. Anim. Sci. 2023, 7, txad054. [Google Scholar] [CrossRef]
- Saleem, F.; Ametaj, B.N.; Bouatra, S.; Mandal, R.; Zebeli, Q.; Dunn, S.M.; Wishart, D.S. A Metabolomics Approach to Uncover the Effects of Grain Diets on Rumen Health in Dairy Cows. J. Dairy Sci. 2012, 95, 6606–6623. [Google Scholar] [CrossRef]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Hua, D.; Zhang, F.; Wang, Y.; Liu, J.; Yao, J.; et al. Dietary Supplementation with Inulin Improves Lactation Performance and Serum Lipids by Regulating the Rumen Microbiome and Metabolome in Dairy Cows. Anim. Nutr. 2021, 7, 1189–1204. [Google Scholar] [CrossRef]
- Calsamiglia, S.; Busquet, M.; Cardozo, P.W.; Castillejos, L.; Ferret, A. Invited Review: Essential Oils as Modifiers of Rumen Microbial Fermentation. J. Dairy Sci. 2007, 90, 2580–2595. [Google Scholar] [CrossRef]
- Benchaar, C.; Greathead, H. Essential Oils and Opportunities to Mitigate Enteric Methane Emissions from Ruminants. Anim. Feed Sci. Technol. 2011, 166–167, 338–355. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; McAllister, T.A.; McGinn, S.M. Dietary Mitigation of Enteric Methane from Cattle. CABI Rev. 2009, 4, 1–18. [Google Scholar] [CrossRef]
- Salinas-Chavira, J.; Arzola-Alvarez, C.; Hume, M.E.; Fonseca, M.; Ruiz-Barrera, O.; Castillo-Castillo, Y.; Ontiveros-Magadan, M.; Jones, B.; Crippen, T.L.; Poole, T.L.; et al. Influence of Medium Chain Fatty Acids on Selected Microbes and on In Vitro Ruminal Fermentation of Air-Exposed Corn Silage. Front. Vet. Sci. 2024, 11, 1416695. [Google Scholar] [CrossRef]
- Busquet, M.; Calsamiglia, S.; Ferret, A.; Carro, M.D.; Kamel, C. Effect of Garlic Oil and Four of Its Compounds on Rumen Microbial Fermentation. J. Dairy Sci. 2005, 88, 4393–4404. [Google Scholar] [CrossRef] [PubMed]
- Cobellis, G.; Trabalza-Marinucci, M.; Yu, Z. Critical Evaluation of Essential Oils as Rumen Modifiers in Ruminant Nutrition: A Review. Sci. Total Environ. 2016, 545–546, 556–568. [Google Scholar] [CrossRef]
- Bach, A.; Calsamiglia, S.; Stern, M.D. Nitrogen Metabolism in the Rumen. J. Dairy Sci. 2005, 88, E9–E21. [Google Scholar] [CrossRef]
- Calsamiglia, S.; Ferret, A.; Reynolds, C.K.; Kristensen, N.B.; van Vuuren, A.M. Strategies for Optimizing Nitrogen Use by Ruminants. Animal 2010, 4, 1184–1196. [Google Scholar] [CrossRef]
- Osorio, J.S.; Ji, P.; Drackley, J.K.; Luchini, D.; Loor, J.J. Supplemental Smartamine M or Metasmart during the Transition Period Benefits Postpartal Cow Performance and Blood Neutrophil Function. J. Dairy Sci. 2013, 96, 6248–6263. [Google Scholar] [CrossRef] [PubMed]
- Van Zijderveld, S.M.; Gerrits, W.J.J.; Dijkstra, J.; Newbold, J.R.; Hulshof, R.B.A.; Perdok, H.B. Persistency of Methane Mitigation by Dietary Nitrate Supplementation in Dairy Cows. J. Dairy Sci. 2011, 94, 4028–4038. [Google Scholar] [CrossRef] [PubMed]
- Aschenbach, J.R.; Kristensen, N.B.; Donkin, S.S.; Hammon, H.M.; Penner, G.B. Gluconeogenesis in Dairy Cows: The Secret of Making Sweet Milk from Sour Dough. IUBMB Life 2010, 62, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Elango, R. Methionine Nutrition and Metabolism: Insights from Animal Studies to Inform Human Nutrition. J. Nutr. 2020, 150, 2518S–2523S. [Google Scholar] [CrossRef] [PubMed]
- Loor, J.J.; Lopreiato, V.; Palombo, V.; D’Andrea, M. Physiological Impact of Amino Acids during Heat Stress in Ruminants. Anim. Front. 2023, 13, 69–80. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Xie, C.; Fang, J. Uridine Metabolism and Its Role in Glucose, Lipid, and Amino Acid Homeostasis. Biomed. Res. Int. 2020, 2020, 7091718. [Google Scholar] [CrossRef]
