Comparison of Adiposomal Lipids between Obese and Non-Obese Individuals
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Enrollment
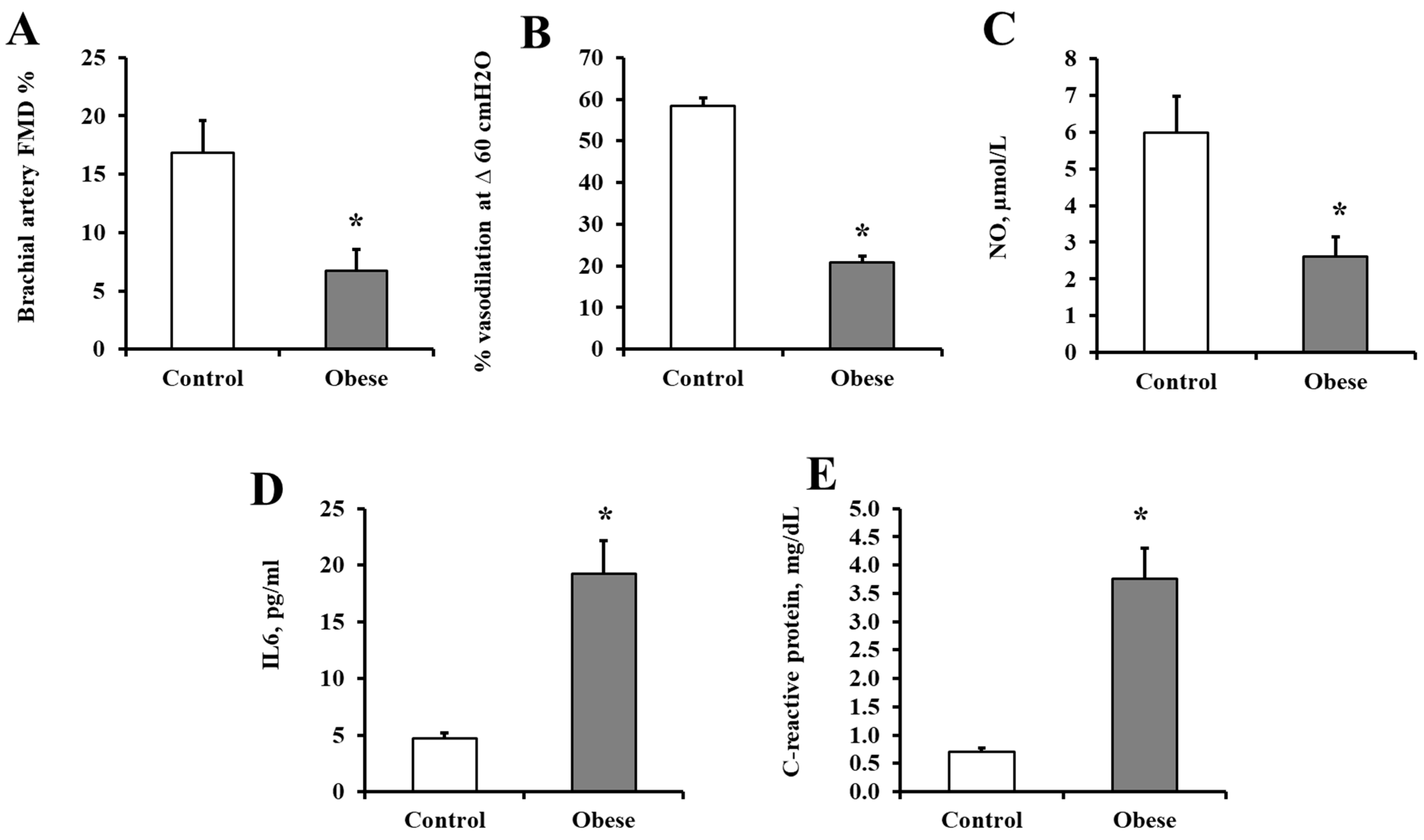
2.2. Cardiometabolic Risk Measurements
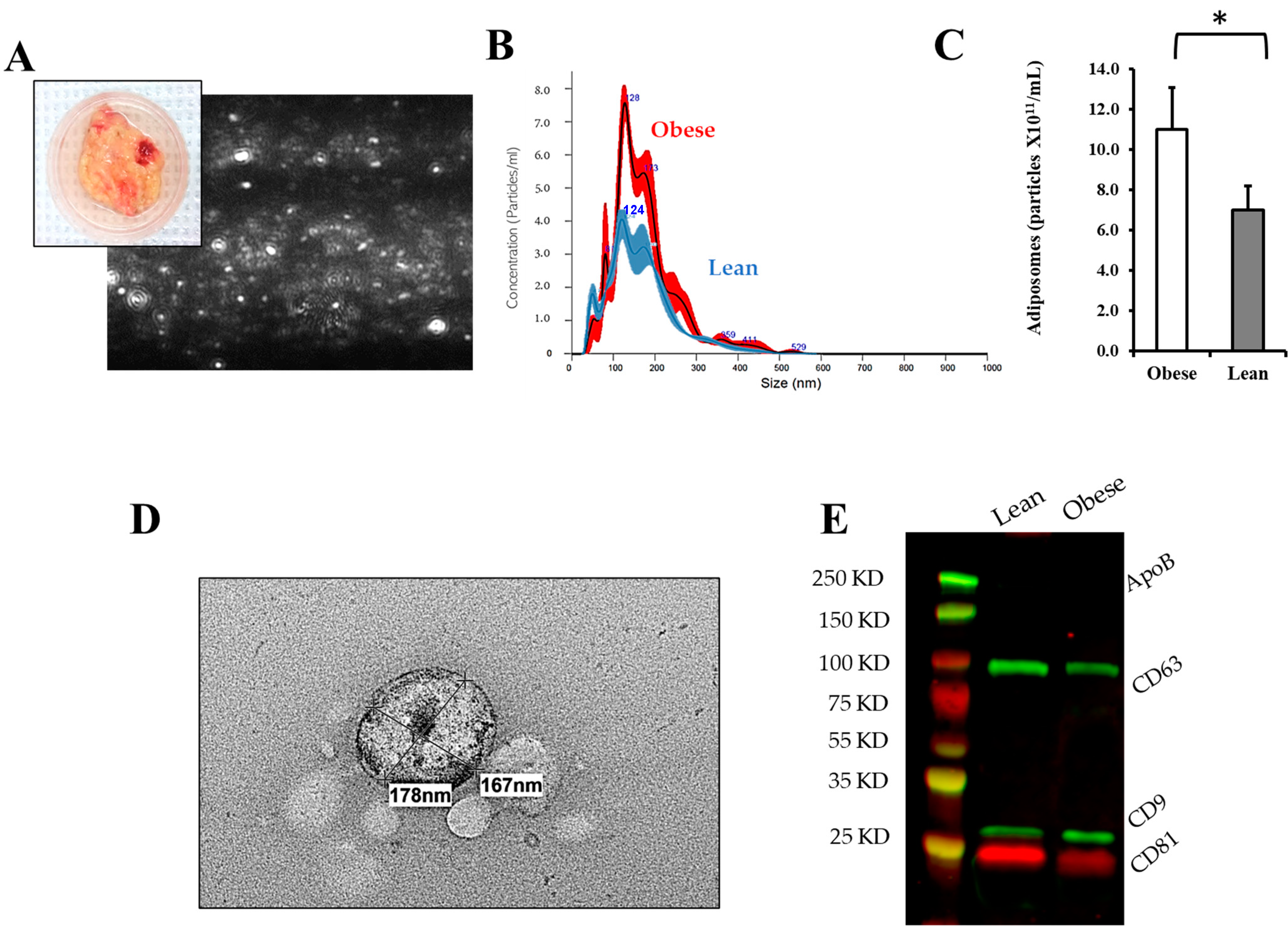
2.3. Adiposome Isolation
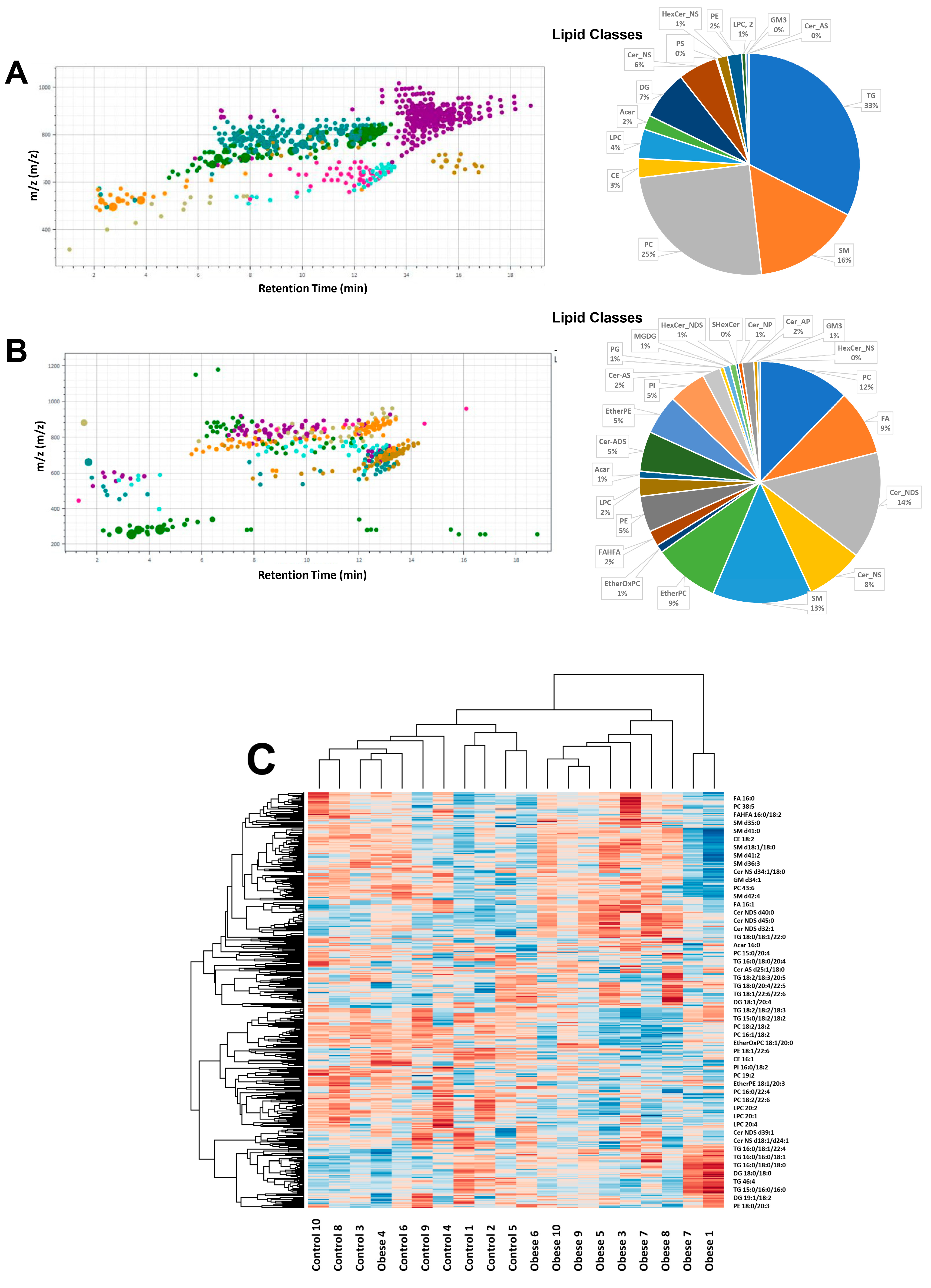
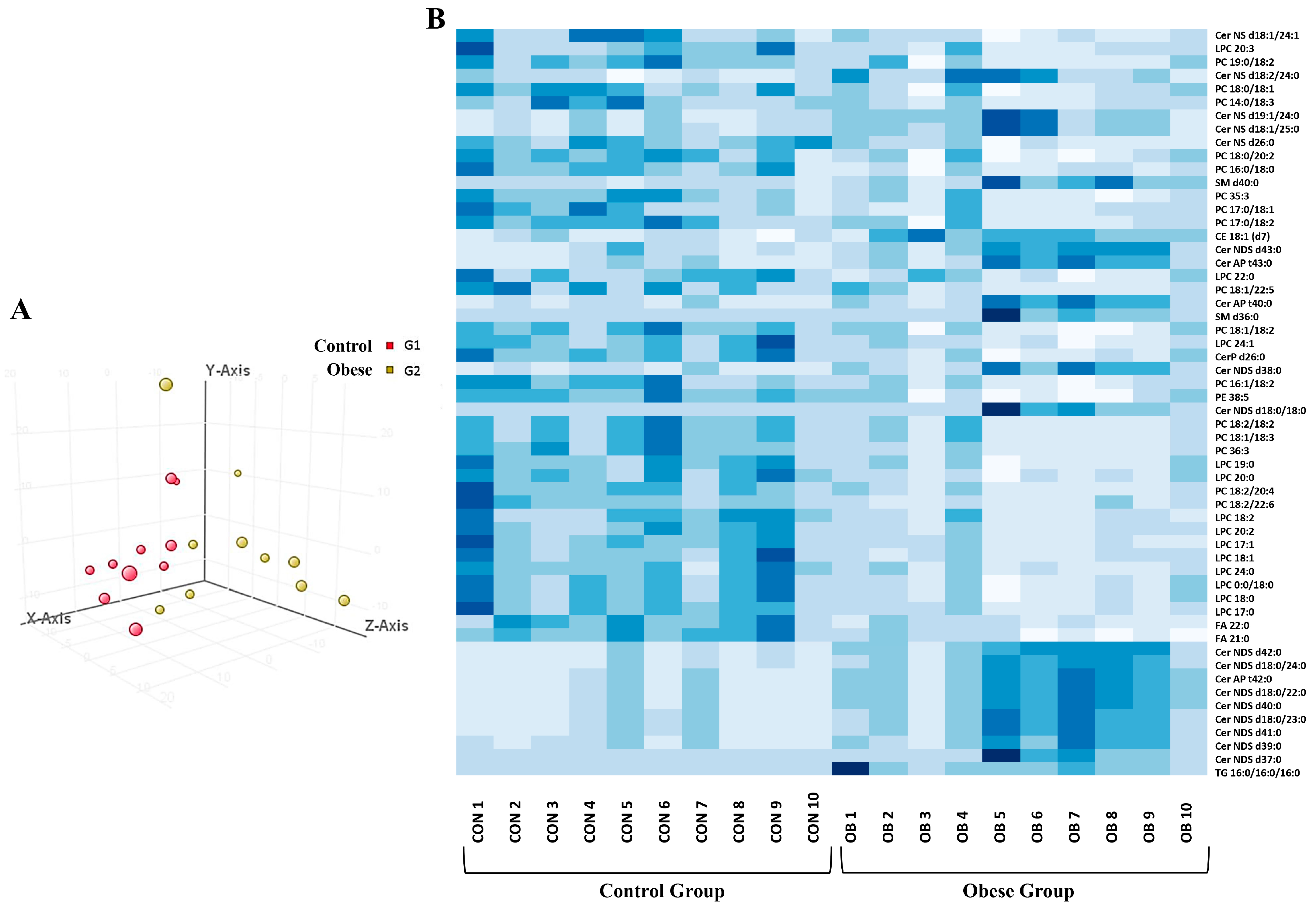
2.4. Adiposomal Lipid Isolation and Analysis
2.5. Flow-Mediated Dilation (FMD)
2.6. Flow-Induced Dilation (FID) in Adipose Tissue Arterioles
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Study Participants
3.2. Adiposomal Characterization
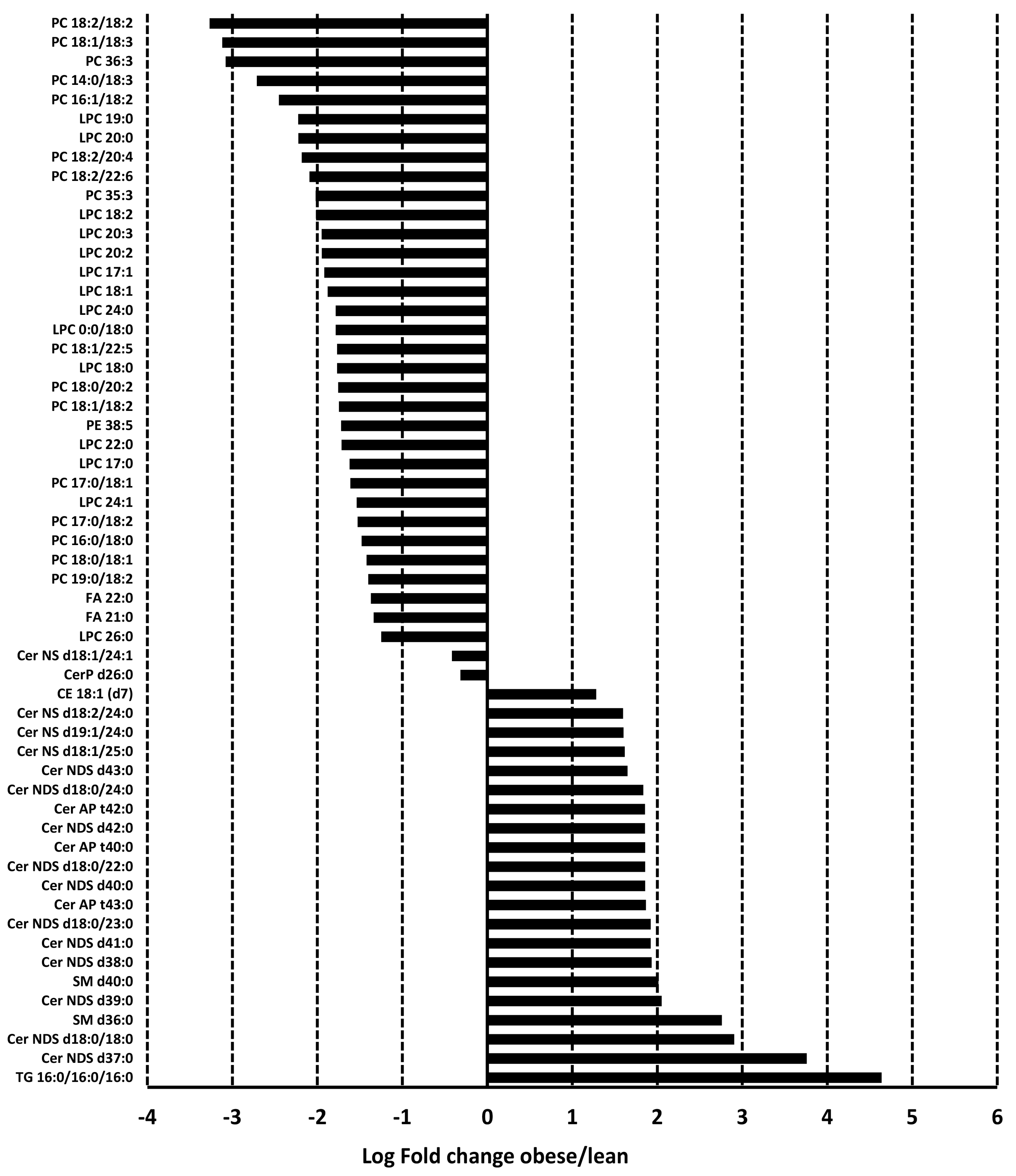
3.3. Adiposomal Lipid Content
3.4. Relationship between the Adiposomal LPCs and Cardiometabolic Risk Factors
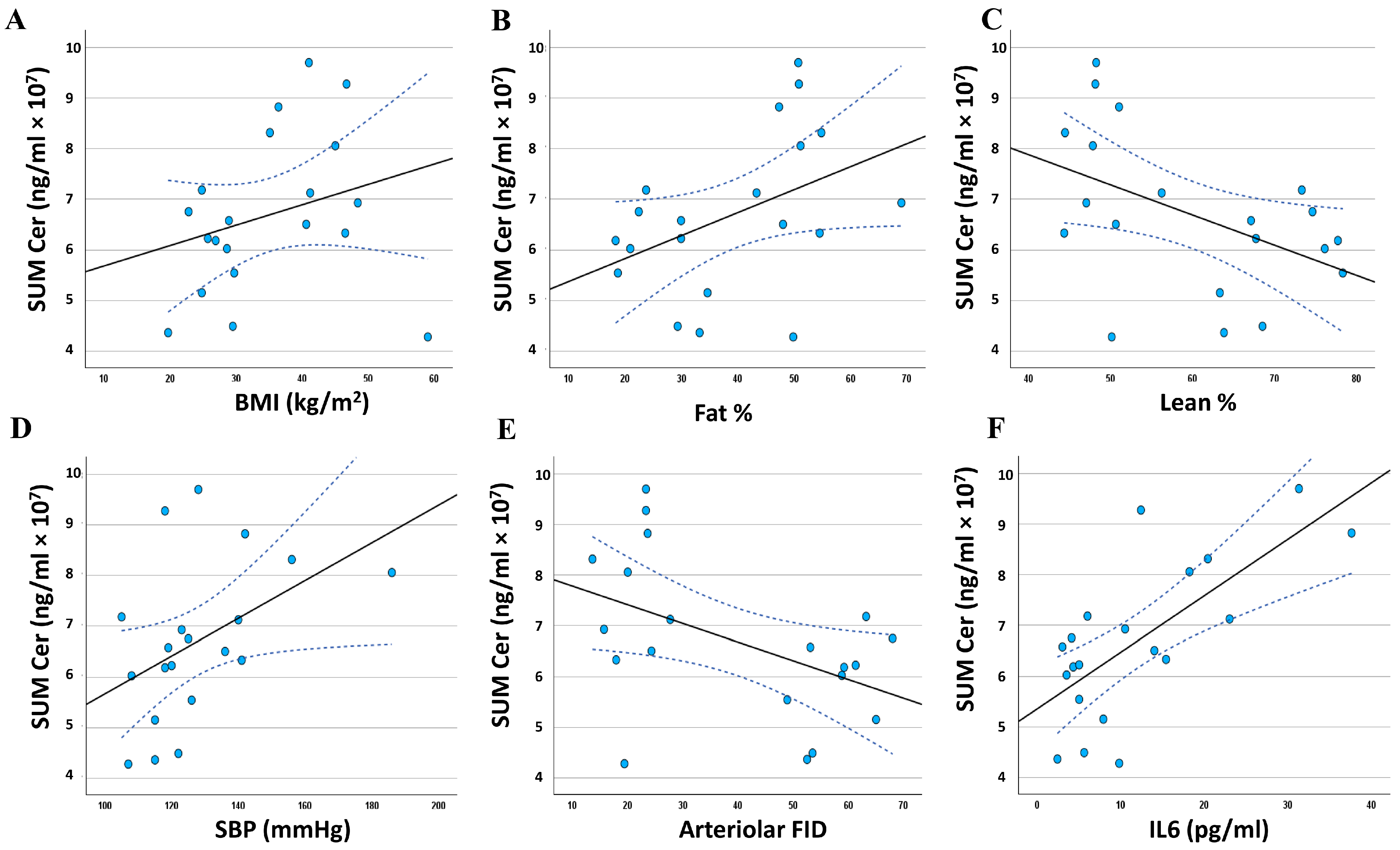
3.5. Relationship between the Adiposomal Ceramides and Cardiometabolic Risk Factors
3.6. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hales, C.M.; Carroll, M.D.; Fryar, C.D.; Ogden, C.L. Prevalence of Obesity and Severe Obesity Among Adults: United States, 2017–2018. NCHS