A Deep Mining Strategy for Peptide Rapid Identification in Lactobacillus reuteri Based on LC–MS/MS Integrated with FBMN and De Novo Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. General Experimental Procedures
2.3. Bacterial Strain and Growth Conditions
2.4. Extraction and Isolation
2.5. Sample Preparation
2.6. Mass Spectral Data Acquisition
2.7. MZmine 2.53 Data-Preprocessing Parameters
2.8. Feature-Based Molecular Networking
2.9. Identification of Peptides Using PEAKS Studio
2.10. In Silico Analysis
2.11. Molecular Docking
3. Results and Discussion
3.1. Fast Determination of Small Peptides Based on LC–MS MS/MS with FBMN and PEAKS Studio
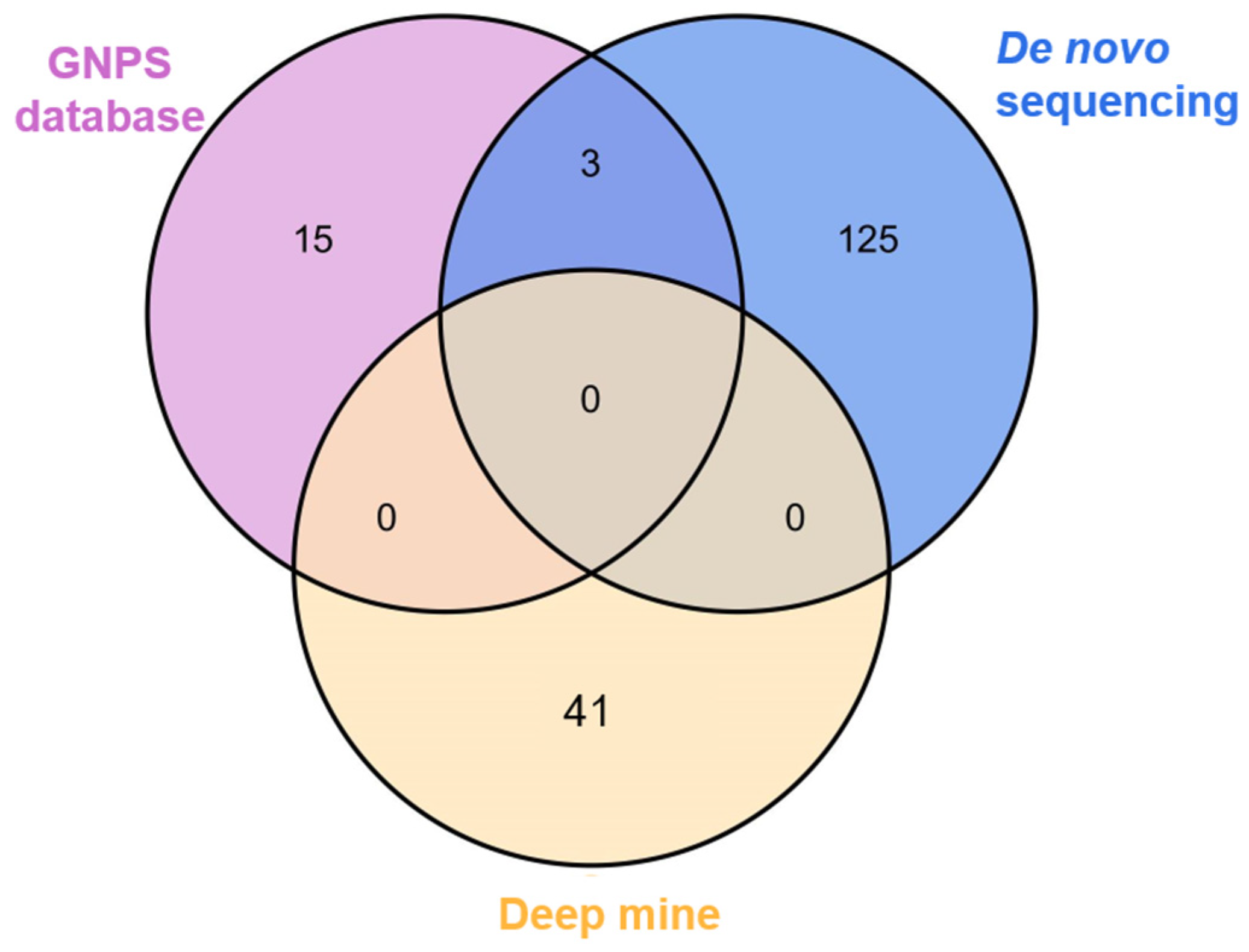
3.2. Thorough Profile of Peptides Based on FBMN
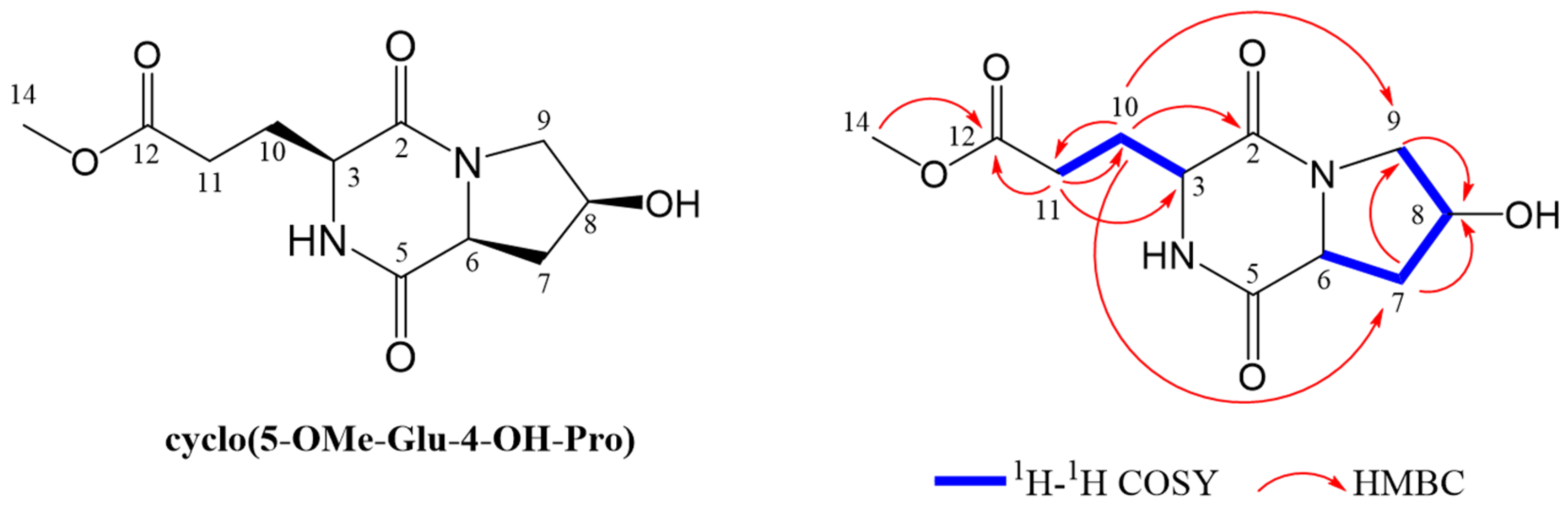
3.3. Isolation and Identification of Novel Peptide
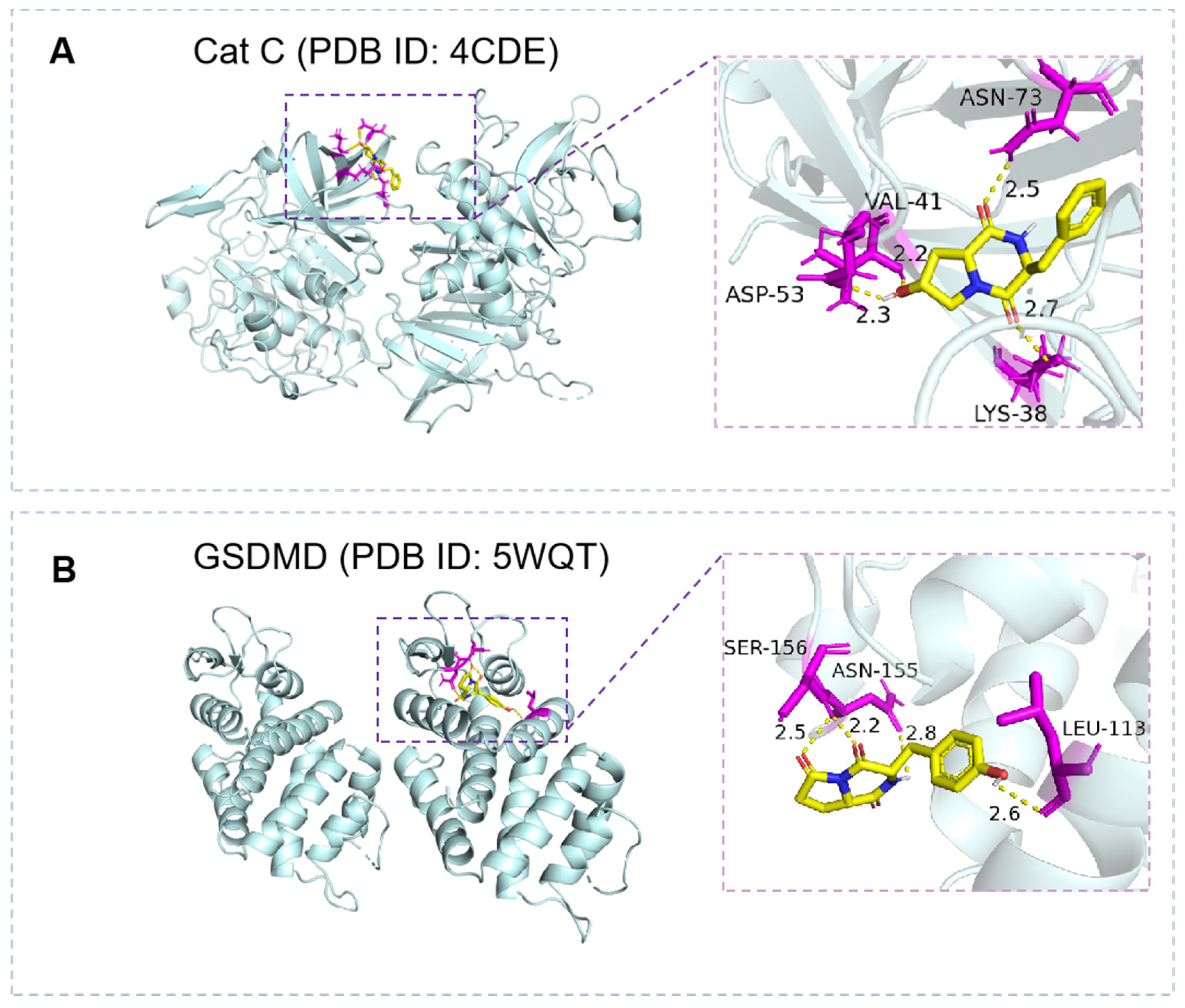
3.4. Prediction of the Potential Anti-Inflammatory Activity of Peptides by Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Luo, Z.; Chen, A.; Xie, A.; Liu, X.; Jiang, S.; Yu, R. Limosilactobacillus reuteri in immunomodulation: Molecular mechanisms and potential applications. Front. Immunol. 2023, 14, 1228754. [Google Scholar] [CrossRef]
- Peng, Y.; Ma, Y.; Luo, Z.; Jiang, Y.; Xu, Z.; Yu, R. Lactobacillus reuteri in digestive system diseases: Focus on clinical trials and mechanisms. Front. Cell. Infect. Microbiol. 2023, 13, 1254198. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Peters, C.; Venkatanarayanan, N.; Goh, Y.Y.; Ho, C.Y.X.; Yeo, W.S. Use of Lactobacillus spp. to prevent recurrent urinary tract infections in females. Med. Hypotheses 2018, 114, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Bell, H.N.; Rebernick, R.J.; Goyert, J.; Singhal, R.; Kuljanin, M.; Kerk, S.A.; Huang, W.; Das, N.K.; Andren, A.; Solanki, S.; et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 2022, 40, 185–200.e186. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8alphaalpha+ T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D‘Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Boahen, A.; Chew, S.Y.; Neela, V.K.; Than, L.T.L. Limosilactobacillus reuteri 29A Cell-Free Supernatant Antibiofilm and Antagonistic Effects in Murine Model of Vulvovaginal Candidiasis. Probiotics Antimicrob. Proteins 2023, 15, 1681–1699. [Google Scholar] [CrossRef] [PubMed]