- Chen, X.B.; Chen, Y.K.; Franklin, M.F.; Ørskov, E.R.; Shand, W.J.; Orskov, E.R.; Shand, W.J. The Effect of Feed Intake and Body Weight on Purine Derivative Excretion and Microbial Protein Supply in Sheep. J. Anim. Sci. 1992, 70, 1534–1542. [Google Scholar] [CrossRef]
- Kim, D.; Kuppusamy, P.; Jung, J.S.; Kim, K.H.; Choi, K.C. Microbial Dynamics and in Vitro Degradation of Plant Secondary Metabolites in Hanwoo Steer Rumen Fluids. Animals 2021, 11, 2350. [Google Scholar] [CrossRef]
- Gladine, C.; Rock, E.; Morand, C.; Bauchart, D.; Durand, D. Bioavailability and Antioxidant Capacity of Plant Extracts Rich in Polyphenols, given as a Single Acute Dose, in Sheep Made Highly Susceptible to Lipoperoxidation. Br. J. Nutr. 2007, 98, 691–701. [Google Scholar] [CrossRef]
- Burr, S.P.; Costa, A.S.H.; Grice, G.L.; Timms, R.T.; Lobb, I.T.; Freisinger, P.; Dodd, R.B.; Dougan, G.; Lehner, P.J.; Frezza, C.; et al. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1α Stability in Aerobic Conditions. Cell Metab. 2016, 24, 740–752. [Google Scholar] [CrossRef]
- Russell, J.B.; Hespell, R.B. Microbial Rumen Fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]
- Ryan, R.P. Cyclic Di-GMP Signalling and the Regulation of Bacterial Virulence. Microbiology 2013, 159, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Waldman, S.A.; Murad, F. Cyclic GMP Synthesis and Function. Pharmacol. Rev. 1987, 39, 163–196. [Google Scholar] [CrossRef]
- Macheboeuf, D.; Morgavi, D.P.; Papon, Y.; Mousset, J.L.; Arturo-Schaan, M. Dose-Response Effects of Essential Oils on In Vitro Fermentation Activity of the Rumen Microbial Population. Anim. Feed Sci. Technol. 2008, 145, 335–350. [Google Scholar] [CrossRef]
- Newbold, C.J.; McIntosh, F.M.; Williams, P.; Losa, R.; Wallace, R.J. Effects of a Specific Blend of Essential Oil Compounds on Rumen Fermentation. Anim. Feed Sci. Technol. 2004, 114, 105–112. [Google Scholar] [CrossRef]
- Nagaraja, T.G.; Titgemeyer, E.C. Ruminal Acidosis in Beef Cattle: The Current Microbiological and Nutritional Outlook. J. Dairy Sci. 2007, 90, E17–E38. [Google Scholar] [CrossRef]
- Benchaar, C.; Calsamiglia, S.; Chaves, A.V.; Fraser, G.R.; Colombatto, D.; McAllister, T.A.; Beauchemin, K.A. A Review of Plant-Derived Essential Oils in Ruminant Nutrition and Production. Anim. Feed Sci. Technol. 2008, 145, 209–228. [Google Scholar] [CrossRef]
- Tao, S.; Dahl, G.E.; Laporta, J.; Bernard, J.K.; Orellana Rivas, R.M.; Marins, T.N. PHYSIOLOGY SYMPOSIUM: Effects of Heat Stress during Late Gestation on the Dam and Its Calf. J. Anim. Sci. 2019, 97, 2245–2257. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Wen, Z.; Cheng, X.; Li, C.; Zhang, P.; Xiao, D.; Xu, Y. Unlocking Gut Health: The Potent Role of Stilbenoids in Intestinal Homeostasis. Animals 2025, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, L.A.; Edwards, R.; Errington, K.A.; Holdcroft, A.M.; Wright, M. Replacement of Grass and Maize Silages with Lucerne Silage: Effects on Performance, Milk Fatty Acid Profile and Digestibility in Holstein-Friesian Dairy Cows. Animal 2015, 9, 1970–1978. [Google Scholar] [CrossRef][Green Version]
- Holeček, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [PubMed]