Data Brief 2020, 1–8. [Google Scholar]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Despres, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 2021, 143, e984–e1010. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Klarqvist, M.D.R.; Diamant, N.; Stanley, T.L.; Ellinor, P.T.; Mehta, N.N.; Philippakis, A.; Ng, K.; Claussnitzer, M.; Grinspoon, S.K.; et al. BMI-adjusted adipose tissue volumes exhibit depot-specific and divergent associations with cardiometabolic diseases. Nat. Commun. 2023, 14, 266. [Google Scholar] [CrossRef]
- Ali, M.M.; Hassan, C.; Masrur, M.; Bianco, F.M.; Naquiallah, D.; Mirza, I.; Frederick, P.; Fernandes, E.T.; Giulianotti, C.P.; Gangemi, A.; et al. Adipose Tissue Hypoxia Correlates with Adipokine Hypomethylation and Vascular Dysfunction. Biomedicines 2021, 9, 1034. [Google Scholar] [CrossRef]
- Ali, M.M.; Phillips, S.A.; Mahmoud, A.M. HIF1alpha/TET1 Pathway Mediates Hypoxia-Induced Adipocytokine Promoter Hypomethylation in Human Adipocytes. Cells 2020, 9, 134. [Google Scholar] [CrossRef]
- Bond, S.T.; Calkin, A.C.; Drew, B.G. Adipose-Derived Extracellular Vesicles: Systemic Messengers and Metabolic Regulators in Health and Disease. Front. Physiol. 2022, 13, 837001. [Google Scholar] [CrossRef]
- Liu, W.; Liu, T.; Zhao, Q.; Ma, J.; Jiang, J.; Shi, H. Adipose Tissue-Derived Extracellular Vesicles: A Promising Biomarker and Therapeutic Strategy for Metabolic Disorders. Stem Cells Int. 2023, 2023, 9517826. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, A. Adipose Extracellular Vesicles in Intercellular and Inter-Organ Crosstalk in Metabolic Health and Diseases. Front. Immunol. 2021, 12, 608680. [Google Scholar] [CrossRef]
- Kumar, M.A.; Baba, S.K.; Sadida, H.Q.; Marzooqi, S.A.; Jerobin, J.; Altemani, F.H.; Algehainy, N.; Alanazi, M.A.; Abou-Samra, A.B.; Kumar, R.; et al. Extracellular vesicles as tools and targets in therapy for diseases. Signal Transduct. Target Ther. 2024, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Zacharia, A.; Saidemberg, D.; Mannully, C.T.; Kogan, N.M.; Shehadeh, A.; Sinai, R.; Zucker, A.; Bruck-Haimson, R.; Goldstein, N.; Haim, Y.; et al. Distinct