- Ozcam, M.; Tocmo, R.; Oh, J.H.; Afrazi, A.; Mezrich, J.D.; Roos, S.; Claesen, J.; van Pijkeren, J.P. Gut Symbionts Lactobacillus reuteri R2lc and 2010 Encode a Polyketide Synthase Cluster That Activates the Mammalian Aryl Hydrocarbon Receptor. Appl. Environ. Microbiol. 2019, 85, e01661-18. [Google Scholar] [CrossRef]
- Hernandez-Granados, M.J.; Franco-Robles, E. Postbiotics in human health: Possible new functional ingredients? Food Res. Int. 2020, 137, 109660. [Google Scholar] [CrossRef]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Extracellular Proteolytic Activity and Amino Acid Production by Lactic Acid Bacteria Isolated from Malaysian Foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef]
- Miao, H.; Wang, Y.N.; Yu, X.Y.; Zou, L.; Guo, Y.; Su, W.; Liu, F.; Cao, G.; Zhao, Y.Y. Lactobacillus species ameliorate membranous nephropathy through inhibiting the aryl hydrocarbon receptor pathway via tryptophan-produced indole metabolites. Br. J. Pharmacol. 2024, 181, 162–179. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, X.; Liu, X.; Wang, X.; Gao, X. Therapeutic and Improving Function of Lactobacilli in the Prevention and Treatment of Cardiovascular-Related Diseases: A Novel Perspective From Gut Microbiota. Front. Nutr. 2021, 8, 693412. [Google Scholar] [CrossRef]
- Song, J.; Peng, S.; Yang, J.; Zhou, F.; Suo, H. Isolation and identification of novel antibacterial peptides produced by Lactobacillus fermentum SHY10 in Chinese pickles. Food Chem. 2021, 348, 129097. [Google Scholar] [CrossRef]
- Mousa, W.K.; Ghemrawi, R.; Abu-Izneid, T.; Ramadan, A.; Al-Marzooq, F. Discovery of Lactomodulin, a Unique Microbiome-Derived Peptide That Exhibits Dual Anti-Inflammatory and Antimicrobial Activity against Multidrug-Resistant Pathogens. Int. J. Mol. Sci. 2023, 24, 6901. [Google Scholar] [CrossRef]
- Bohle, L.A.; Brede, D.A.; Diep, D.B.; Holo, H.; Nes, I.F. Specific degradation of the mucus adhesion-promoting protein (MapA) of Lactobacillus reuteri to an antimicrobial peptide. Appl. Environ. Microbiol. 2010, 76, 7306–7309. [Google Scholar] [CrossRef] [PubMed]