- Lambert, I.H.; Kristensen, D.M.; Holm, J.B.; Mortensen, O.H. Physiological Role of Taurine—From Organism to Organelle. Acta Physiol. 2015, 213, 191–212. [Google Scholar] [CrossRef]
- Garsin, D.A. Ethanolamine Utilization in Bacterial Pathogens: Roles and Regulation. Nat. Rev. Microbiol. 2010, 8, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Kaval, K.G.; Garsin, D.A. Ethanolamine Utilization in Bacteria. mBio 2018, 9, e00066-18. [Google Scholar] [CrossRef]
- Roof, D.M.; Roth, J.R. Ethanolamine Utilization in Salmonella typhimurium. J. Bacteriol. 1988, 170, 3855–3863. [Google Scholar] [CrossRef]
- Chapman, K.D. Occurrence, Metabolism, and Prospective Functions of N-Acylethanolamines in Plants. Prog. Lipid Res. 2004, 43, 302–327. [Google Scholar] [CrossRef]
- Narvaez, N.; Wang, Y.; Mcallister, T. Effects of Extracts of Humulus lupulus (Hops) and Yucca schidigera Applied Alone or in Combination with Monensin on Rumen Fermentation and Microbial Populations in Vitro. J. Sci. Food Agric. 2013, 93, 2517–2522. [Google Scholar] [CrossRef]
- Chen, G.; Sniffen, C.J.; Russell, J.B. Concentration and Estimated Flow of Peptides from the Rumen of Dairy Cattle: Effects of Protein Quantity, Protein Solubility, and Feeding Frequency. J. Dairy Sci. 1987, 70, 983–992. [Google Scholar] [CrossRef]
- Russell, J.B.; O’Connor, J.D.; Fox, D.G.; Van Soest, P.J.; Sniffen, C.J. A Net Carbohydrate and Protein System for Evaluating Cattle Diets: I. Ruminal Fermentation. J. Anim. Sci. 1992, 70, 3551–3561. [Google Scholar] [CrossRef]
- Schwab, C.G. Rumen-Protected Amino Acids for Dairy Cattle: Progress towards Determining Lysine and Methionine Requirements. Anim. Feed Sci. Technol. 1996, 59, 87–101. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Dairy Cattle; National Academies Press: Washington, DC, USA, 2001; ISBN 978-0-309-06997-7. [Google Scholar]
- Allison, M.J. Biosynthesis of Amono Acids by Ruminal Microorganisms. J. Anim. Sci. 1969, 29, 797–807. [Google Scholar] [CrossRef]
- Hultquist, K.M.; Casper, D.P. Effects of Feeding Rumen-Degradable Valine on Milk Production in Late-Lactating Dairy Cows. J. Dairy Sci. 2016, 99, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M.; Pandhare, J.; Liu, Y. The Metabolism of Proline as Microenvironmental Stress Substrate. J. Nutr. 2008, 138, 2008S–2015S. [Google Scholar] [CrossRef] [PubMed]
- Talley, J.T.; Mohiuddin, S.S. Biochemistry, Fatty Acid Oxidation; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Friedman, M.; Xu, A.; Lee, R.; Nguyen, D.N.; Phan, T.A.; Hamada, S.M.; Panchel, R.; Tam, C.C.; Kim, J.H.; Cheng, L.W.; et al. The Inhibitory Activity of Anthraquinones against Pathogenic Protozoa, Bacteria, and Fungi and the Relationship to Structure. Molecules 2020, 25, 3101. [Google Scholar] [CrossRef]
- Solmonson, A.; DeBerardinis, R.J. Lipoic Acid Metabolism and Mitochondrial Redox Regulation. J. Biol. Chem. 2018, 293, 7522–7530. [Google Scholar] [CrossRef] [PubMed]
- Benchaar, C.; Petit, H.V.; Berthiaume, R.; Ouellet, D.R.; Chiquette, J.; Chouinardt, P.Y. Effects of Essential Oils on Digestion, Ruminal Fermentation, Rumen Microbial Populations, Milk Production, and Milk Composition in Dairy Cows Fed Alfalfa Silage or Corn Silage. J. Dairy Sci. 2007, 90, 886–897. [Google Scholar] [CrossRef]
- Ungerfeld, E.M.; Kohn, R.A.; Wallace, R.J.; Newbold, C.J. A Meta-Analysis of Fumarate Effects on Methane Production in Ruminal Batch Cultures. J. Anim. Sci. 2007, 85, 2556–2563. [Google Scholar] [CrossRef]