infrastructure of lipid networks in visceral and subcutaneous adipose tissues in overweight humans. Am. J. Clin. Nutr. 2020, 112, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.; De Filippo, E.; Goddeke, S.; Knebel, B.; Kotzka, J.; Al-Hasani, H.; Roden, M.; Lehr, S.; Sell, H. Exosomal proteins constitute an essential part of the human adipose tissue secretome. Biochim. Biophys. Acta Proteins Proteom 2019, 1867, 140172. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Mirza, I.; Haloul, M.; Hassan, C.; Masrur, M.; Mostafa, A.; Bianco, F.M.; Ali, M.M.; Minshall, R.D.; Mahmoud, A.M. Adiposomes from Obese-Diabetic Individuals Promote Endothelial Dysfunction and Loss of Surface Caveolae. Cells 2023, 12, 2453. [Google Scholar] [CrossRef]
- Mirza, I.; Hassan, C.; Masrur, M.; Bianco, F.M.; Ali, M.M.; Mahmoud, A.M. The Role of Adipocyte-derived Extracellular Vesicles in Diabetes-associated Endothelial Dysfunction. FASEB J. 2022, 36 (Suppl. S1). [Google Scholar] [CrossRef]
- Durcin, M.; Fleury, A.; Taillebois, E.; Hilairet, G.; Krupova, Z.; Henry, C.; Truchet, S.; Trotzmuller, M.; Kofeler, H.; Mabilleau, G.; et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1305677. [Google Scholar] [CrossRef]
- Eguchi, A.; Lazic, M.; Armando, A.M.; Phillips, S.A.; Katebian, R.; Maraka, S.; Quehenberger, O.; Sears, D.D.; Feldstein, A.E. Circulating adipocyte-derived extracellular vesicles are novel markers of metabolic stress. J. Mol. Med. 2016, 94, 1241–1253. [Google Scholar] [CrossRef]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef]
- Eguchi, A.; Mulya, A.; Lazic, M.; Radhakrishnan, D.; Berk, M.P.; Povero, D.; Gornicka, A.; Feldstein, A.E. Microparticles release by adipocytes act as “find-me” signals to promote macrophage migration. PLoS ONE 2015, 10, e0123110. [Google Scholar] [CrossRef]
- Wadey, R.M.; Connolly, K.D.; Mathew, D.; Walters, G.; Rees, D.A.; James, P.E. Inflammatory adipocyte-derived extracellular vesicles promote leukocyte attachment to vascular endothelial cells. Atherosclerosis 2019, 283, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Haloul, M.; Vinjamuri, S.J.; Naquiallah, D.; Mirza, M.I.; Qureshi, M.; Hassan, C.; Masrur, M.; Bianco, F.M.; Frederick, P.; Cristoforo, G.P.; et