- Sobanbua, S.; Dolkittikul, S.; Nakphaichit, M.; Keawsompong, S.; Nitisinprasert, S. Antimicrobial peptide presenting potential strain-specific real time polymerase chain reaction assay for detecting the probiotic Lactobacillus reuteri KUB-AC5 in chicken intestine. Poult. Sci. 2020, 99, 526–535. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365. [Google Scholar] [CrossRef]
- Zhu, F.; Cao, J.; Song, Y.; Yu, P.; Su, E. Plant Protein-Derived Active Peptides: A Comprehensive Review. J. Agric. Food Chem. 2023, 71, 20479–20499. [Google Scholar] [CrossRef]
- Chen, C.J.; Lee, D.Y.; Yu, J.; Lin, Y.N.; Lin, T.M. Recent advances in LC-MS-based metabolomics for clinical biomarker discovery. Mass Spectrom. Rev. 2023, 42, 2349–2378. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Chen, S.; Huang, G.; Liao, W.; Gong, S.; Xiao, J.; Bai, J.; Wendy Hsiao, W.L.; Li, N.; Wu, J.L. Discovery of the bioactive peptides secreted by Bifidobacterium using integrated MCX coupled with LC-MS and feature-based molecular networking. Food Chem. 2021, 347, 129008. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Khatun, M.S.; Hasan, M.M.; Kurata, H. PreAIP: Computational Prediction of Anti-inflammatory Peptides by Integrating Multiple Complementary Features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef]
- Ngoh, Y.Y.; Gan, C.Y. Identification of Pinto bean peptides with inhibitory effects on α-amylase and angiotensin converting enzyme (ACE) activities using an integrated bioinformatics-assisted approach. Food Chem. 2018, 267, 124–131. [Google Scholar] [CrossRef]
- Ilisz, I.; Aranyi, A.; Peter, A. Chiral derivatizations applied for the separation of unusual amino acid enantiomers by liquid chromatography and related techniques. J. Chromatogr. A 2013, 1296, 119–139. [Google Scholar] [CrossRef]
- Funasaki, N.; Hada, S.; Neya, S. Conformational Effects in Reversed-Phase Liquid-Chromatographic Separation of Diastereomers of Cyclic Dipeptides. Anal. Chem. 1993, 65, 1861–1867. [Google Scholar] [CrossRef]
- Liu, K.X.; Kato, Y.; Kaku, T.I.; Santa, T.; Imai, K.; Yagi, A.; Ishizu, T.; Sugiyama, Y. Hydroxyprolylserine derivatives JBP923 and JBP485 exhibit the antihepatitis activities after gastrointestinal absorption in rats. J. Pharmacol. Exp. Ther. 2000, 294, 510–515. [Google Scholar]