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane Emissions from Beef Cattle: Effects of Monensin, Sunflower Oil, Enzymes, Yeast, and Fumaric Acid. J. Anim. Sci. 2004, 82, 3346–3356. [Google Scholar] [CrossRef]
- Castillejos, L.; Calsamiglia, S.; Ferret, A.; Losa, R. Effects of Dose and Adaptation Time of a Specific Blend of Essential Oil Compounds on Rumen Fermentation. Anim. Feed Sci. Technol. 2007, 132, 186–201. [Google Scholar] [CrossRef]
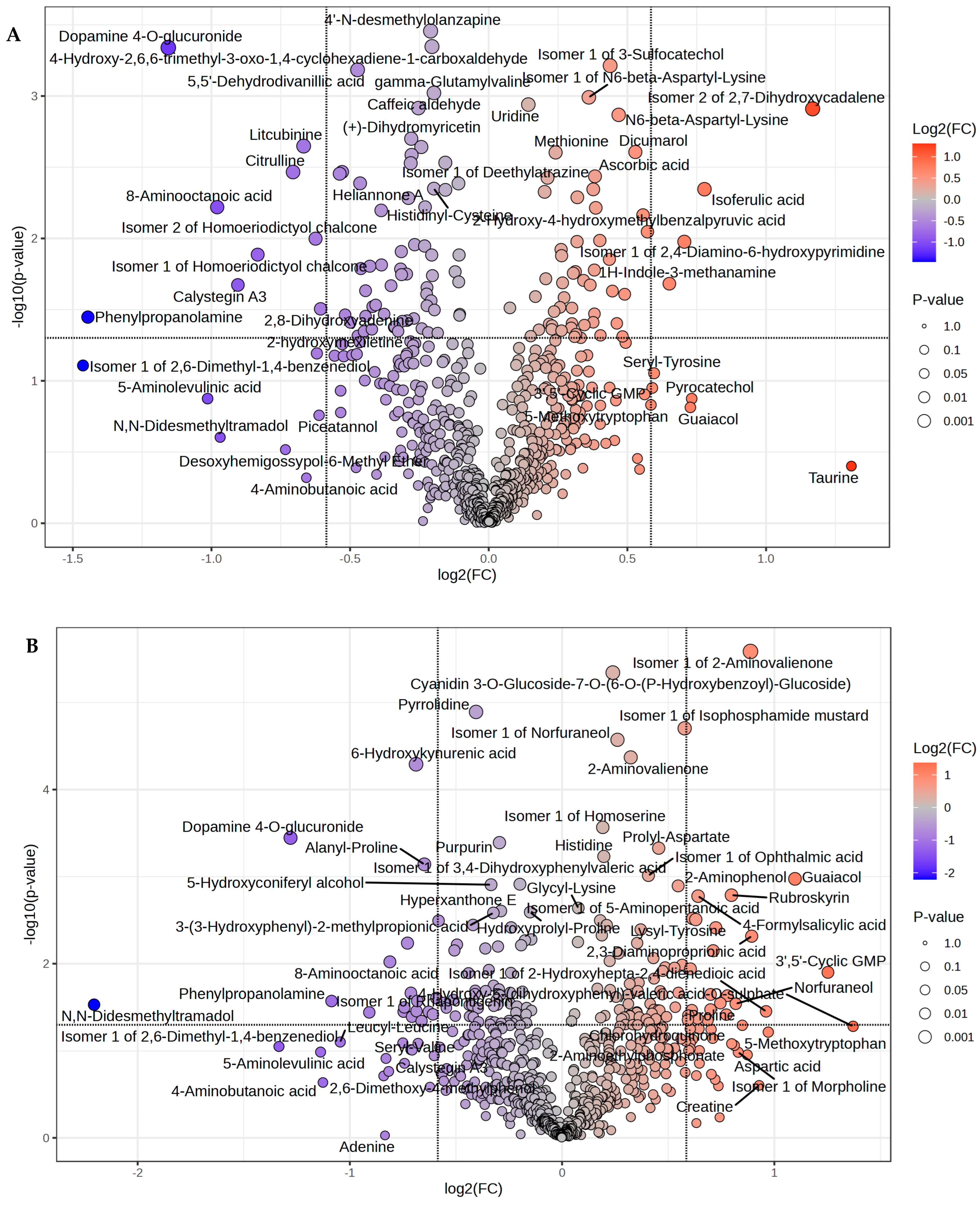


| Metabolites | Fold Change | Log2(FC) | FDR |
|---|---|---|---|
| Taurine | 2.4752 | 1.3075 | 0.0110 |
| Isomer 2 of 2,7-Dihydroxycadalene | 2.2466 | 1.1678 | 0.0110 |
| Isoferulic acid | 1.7146 | 0.77787 | 0.0134 |
| Pyrocatechol | 1.6607 | 0.73178 | 0.0151 |
| Guaiacol | 1.6553 | 0.72708 | 0.0151 |
| Isomer 1 of 2,4-Diamino-6-hydroxypyrimidine | 1.6304 | 0.70524 | 0.0151 |
| 1H-Indole-3-methanamine | 1.5707 | 0.65137 | 0.0151 |
| Seryl-Tyrosine | 1.5119 | 0.59634 | 0.0163 |
| 3′,5′-Cyclic GMP | 1.5049 | 0.58971 | 0.0163 |
| 2,8-Dihydroxyadenine | 0.65687 | −0.60632 | 0.0151 |
| Piceatannol | 0.65434 | −0.61189 | 0.0151 |
| 2-hydroxymexiletine | 0.65078 | −0.61976 | 0.0163 |
| Isomer 2 of Homoeriodictyol chalcone | 0.64859 | −0.62462 | 0.0163 |
| 4-Aminobutanoic acid | 0.63386 | −0.65777 | 0.0165 |
| Litcubinine | 0.62962 | −0.66744 | 0.0168 |
| Citrulline | 0.61313 | −0.70573 | 0.0172 |
| Desoxyhemigossypol-6-Methyl Ether | 0.60158 | −0.73317 | 0.0183 |
| Isomer 1 of Homoeriodictyol chalcone | 0.56125 | −0.83328 | 0.0185 |
| Calystegin A3 | 0.53417 | −0.90462 | 0.0188 |
| N,N-Didesmethyltramadol | 0.51099 | −0.96864 | 0.0191 |
| 8-Aminooctanoic acid | 0.50719 | −0.9794 | 0.0193 |
| 5-Aminolevulinic acid | 0.49522 | −1.0139 | 0.0213 |
| Dopamine 4-O-glucuronide | 0.44884 | −1.1557 | 0.0213 |
| Phenylpropanolamine | 0.36712 | −1.4457 | 0.0240 |
| Isomer 1 of 2,6-Dimethyl-1,4-benzenediol | 0.36263 | −1.4634 | 0.0498 |
| Metabolites | Fold Change | Log2(FC) | FDR |
|---|---|---|---|
| 4-Hydroxy-5-(dihydroxyphenyl)-valeric acid-O-sulphate | 2.5864 | 1.371 | 0.0003 |
| 3′,5′-Cyclic GMP | 2.3826 | 1.2525 | 0.0005 |
| Guaiacol | 2.1393 | 1.0972 | 0.0015 |
| 5-Methoxytryptophan | 1.9607 | 0.97139 | 0.0066 |
| Isomer 1 of 2-Hydroxyhepta-2,4-dienedioic acid | 1.9465 | 0.96086 | 0.0079 |
| Creatine | 1.9034 | 0.92855 | 0.0096 |
| 2,3-Diaminoproprionic acid | 1.8573 | 0.89323 | 0.0020 |
| Isomer 1 of 2-Aminovalienone | 1.85 | 0.88752 | 0.0023 |
| Aspartic acid | 1.8313 | 0.87285 | 0.0027 |
| Proline | 1.8013 | 0.84902 | 0.0035 |
| Isomer 1 of Morpholine | 1.7777 | 0.82999 | 0.0042 |
| Norfuraneol | 1.7633 | 0.81827 | 0.0046 |
| 2-Aminoethylphosphonate | 1.7554 | 0.81178 | 0.0047 |
| Chlorohydroquinone | 1.7399 | 0.79898 | 0.0049 |
| Rubroskyrin | 1.7387 | 0.79799 | 0.0049 |
| 2,3-Diaminosalicylic acid | 1.7156 | 0.77871 | 0.0053 |
| 4-Guanidinobutanal | 1.7100 | 0.77401 | 0.0055 |
| Isomer 1 of 3-Cyano-l-alanine | 1.6767 | 0.74563 | 0.0056 |
| (+/−)-N-Methylsalsolinol | 1.6728 | 0.74225 | 0.0056 |
| N2-Acetyl-5-Phosphooxy-l-lysine | 1.6661 | 0.73647 | 0.0057 |
| 4-Fluorocatechol | 1.6518 | 0.72408 | 0.0057 |
| 2-Hydroxy-6-oxonona-2,4-diene-1,9-dioic acid | 1.6486 | 0.72127 | 0.0061 |
| 1H-Indole-3-methanamine | 1.6378 | 0.71177 | 0.0064 |
| Isomer 2 of Seryl-Valine | 1.6353 | 0.70955 | 0.0067 |
| 2-Methylbutylamine | 1.6281 | 0.70317 | 0.0068 |
| 1-Methylguanosine | 1.6266 | 0.7019 | 0.0078 |
| Isomer 1 of Hulupinic acid | 1.6238 | 0.69941 | 0.0101 |
| Isomer 1 of 3-Hydroxyaminophenol | 1.6237 | 0.69928 | 0.0127 |
| N2-Acetyl-N6-(l-1,3-Dicarboxypropyl)-l-lysine | 1.6086 | 0.68583 | 0.0139 |
| Glycine | 1.5659 | 0.64698 | 0.0139 |
| l-trans-4-Methyl-2-pyrrolidinecarboxylic acid | 1.5658 | 0.64693 | 0.0161 |
| N6-beta-Aspartyl-Lysine | 1.5638 | 0.64509 | 0.0165 |
| 4-Formylsalicylic acid | 1.5592 | 0.64081 | 0.0188 |
| (3S,5S)-3,5-Diaminohexanoic acid | 1.5559 | 0.63774 | 0.0200 |
| Taurine | 1.5505 | 0.63276 | 0.0202 |
| Ethanolamine | 1.5503 | 0.6325 | 0.0213 |
| Lysyl-Tyrosine | 1.5474 | 0.62989 | 0.0215 |
| Isomer 1 of 5-Aminopentanoic acid | 1.5316 | 0.61501 | 0.0218 |
| 4-Aminobutyraldehyde | 1.5307 | 0.6142 | 0.0221 |
| 4-Hydroxypiperidine | 1.5275 | 0.6112 | 0.0243 |
| Isomer 2 of 2,7-Dihydroxycadalene | 1.5207 | 0.60475 | 0.0334 |
| Sarcosine | 1.5200 | 0.60405 | 0.0334 |
| Seryl-Tyrosine | 1.5196 | 0.60373 | 0.0351 |
| Ascorbic acid | 1.5195 | 0.60358 | 0.0368 |
| N(6)-Methyllysine | 1.5125 | 0.59693 | 0.0373 |
| Aspartyl-Proline | 1.5101 | 0.59462 | 0.0401 |
| Isomer 1 of Lysyl-Glutamate | 1.5082 | 0.59281 | 0.0432 |
| Lysyl-Valine | 0.66326 | −0.59235 | 0.0002 |
| dl-2-Aminooctanoic acid | 0.65867 | −0.60238 | 0.0003 |
| 2,4-Diamino-6-hydroxypyrimidine | 0.65579 | −0.6087 | 0.0004 |
| 2-Aminomuconic acid | 0.64948 | −0.62265 | 0.0004 |
| 1,4-diaminobutane | 0.64692 | −0.62833 | 0.0005 |
| Alanyl-Proline | 0.63768 | −0.6491 | 0.0006 |
| Kuwanol D | 0.63195 | −0.66212 | 0.0006 |
| Alanyl-Lysine | 0.62479 | −0.67856 | 0.0007 |
| Isomer 1 of Homoeriodictyol chalcone | 0.62326 | −0.6821 | 0.0007 |
| 3-Hydroxyaminophenol | 0.62162 | −0.6859 | 0.0008 |
| 6-Hydroxykynurenic acid | 0.62068 | −0.68809 | 0.0010 |
| Valyl-Valine | 0.61277 | −0.70659 | 0.0013 |
| Citrulline | 0.61247 | −0.7073 | 0.0015 |
| Isomer 1 of Monodehydroascorbic acid | 0.61032 | −0.71236 | 0.0015 |
| Glutamyl-Leucine | 0.61024 | −0.71256 | 0.0027 |
| 3-Formyl-6-hydroxyindole | 0.60358 | −0.72837 | 0.0057 |
| Leucyl-Valine | 0.59776 | −0.74237 | 0.0057 |
| Litcubinine | 0.59736 | −0.74333 | 0.0078 |
| Isoleucyl-Lysine | 0.59598 | −0.74667 | 0.0101 |
| Threoninyl-Valine | 0.59453 | −0.75019 | 0.0127 |
| 8-Aminooctanoic acid | 0.56984 | −0.81136 | 0.0139 |
| Calystegin A3 | 0.56768 | −0.81685 | 0.0139 |
| Seryl-Valine | 0.56261 | −0.8298 | 0.0161 |
| Adenine | 0.56078 | −0.83449 | 0.0165 |
| 2,6-Dimethoxy-4-methylphenol | 0.55841 | −0.84062 | 0.0188 |
| Isomer 1 of Rhapontigenin | 0.53274 | −0.90849 | 0.0200 |
| Leucyl-Leucine | 0.48451 | −1.0454 | 0.0221 |
| Phenylpropanolamine | 0.47098 | −1.0863 | 0.0243 |
| 4-Aminobutanoic acid | 0.45787 | −1.127 | 0.0334 |
| 5-Aminolevulinic acid | 0.45439 | −1.138 | 0.0334 |
| Dopamine 4-O-glucuronide | 0.41192 | −1.2796 | 0.0351 |
| Isomer 1 of 2,6-Dimethyl-1,4-benzenediol | 0.39663 | −1.3341 | 0.0373 |
| N,N-Didesmethyltramadol | 0.21681 | −2.2055 | 0.0401 |
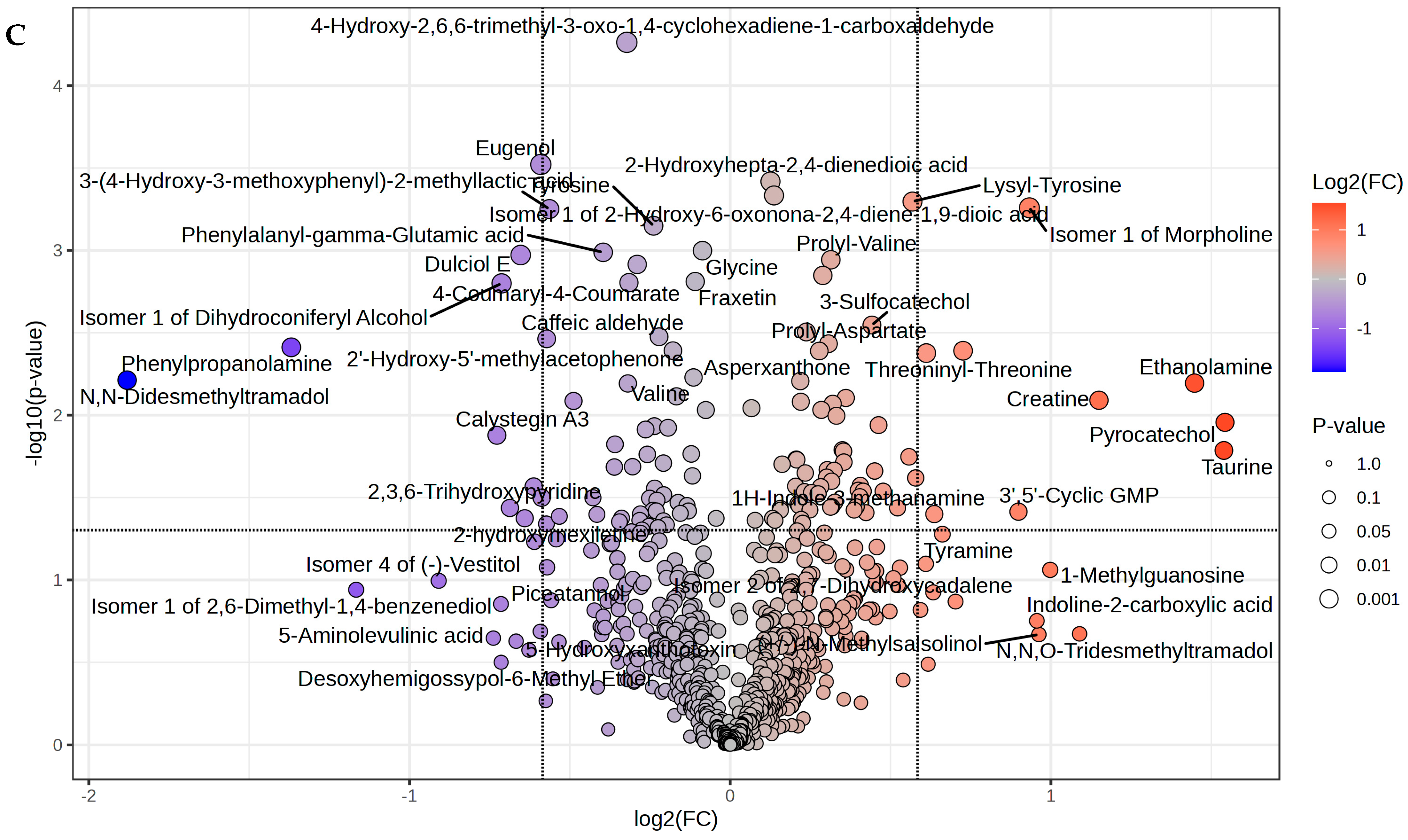
| Metabolites | Fold Change | Log2(FC) | FDR |
|---|---|---|---|
| Pyrocatechol | 2.9137 | 1.5428 | 0.0020 |
| Taurine | 2.9068 | 1.5394 | 0.0023 |
| Ethanolamine | 2.7283 | 1.448 | 0.0023 |
| Creatine | 2.2189 | 1.1498 | 0.0027 |
| N,N,O-Tridesmethyltramadol | 2.1277 | 1.0893 | 0.0035 |
| 1-Methylguanosine | 1.997 | 0.99785 | 0.0056 |
| (+/−)-N-Methylsalsolinol | 1.9487 | 0.96248 | 0.0056 |
| Indoline-2-carboxylic acid | 1.9400 | 0.95608 | 0.0083 |
| Isomer 1 of Morpholine | 1.9090 | 0.93285 | 0.0096 |