- Strom, K.; Sjogren, J.; Broberg, A.; Schnurer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Mizushige, T.; Mori, Y.; Shimmura, Y.; Fukutomi, R.; Kanamoto, R.; Ohinata, K. Antidepressant-like effect of food-derived pyroglutamyl peptides in mice. Neuropeptides 2015, 51, 25–29. [Google Scholar] [CrossRef]
- Trigos, A.; Reyna, S.; Cervantes, L. Three diketopiperazines from the cultivated fungus Fusarium oxysporum. Nat. Prod. Lett. 1995, 6, 241–246. [Google Scholar] [CrossRef]
- Cronan, J.M.; Davidson, T.R.; Singleton, F.L.; Colwell, R.R.; Cardellina, J.H. Plant Growth Promoters Isolated from a Marine Bacterium Associated with Palythoa sp. Nat. Prod. Lett. 1998, 11, 271–278. [Google Scholar] [CrossRef]
- Yu, W.; Zhang, G.; Wang, W.; Jiang, C.; Cao, L. Identification and comparison of proteomic and peptide profiles of mung bean seeds and sprouts. BMC Chem. 2020, 14, 46. [Google Scholar] [CrossRef]
- Fadimu, G.J.; Farahnaky, A.; Gill, H.; Olalere, O.A.; Gan, C.Y.; Truong, T. In-Silico Analysis and Antidiabetic Effect of alpha-Amylase and alpha-Glucosidase Inhibitory Peptides from Lupin Protein Hydrolysate: Enzyme-Peptide Interaction Study Using Molecular Docking Approach. Foods 2022, 11, 3375. [Google Scholar] [CrossRef]
- de Fatima Garcia, B.; de Barros, M.; de Souza Rocha, T. Bioactive peptides from beans with the potential to decrease the risk of developing noncommunicable chronic diseases. Crit. Rev. Food Sci. Nutr. 2021, 61, 2003–2021. [Google Scholar] [CrossRef]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-Based Prediction of Anti-inflammatory Peptides Using Random Forest. Front. Pharmacol. 2018, 9, 276. [Google Scholar] [CrossRef]
- Vismara, I.; Papa, S.; Veneruso, V.; Mauri, E.; Mariani, A.; De Paola, M.; Affatato, R.; Rossetti, A.; Sponchioni, M.; Moscatelli, D.; et al. Selective Modulation of A1 Astrocytes by Drug-Loaded Nano-Structured Gel in Spinal Cord Injury. ACS Nano 2020, 14, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, J.; Chen, X.; Jiang, Y.; Pan, Z.K. Rolipram Protects Mice from Gram-negative Bacterium Escherichia coli-induced Inflammation and Septic Shock. Sci. Rep. 2020, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.E.; Wang, Y.; Bian, S.; Qin, S.; Zhao, H.; Wen, J.; Liu, T.; Ren, L.; Li, Q.; Shi, W.; et al. Helenine blocks NLRP3 activation by disrupting the NEK7-NLRP3 interaction and ameliorates inflammatory diseases. Phytomedicine 2024, 122, 155159. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Caughey, G.H.; Chapple, I.; Gauthier, F.; Hirschfeld, J.; Jenne, D.E.; Kettritz, R.; Lalmanach, G.; Lamort, A.S.; Lauritzen, C.; et al. Therapeutic targeting of cathepsin C: From pathophysiology to treatment. Pharmacol. Ther. 2018, 190, 202–236. [Google Scholar] [CrossRef]
- Li, Y.; Tu, Z.; Chen, F.; Yang, X.; Deng, R.; Su, F.; Cheng, Z.; Li, S.; Ong, S.B.; Wang, D.; et al. Anti-inflammatory effect of Danhong injection through inhibition of GSDMD-mediated pyroptosis. Phytomedicine 2023, 113, 154743. [Google Scholar] [CrossRef]
- Burdette, B.E.; Esparza, A.N.; Zhu, H.; Wang, S. Gasdermin D in pyroptosis. Acta Pharm. Sin. B 2021, 11, 2768–2782. [Google Scholar] [CrossRef]
- Gao, R.C.; Shu, W.H.; Shen, Y.; Sun, Q.C.; Jin, W.G.; Li, D.J.; Li, Y.; Yuan, L. Peptide fraction from sturgeon muscle by pepsin hydrolysis exerts anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages via MAPK and NF-κB pathways. Food Sci. Hum. Wellness 2021, 10, 103–111. [Google Scholar] [CrossRef]
- Yang, T.; Wu, J.; Wang, C.; Liu, Q.; Ma, X.; Peng, J.; Kaku, T.; Liu, K. Protective effect of JBP485 on concanavalin A-induced liver injury in mice. J. Pharm. Pharmacol. 2009, 61, 767–774. [Google Scholar] [CrossRef]
- Nalli, Y.; Gupta, S.; Khajuria, V.; Singh, V.P.; Sajgotra, M.; Ahmed, Z.; Thakur, N.L.; Ali, A. TNF-α and IL-6 inhibitory effects of cyclic dipeptides isolated from marine bacteria Streptomyces sp. Med. Chem. Res. 2017, 26, 93–100. [Google Scholar] [CrossRef]
- Ferro, J.N.; de Aquino, F.L.; de Brito, R.G.; dos Santos, P.L.; Quintans Jde, S.; de Souza, L.C.; de Araujo, A.F.; Diaz, B.L.; Lucca-Junior, W.; Quintans-Junior, L.J.; et al. Cyclo-Gly-Pro, a cyclic dipeptide, attenuates nociceptive behaviour and inflammatory response in mice. Clin. Exp. Pharmacol. Physiol. 2015, 42, 1287–1295. [Google Scholar] [CrossRef]
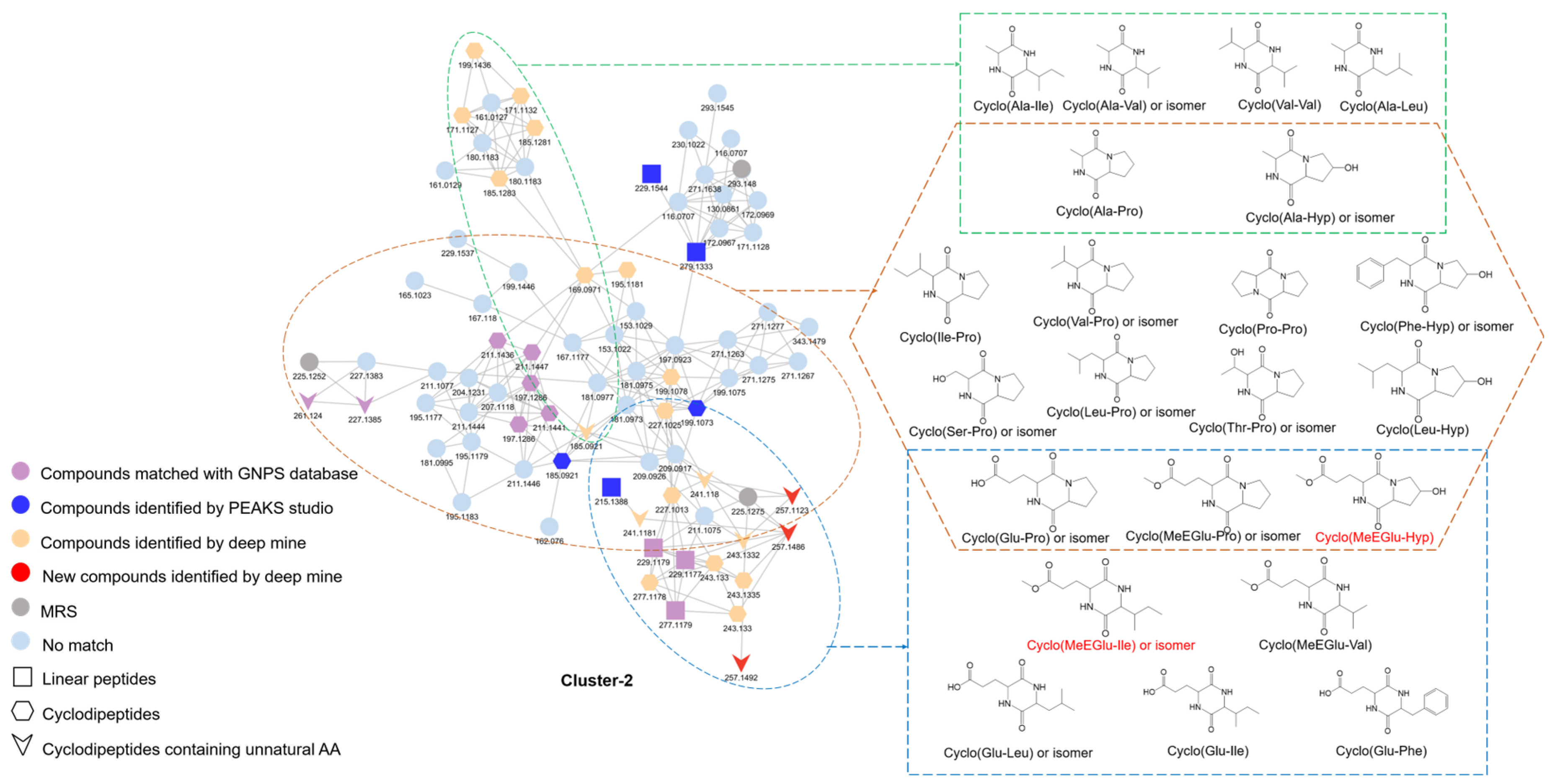