| 3′,5′-Cyclic GMP | 1.8645 | 0.89878 | 0.0101 |
| Isomer 1 of (2R,3R,4R)-2-Amino-4-hydroxy-3-methylpentanoic acid | 1.6542 | 0.72614 | 0.0127 |
| Isomer 2 of 2,7-Dihydroxycadalene | 1.6274 | 0.70261 | 0.0150 |
| Tyramine | 1.5818 | 0.66161 | 0.0178 |
| 1H-Indole-3-methanamine | 1.5545 | 0.63645 | 0.0191 |
| 2-Isopropyl-1,4-benzenediol | 1.5507 | 0.63296 | 0.0223 |
| 2-Phenylethylamine | 1.5339 | 0.6172 | 0.0223 |
| Threoninyl-Threonine | 1.5278 | 0.61145 | 0.0256 |
| Methylguanidine | 1.5261 | 0.60987 | 0.0278 |
| 2,6-Dimethyl-1,4-benzenediol | 1.5086 | 0.59324 | 0.0284 |
| Isomer 2 of Homoeriodictyol chalcone | 0.6653 | −0.58792 | 0.0064 |
| Eugenol | 0.6641 | −0.59053 | 0.0083 |
| Litcubinine | 0.66344 | −0.59196 | 0.0096 |
| Isomer 1 of Pyridoxamine | 0.65486 | −0.61074 | 0.0101 |
| Sinapyl alcohol | 0.65398 | −0.61267 | 0.0127 |
| 2-Aminomuconic acid | 0.64724 | −0.62762 | 0.0150 |
| 2-hydroxymexiletine | 0.64141 | −0.64068 | 0.0178 |
| Dulciol E | 0.6358 | −0.65335 | 0.0191 |
| 5-Hydroxyxanthotoxin | 0.62955 | −0.6676 | 0.0223 |
| 2,3,6-Trihydroxypyridine | 0.62115 | −0.68699 | 0.0223 |
| Isomer 1 of Dihydroconiferyl Alcohol | 0.61012 | −0.71283 | 0.0256 |
| Desoxyhemigossypol-6-Methyl Ether | 0.60952 | −0.71426 | 0.0278 |
| Piceatannol | 0.60926 | −0.71488 | 0.0284 |
| Calystegin A3 | 0.60395 | −0.72749 | 0.0313 |
| 5-Aminolevulinic acid | 0.59936 | −0.73849 | 0.0320 |
| Isomer 4 of (−)-Vestitol | 0.53257 | −0.90896 | 0.0337 |
| Isomer 1 of 2,6-Dimethyl-1,4-benzenediol | 0.44556 | −1.1663 | 0.0361 |
| Phenylpropanolamine | 0.38726 | −1.3686 | 0.0365 |
| N,N-Didesmethyltramadol | 0.2716 | −1.8805 | 0.0430 |
| Treatments * | Measure | 1 Comp | 2 Comps | 3 Comps | 4 Comps |
|---|---|---|---|---|---|
| EFA1 | Accuracy | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| R2 | 0.99878 | 0.99882 | 0.99886 | 0.99981 | |
| Q2 | 0.56579 | 0.8953 | 0.99254 | 0.9638 | |
| EFA2 | Accuracy | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| R2 | 0.96769 | 0.99887 | 0.9989 | 0.9998 | |
| Q2 | 0.71056 | 0.91829 | 0.99026 | 0.9720 | |
| EFA3 | Accuracy | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| R2 | 0.99447 | 0.99798 | 0.99815 | 0.99898 | |
| Q2 | 0.59845 | 0.89773 | 0.98844 | 0.9387 |
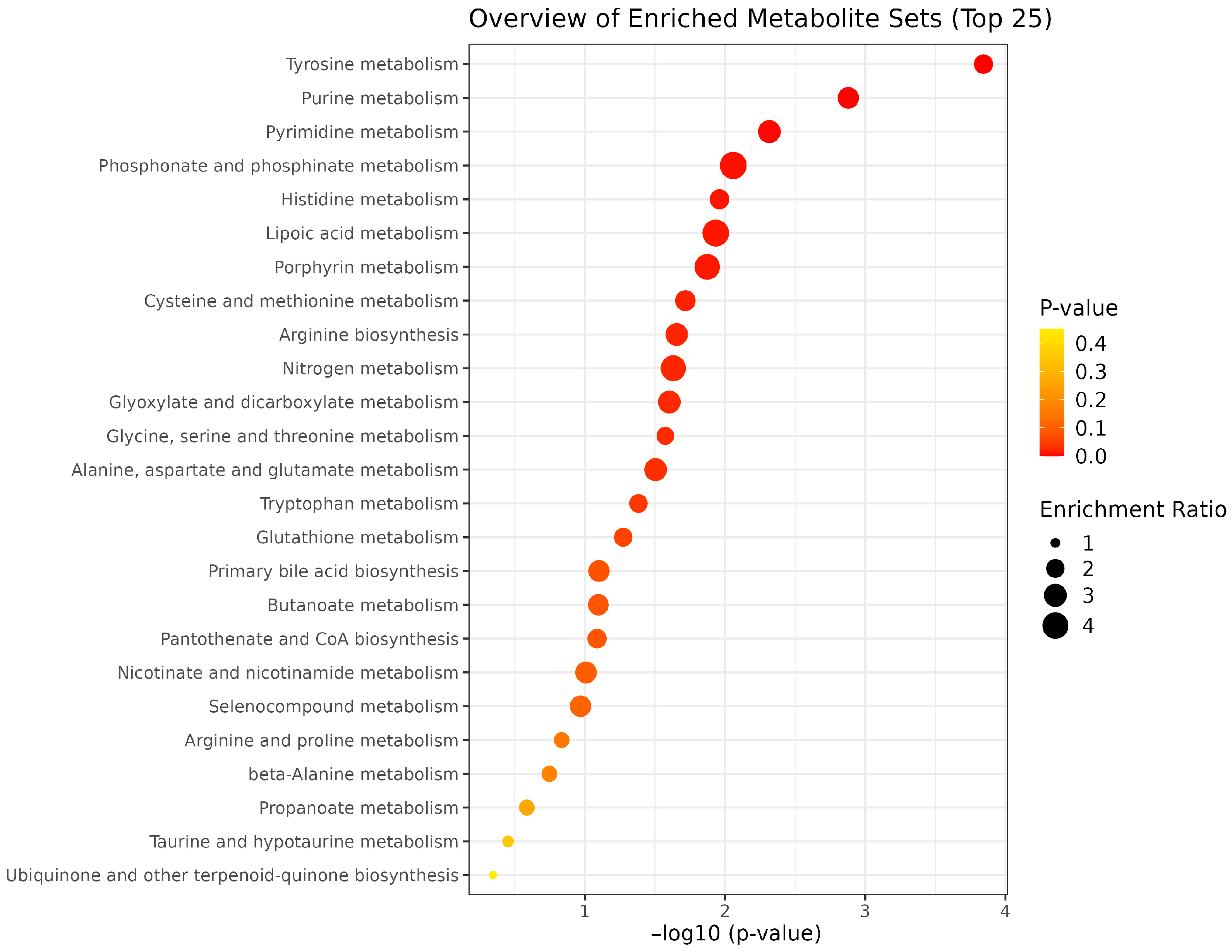
| Pathway Name | Total Metabolites | Hits | Impact Value | p-Value | FDR |
|---|---|---|---|---|---|
| Tyrosine metabolism | 42 | 13 | 0.310 | 0.0001 | 5.74 × 10−6 |
| Purine metabolism | 70 | 11 | 0.157 | 0.0013 | 1.06 × 10−4 |
| Pyrimidine metabolism | 39 | 6 | 0.154 | 0.0048 | 5.79 × 10−4 |
| Phosphonate and phosphinate metabolism | 6 | 1 | 0.167 | 0.0087 | 0.0014 |
| Histidine metabolism | 16 | 6 | 0.375 | 0.0110 | 0.0022 |
| Lipoic acid metabolism | 28 | 1 | 0.036 | 0.0117 | 0.0028 |
| Porphyrin metabolism | 31 | 3 | 0.097 | 0.0134 | 0.0037 |
| Cysteine and methionine metabolism | 33 | 5 | 0.152 | 0.0192 | 0.0061 |
| Arginine biosynthesis | 14 | 6 | 0.429 | 0.0221 | 0.0080 |
| Nitrogen metabolism | 6 | 2 | 0.333 | 0.0234 | 0.0094 |
| Glyoxylate and dicarboxylate metabolism | 31 | 4 | 0.129 | 0.0250 | 0.0110 |
| Glycine, serine and threonine metabolism | 33 | 9 | 0.273 | 0.0267 | 0.0128 |
| Alanine, aspartate and glutamate metabolism | 28 | 6 | 0.214 | 0.0313 | 0.0163 |
| Tryptophan metabolism | 41 | 6 | 0.146 | 0.0414 | 0.0232 |
| Glutathione metabolism | 28 | 6 | 0.214 | 0.0415 | 0.0249 |
| Primary bile acid biosynthesis | 46 | 2 | 0.043 | 0.0417 | 0.0267 |
| Butanoate metabolism | 15 | 2 | 0.133 | 0.0418 | 0.0284 |
| Pantothenate and CoA biosynthesis | 20 | 6 | 0.300 | 0.0419 | 0.0302 |
| Nicotinate and nicotinamide metabolism | 15 | 1 | 0.067 | 0.0421 | 0.0320 |
| Selenocompound metabolism | 20 | 1 | 0.050 | 0.0422 | 0.0338 |
| Arginine and proline metabolism | 36 | 15 | 0.417 | 0.0423 | 0.0356 |
| beta-Alanine metabolism | 21 | 7 | 0.333 | 0.0532 | 0.0468 |
| Propanoate metabolism | 21 | 1 | 0.048 | 0.0792 | 0.0697 |
| Taurine and hypotaurine metabolism | 8 | 1 | 0.125 | 0.0800 | 0.0736 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 18 | 3 | 0.167 | 0.0817 | 0.0785 |
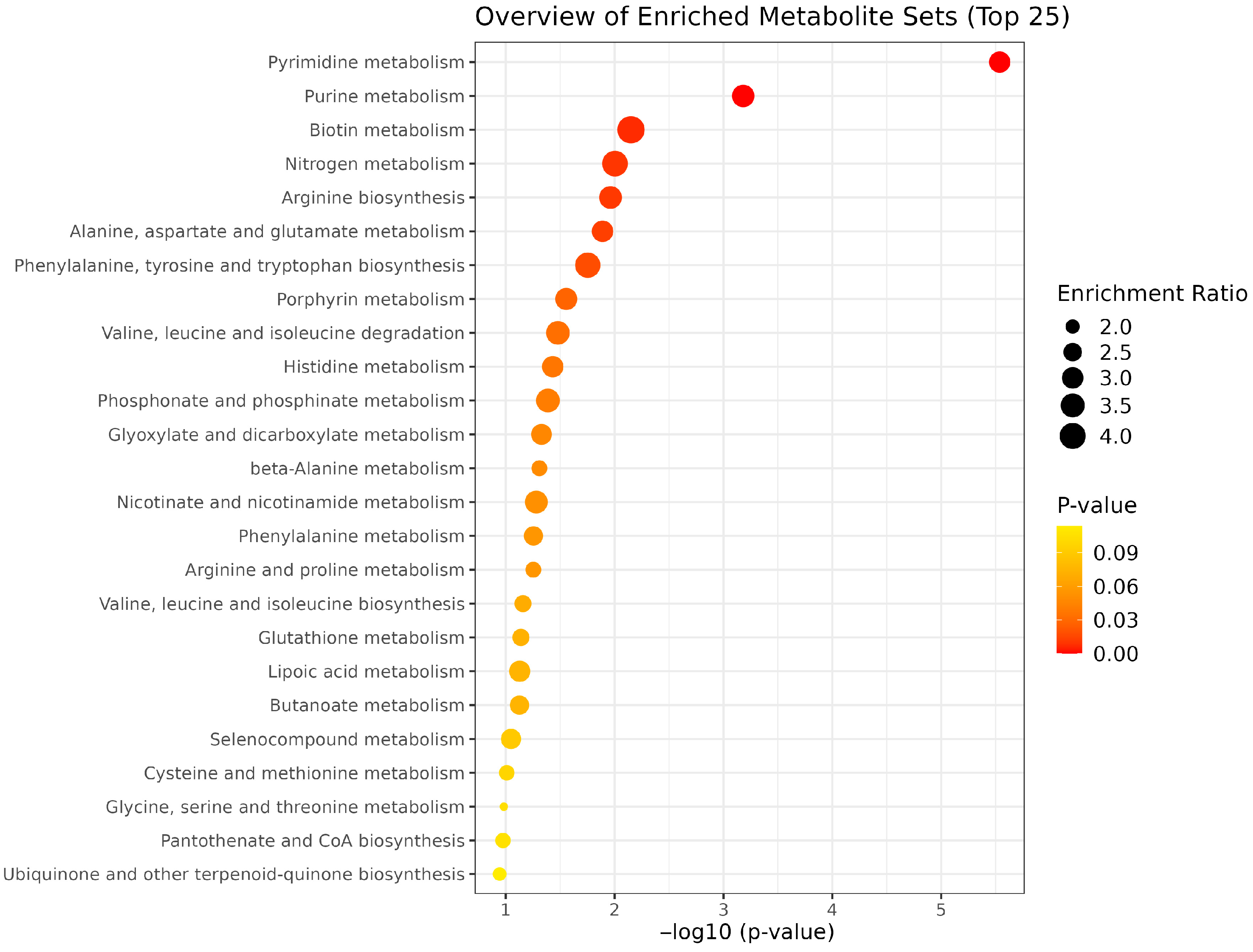
| Pathway Name | Total Metabolites | Hits | Impact Value | p-Value | FDR |
|---|---|---|---|---|---|
| Pyrimidine metabolism | 39 | 6 | 0.154 | 2.90 × 10−5 | 1.94 × 10−7 |
| Purine metabolism | 70 | 11 | 0.157 | 6.59 × 10−4 | 8.79 × 10−5 |
| Biotin metabolism | 10 | 1 | 0.100 | 0.00708 | 0.00142 |
| Nitrogen metabolism | 6 | 2 | 0.333 | 0.00993 | 0.00265 |
| Arginine biosynthesis | 14 | 6 | 0.429 | 0.01091 | 0.00364 |
| Alanine, aspartate and glutamate metabolism | 28 | 6 | 0.214 | 0.01291 | 0.00516 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 4 | 2 | 0.500 | 0.01766 | 0.00824 |
| Porphyrin metabolism | 31 | 3 | 0.097 | 0.02781 | 0.01483 |
| Valine, leucine and isoleucine degradation | 39 | 2 | 0.051 | 0.03321 | 0.01993 |
| Histidine metabolism | 16 | 6 | 0.375 | 0.03701 | 0.02467 |
| Phosphonate and phosphinate metabolism | 6 | 1 | 0.167 | 0.04099 | 0.03006 |
| Glyoxylate and dicarboxylate metabolism | 31 | 4 | 0.129 | 0.04695 | 0.03756 |
| β-Alanine metabolism | 21 | 7 | 0.333 | 0.04912 | 0.04257 |
| Nicotinate and nicotinamide metabolism | 15 | 1 | 0.067 | 0.05245 | 0.04895 |
| Phenylalanine metabolism | 8 | 3 | 0.375 | 0.05563 | 0.05563 |
| Arginine and proline metabolism | 36 | 15 | 0.417 | 0.05585 | 0.05957 |
| Valine, leucine and isoleucine biosynthesis | 8 | 3 | 0.375 | 0.06948 | 0.07874 |
| Glutathione metabolism | 28 | 6 | 0.214 | 0.07261 | 0.08713 |
| Lipoic acid metabolism | 28 | 1 | 0.036 | 0.07442 | 0.09427 |
| Butanoate metabolism | 15 | 2 | 0.133 | 0.07483 | 0.09977 |
| Selenocompound metabolism | 20 | 1 | 0.050 | 0.08941 | 0.12517 |
| Cysteine and methionine metabolism | 33 | 5 | 0.152 | 0.09789 | 0.14357 |
| Glycine, serine and threonine metabolism | 33 | 9 | 0.273 | 0.10412 | 0.15965 |
| Pantothenate and CoA biosynthesis | 20 | 6 | 0.300 | 0.10601 | 0.16962 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 18 | 3 | 0.167 | 0.11376 | 0.18960 |
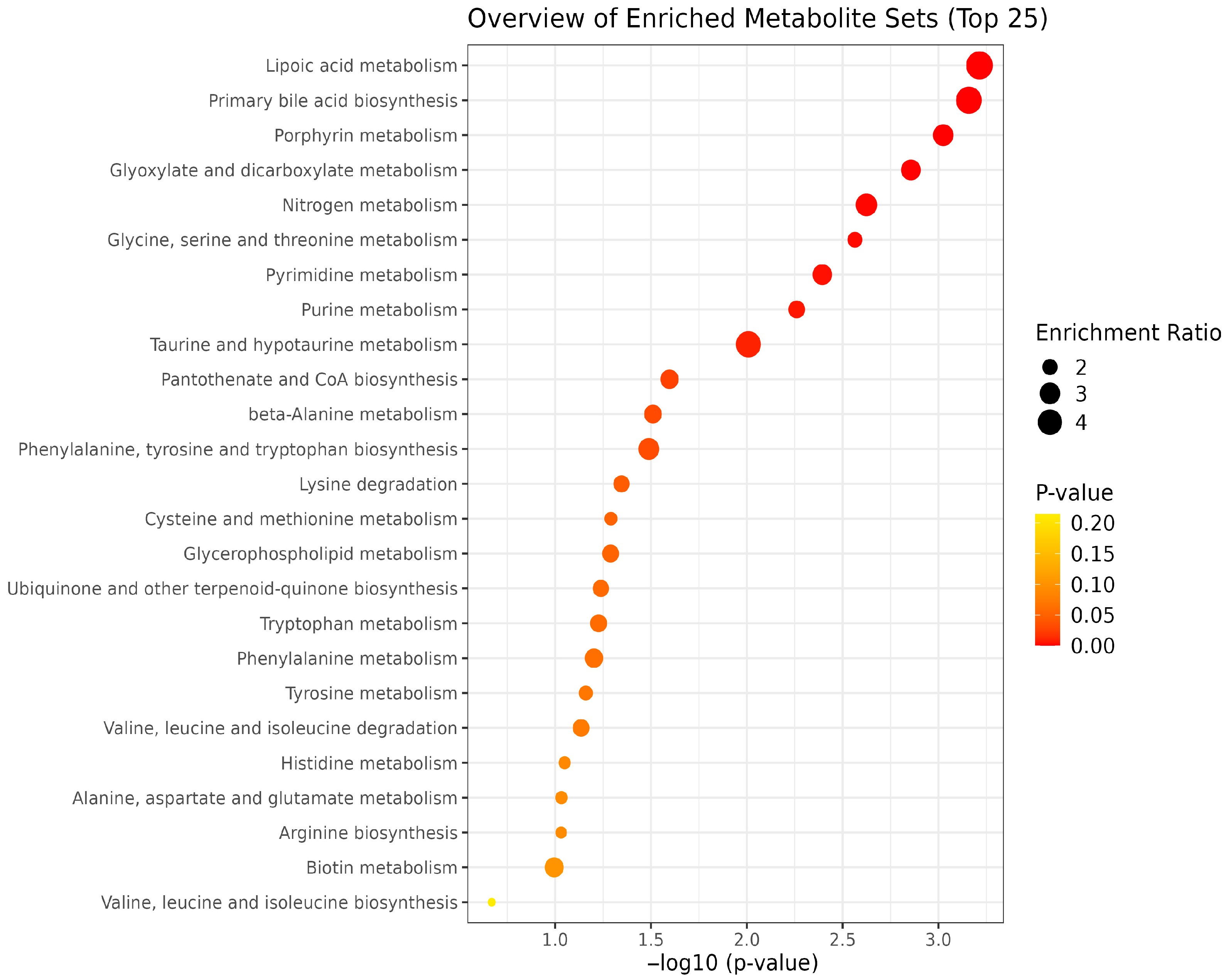
| Pathway Name | Total Metabolites | Hits | Impact Value | p-Value | FDR |
|---|---|---|---|---|---|
| Lipoic acid metabolism | 28 | 1 | 0.036 | 6.11 × 10−4 | 2.54 × 10−5 |
| Primary bile acid biosynthesis | 46 | 2 | 0.043 | 6.94 × 10−4 | 5.78 × 10−5 |
| Porphyrin metabolism | 31 | 3 | 0.097 | 3.76 × 10−4 | 4.70 × 10−5 |
| Glyoxylate and dicarboxylate metabolism | 31 | 4 | 0.129 | 7.28 × 10−4 | 1.21 × 10−4 |
| Nitrogen metabolism | 6 | 2 | 0.333 | 9.44 × 10−4 | 1.97 × 10−4 |
| Glycine, serine and threonine metabolism | 33 | 9 | 0.273 | 0.0014 | 3.48 × 10−4 |
| Pyrimidine metabolism | 39 | 6 | 0.154 | 0.0020 | 5.94 × 10−4 |
| Purine metabolism | 70 | 11 | 0.157 | 0.0035 | 0.0012 |
| Taurine and hypotaurine metabolism | 8 | 1 | 0.125 | 0.0078 | 0.0029 |