| No. | Compound Name | Molecular Formula | m/z | RT (min) |
|---|---|---|---|---|
| 1 | cyclo(Gly-Pro) | C7H10N2O2 | 155.0817 | 3.62 |
| 2 | cyclo(Ala-Pro) | C8H12N2O2 | 169.0971 | 5.05 |
| 3 | cyclo(Ala-Val) or isomer | C8H14N2O2 | 171.1127 | 7.57 |
| 4 | cyclo(Ala-Val) or isomer | C8H14N2O2 | 171.1132 | 9.68 |
| 5 | cyclo(Ser-Pro) or isomer | C8H12N2O3 | 185.0918 | 1.20 |
| 6 | cyclo(Ser-Pro) or isomer | C8H12N2O3 | 185.0921 | 2.85 |
| 7 | cyclo(Ala-Hyp) or isomer | C8H12N2O3 | 185.0921 | 2.20 |
| 8 | cyclo(Ala-Hyp) or isomer | C8H12N2O3 | 185.0921 | 2.52 |
| 9 | cyclo(Ala-Leu) | C9H16N2O2 | 185.1281 | 13.34 |
| 10 | cyclo(Ala-Ile) | C9H16N2O2 | 185.1283 | 12.02 |
| 11 | cyclo(Pro-Pro) | C10H14N2O2 | 195.1181 | 10.03 |
| 12 | cyclo(Val-Pro) or isomer | C10H16N2O2 | 197.1286 | 12.71 |
| 13 | cyclo(Val-Pro) or isomer | C10H16N2O2 | 197.1286 | 13.11 |
| 14 | cyclo(Thr-Pro) or isomer | C9H14N2O3 | 199.1073 | 3.15 |
| 15 | cyclo(Thr-Pro) or isomer | C9H14N2O3 | 199.1078 | 4.21 |
| 16 | cyclo(Val-Val) | C10H18N2O2 | 199.1436 | 18.61 |
| 17 | cyclo(Ser-Leu) | C9H16N2O3 | 201.1222 | 6.32 |
| 18 | cyclo(Leu-Pro) or isomer | C11H18N2O2 | 211.1436 | 18.79 |
| 19 | cyclo(Ile-Pro) | C11H18N2O2 | 211.1441 | 17.99 |
| 20 | cyclo(Leu-Pro) or isomer | C11H18N2O2 | 211.1447 | 19.16 |
| 21 | cyclo(Asn-Pro) | C9H13N3O3 | 212.1040 | 2.24 |
| 22 | cyclo(Val-Leu) | C11H20N2O2 | 213.1594 | 23.37 |
| 23 | cyclo(Asn-Val) | C9H15N3O3 | 214.1181 | 2.90 |
| 24 | cyclo(Asp-Val) or isomer | C9H14N2O4 | 215.1020 | 7.26 |
| 25 | cyclo(Asp-Val) or isomer | C9H14N2O4 | 215.1030 | 9.41 |
| 26 | cyclo(Glu-Pro) or isomer | C10H14N2O4 | 227.1013 | 6.47 |
| 27 | cyclo(Glu-Pro) or isomer | C10H14N2O4 | 227.1025 | 7.64 |
| 28 | cyclo(Leu-Hyp) or isomer | C11H18N2O3 | 227.1385 | 14.50 |
| 29 | cyclo(Leu-Hyp) or isomer | C11H18N2O3 | 227.1387 | 15.15 |
| 30 | cyclo(Asn-Ile) | C10H17N3O3 | 228.1343 | 6.09 |
| 31 | cyclo(Asn-Leu) | C10H17N3O3 | 228.1343 | 6.70 |
| 32 | cyclo(Asp-Ile) or isomer | C10H16N2O4 | 229.1171 | 12.13 |
| 33 | cyclo(Asp-Ile) or isomer | C10H16N2O4 | 229.1174 | 11.53 |
| 34 | cyclo(MeEGlu-Pro) or isomer | C11H16N2O4 | 241.1180 | 11.88 |
| 35 | cyclo(MeEGlu-Pro) or isomer | C11H16N2O4 | 241.1181 | 13.12 |
| 36 | cyclo(Glu-Leu) or isomer | C11H18N2O4 | 243.1330 | 17.17 |
| 37 | cyclo(Glu-Leu) or isomer | C11H18N2O4 | 243.1330 | 18.51 |
| 38 | cyclo(MeEGlu-Val) | C11H18N2O4 | 243.1332 | 16.45 |
| 39 | cyclo(Glu-Ile) | C11H18N2O4 | 243.1335 | 14.83 |
| 40 | cyclo(Phe-Pro) | C14H16N2O2 | 245.1280 | 22.23 |
| 41 | cyclo(MeEGlu-Hyp) * | C11H16N2O3 | 257.1123 | 8.80 |
| 42 | cyclo(MeEGlu-Ile) or isomer * | C12H20N2O4 | 257.1486 | 23.26 |
| 43 | cyclo(MeEGlu-Ile) or isomer * | C12H20N2O4 | 257.1492 | 20.82 |
| 44 | cyclo(Phe-Hyp) or isomer | C11H18N2O3 | 261.1230 | 16.84 |
| 45 | cyclo(Tyr-Pro) | C14H16N2O3 | 261.1237 | 13.31 |
| 46 | cyclo(Phe-Hyp) or isomer | C11H18N2O3 | 261.1240 | 17.80 |
| 47 | cyclo(PyroGlu-Tyr) * | C14H14N2O4 | 275.1037 | 11.45 |
| 48 | cyclo(Tyr-Hyp) or isomer | C14H16N2O4 | 277.1171 | 12.43 |
| 49 | cyclo(Tyr-Hyp) or isomer | C14H16N2O4 | 277.1178 | 12.10 |
| 50 | cyclo(Glu-Phe) | C14H16N2O4 | 277.1178 | 20.30 |
| 51 | cyclo(Tyr-Asp) | C13H14N2O5 | 279.1332 | 10.06 |
| 52 | cyclo(Glu-Tyr) or isomer | C14H16N2O5 | 293.1119 | 10.26 |
| 53 | cyclo(Glu-Tyr) or isomer | C14H16N2O5 | 293.1122 | 11.43 |
| Binding Energy (kcal/mol) | ||||
|---|---|---|---|---|
| No. | Ligand | NEK7 | Cat C | GSDMD |
| 1 | cyclo(Glu-Phe) | −7.6 | −7.6 | −6.2 |
| 2 | cyclo(Phe-Pro) | −7.5 | −7.6 | −6.6 |
| 3 | cyclo(Tyr-Hyp) | −7.4 | −7.6 | −6.8 |
| 4 | cyclo(Phe-Hyp) | −7.3 | −8.0 | −6.5 |
| 5 | PyroGlu-Phe | −7.2 | −7.8 | −6.4 |
| 6 | Rolipram | −7.2 | −7.7 | −6.3 |
| 7 | cyclo(Tyr-Asp) | −7.1 | −7.7 | −6.6 |
| 8 | cyclo(PyroGlu-Tyr) | −7.0 | −7.4 | −7.1 |
| 9 | cyclo(Tyr-Pro) | −7.0 | −7.2 | −6.6 |
| 10 | LPNLP | −7.0 | −6.5 | −6.5 |
| 11 | VYPFPGPLPQ | −7.0 | −7.8 | −6.3 |
| 12 | YPFELP | −7.0 | −7.1 | −6.7 |
| 13 | cyclo(Glu-Tyr) | −6.9 | −7.6 | −6.4 |
| 14 | WS(+14.02) | −6.8 | −6.8 | −5.9 |
| 15 | PLLLP | −6.8 | −6.5 | −7.1 |
| 16 | RMPPSP | −6.8 | −6.9 | −6.0 |
| 17 | VYPFPGPLPE | −6.7 | −6.8 | −6.2 |
| 18 | VYPFPGPLPN | −6.7 | −6.7 | −7.1 |
| 19 | cyclo(MeEGlu-Hyp) | −6.6 | −6.8 | −5.6 |
| 20 | cyclo(Glu-Leu) | −6.6 | −6.5 | −5.8 |
| 21 | YPFPALP | −6.6 | −7.7 | −6.4 |
| 22 | cyclo(Glu-Pro) | −6.5 | −6.6 | −5.6 |
| 23 | L(+72.02)A | −6.5 | −6.2 | −5.0 |
| 24 | LPLLP | −6.5 | −7.3 | −6.8 |