| Pantothenate and CoA biosynthesis | 20 | 6 | 0.300 | 0.0233 | 0.0097 |
| beta-Alanine metabolism | 21 | 7 | 0.333 | 0.0288 | 0.0132 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 4 | 2 | 0.500 | 0.0304 | 0.0152 |
| Lysine degradation | 30 | 4 | 0.133 | 0.0430 | 0.0233 |
| Cysteine and methionine metabolism | 33 | 5 | 0.152 | 0.0482 | 0.0281 |
| Glycerophospholipid metabolism | 36 | 2 | 0.056 | 0.0484 | 0.0302 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 18 | 3 | 0.167 | 0.0547 | 0.0365 |
| Tryptophan metabolism | 41 | 6 | 0.146 | 0.0563 | 0.0399 |
| Phenylalanine metabolism | 8 | 3 | 0.375 | 0.0597 | 0.0448 |
| Tyrosine metabolism | 42 | 13 | 0.310 | 0.0662 | 0.0524 |
| Valine, leucine and isoleucine degradation | 39 | 2 | 0.051 | 0.0702 | 0.0585 |
| Histidine metabolism | 16 | 6 | 0.375 | 0.0862 | 0.0755 |
| Alanine, aspartate and glutamate metabolism | 28 | 6 | 0.214 | 0.0897 | 0.0822 |
| Arginine biosynthesis | 14 | 6 | 0.429 | 0.0900 | 0.0863 |
| Biotin metabolism | 10 | 1 | 0.100 | 0.0981 | 0.0981 |
| Valine, leucine and isoleucine biosynthesis | 8 | 3 | 0.375 | 0.2143 | 0.2232 |
| Selenocompound metabolism | 20 | 1 | 0.050 | 0.2747 | 0.2976 |
| Butanoate metabolism | 15 | 2 | 0.133 | 0.2784 | 0.3131 |
| Arginine and proline metabolism | 36 | 15 | 0.417 | 0.2800 | 0.3266 |
| Nicotinate and nicotinamide metabolism | 15 | 1 | 0.067 | 0.2836 | 0.3426 |
| Glutathione metabolism | 28 | 6 | 0.214 | 0.3063 | 0.3829 |
| Propanoate metabolism | 21 | 1 | 0.048 | 0.4600 | 0.5942 |
| Vitamin B6 metabolism | 9 | 3 | 0.333 | 0.4922 | 0.6563 |
| Phosphonate and phosphinate metabolism | 6 | 1 | 0.167 | 0.8493 | 1.1678 |
| D-Amino acid metabolism | 15 | 1 | 0.067 | 0.8959 | 1.2692 |
| Sphingolipid metabolism | 32 | 1 | 0.031 | 0.8959 | 1.3066 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alabi, J.O.; Okedoyin, D.O.; Wuaku, M.; Anotaenwere, C.C.; Adelusi, O.O.; Ike, K.A.; Gray, D.; Oderinwale, O.A.; Enikuomehin, J.M.; Ekwemalor, K.A.; et al. Untargeted Metabolomics of Dairy Cows as Influenced by the Combinations of Essential Oil Blends and Fumaric Acid as Natural Feed Additives Using RUSITEC. Metabolites 2025, 15, 681. https://doi.org/10.3390/metabo15100681
Alabi JO, Okedoyin DO, Wuaku M, Anotaenwere CC, Adelusi OO, Ike KA, Gray D, Oderinwale OA, Enikuomehin JM, Ekwemalor KA, et al. Untargeted Metabolomics of Dairy Cows as Influenced by the Combinations of Essential Oil Blends and Fumaric Acid as Natural Feed Additives Using RUSITEC. Metabolites. 2025; 15(10):681. https://doi.org/10.3390/metabo15100681
Chicago/Turabian StyleAlabi, Joel O., Deborah O. Okedoyin, Michael Wuaku, Chika C. Anotaenwere, Oludotun O. Adelusi, Kelechi A. Ike, DeAndrea Gray, Olatunde A. Oderinwale, James M. Enikuomehin, Kingsley A. Ekwemalor, and et al. 2025. "Untargeted Metabolomics of Dairy Cows as Influenced by the Combinations of Essential Oil Blends and Fumaric Acid as Natural Feed Additives Using RUSITEC" Metabolites 15, no. 10: 681. https://doi.org/10.3390/metabo15100681
APA StyleAlabi, J. O., Okedoyin, D. O., Wuaku, M., Anotaenwere, C. C., Adelusi, O. O., Ike, K. A., Gray, D., Oderinwale, O. A., Enikuomehin, J. M., Ekwemalor, K. A., Fasina, Y. O., Ismail, H. D., Kholif, A. E., & Anele, U. Y. (2025). Untargeted Metabolomics of Dairy Cows as Influenced by the Combinations of Essential Oil Blends and Fumaric Acid as Natural Feed Additives Using RUSITEC. Metabolites, 15(10), 681. https://doi.org/10.3390/metabo15100681