| 25 | cyclo(Glu-Ile) | −6.4 | −7.1 | −5.3 |
| 26 | cyclo(Val-Leu) | −6.4 | −6.6 | −5.9 |
| 27 | cyclo(Leu-Pro) | −6.4 | −6.4 | −5.5 |
| 28 | LPPL | −6.4 | −6.3 | −6.8 |
| 29 | cyclo(Asn-Pro) | −6.4 | −6.5 | −5.4 |
| 30 | VYPFPGPLEP | −6.4 | −7.1 | −6.3 |
| 31 | cyclo(Leu-Hyp) | −6.3 | −6.6 | −5.8 |
| 32 | cyclo(MeEGlu-Pro) | −6.3 | −6.4 | −5.9 |
| 33 | cyclo(Val-Val) | −6.3 | −6.3 | −5.5 |
| 34 | VAPFPEVFA | −6.3 | −6.6 | −7.7 |
| 35 | YVPL | −6.2 | −7.5 | −6.7 |
| 36 | cyclo(MeGlu-Ile) | −6.2 | −6.6 | −5.4 |
| 37 | PLEFP | −6.2 | −7.6 | −6.5 |
| 38 | TLEQLFPPVLVPVPNTPLP | −6.2 | −6.4 | −6.0 |
| 39 | YPVEPF | −6.2 | −7.7 | −7.1 |
| 40 | D(−18.01)APL | −6.1 | −7.0 | −5.9 |
| 41 | cyclo(Asp-Val) | −6.1 | −6.4 | −5.5 |
| 42 | cyclo(Ile-Pro) | −6.0 | −6.9 | −5.6 |
| 43 | cyclo(MeEGlu-Val) | −6.0 | −6.4 | −5.9 |
| 44 | P(+27.99)VSY | −6.0 | −6.2 | −6.0 |
| 45 | E(+14.02)L | −6.0 | −5.9 | −4.9 |
| 46 | cyclo(Asn-Val) | −6.0 | −6.5 | −5.7 |
| 47 | cyclo(Asn-Leu) | −6.0 | −6.6 | −5.8 |
| 48 | LPLPL | −6.0 | −6.1 | −6.0 |
| 49 | YVPFPGPLEP | −6.0 | −7.4 | −7.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Y.; Gong, S.; Zhang, L.; Zhou, J.; Wu, J.-L.; Li, N. A Deep Mining Strategy for Peptide Rapid Identification in Lactobacillus reuteri Based on LC–MS/MS Integrated with FBMN and De Novo Sequencing. Metabolites 2024, 14, 467. https://doi.org/10.3390/metabo14090467
Zuo Y, Gong S, Zhang L, Zhou J, Wu J-L, Li N. A Deep Mining Strategy for Peptide Rapid Identification in Lactobacillus reuteri Based on LC–MS/MS Integrated with FBMN and De Novo Sequencing. Metabolites. 2024; 14(9):467. https://doi.org/10.3390/metabo14090467
Chicago/Turabian StyleZuo, Yilang, Shilin Gong, Li Zhang, Jie Zhou, Jian-Lin Wu, and Na Li. 2024. "A Deep Mining Strategy for Peptide Rapid Identification in Lactobacillus reuteri Based on LC–MS/MS Integrated with FBMN and De Novo Sequencing" Metabolites 14, no. 9: 467. https://doi.org/10.3390/metabo14090467
APA StyleZuo, Y., Gong, S., Zhang, L., Zhou, J., Wu, J.-L., & Li, N. (2024). A Deep Mining Strategy for Peptide Rapid Identification in Lactobacillus reuteri Based on LC–MS/MS Integrated with FBMN and De Novo Sequencing. Metabolites, 14(9), 467. https://doi.org/10.3390/metabo14090467