al. Hyperhomocysteinemia and Low Folate and Vitamin B12 Are Associated with Vascular Dysfunction and Impaired Nitric Oxide Sensitivity in Morbidly Obese Patients. Nutrients 2020, 12, 2014. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Naquiallah, D.; Qureshi, M.; Mirza, M.I.; Hassan, C.; Masrur, M.; Bianco, F.M.; Frederick, P.; Cristoforo, G.P.; Gangemi, A.; et al. DNA methylation profile of genes involved in inflammation and autoimmunity correlates with vascular function in morbidly obese adults. Epigenetics 2021, 17, 93–109. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Szczurek, M.R.; Blackburn, B.K.; Mey, J.T.; Chen, Z.; Robinson, A.T.; Bian, J.T.; Unterman, T.G.; Minshall, R.D.; Brown, M.D.; et al. Hyperinsulinemia augments endothelin-1 protein expression and impairs vasodilation of human skeletal muscle arterioles. Physiol. Rep. 2016, 4, e12895. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Szczurek, M.; Hassan, C.; Masrur, M.; Gangemi, A.; Phillips, S.A. Vitamin D Improves Nitric Oxide-Dependent Vasodilation in Adipose Tissue Arterioles from Bariatric Surgery Patients. Nutrients 2019, 11, 2521. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Hwang, C.L.; Szczurek, M.R.; Bian, J.T.; Ranieri, C.; Gutterman, D.D.; Phillips, S.A. Low-Fat Diet Designed for Weight Loss But Not Weight Maintenance Improves Nitric Oxide-Dependent Arteriolar Vasodilation in Obese Adults. Nutrients 2019, 11, 1339. [Google Scholar] [CrossRef]
- Ali, M.M.; Naquiallah, D.; Hassan, C.; Masrur, M.; Bianco, F.M.; Frederick, P.; Cristoforo, G.; Gangemi, A.; Phillips, S.A.; Mahmoud, A.M. Obesity-associated Hypoxia Contributes to Aberrant Methylation of Genes Implicated in Inflammation and vascular Function. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Kranendonk, M.E.; Visseren, F.L.; van Balkom, B.W.; Nolte-‘t Hoen, E.N.; van Herwaarden, J.A.; de Jager, W.; Schipper, H.S.; Brenkman, A.B.; Verhaar, M.C.; Wauben, M.H.; et al. Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity 2014, 22, 1296–1308. [Google Scholar] [CrossRef]
- Kreimer, S.; Belov, A.M.; Ghiran, I.; Murthy, S.K.; Frank, D.A.; Ivanov, A.R. Mass-spectrometry-based molecular characterization of extracellular vesicles: Lipidomics and proteomics. J. Proteome Res. 2015, 14, 2367–2384. [Google Scholar] [CrossRef]
- Barber, M.N.; Risis, S.; Yang, C.; Meikle, P.J.; Staples, M.; Febbraio, M.A.; Bruce, C.R. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE 2012, 7, e41456. [Google Scholar] [CrossRef]
- Bachlechner, U.; Floegel, A.; Steffen, A.; Prehn, C.; Adamski, J.; Pischon, T.; Boeing, H. Associations of anthropometric markers with serum metabolites using a targeted metabolomics approach: Results of the EPIC-potsdam study. Nutr. Diabetes 2016, 6, e215. [Google Scholar] [CrossRef]
- Bagheri, M.; Djazayery, A.; Farzadfar, F.; Qi, L.; Yekaninejad, M.S.; Aslibekyan, S.; Chamari, M.; Hassani, H.; Koletzko, B.; Uhl, O. Plasma metabolomic profiling of amino acids and polar lipids in Iranian obese adults. Lipids Health Dis. 2019, 18, 94. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.; Morris, C.; O’Grada, C.M.; Ryan, M.; Dillon, E.T.; Coleman, E.; Gibney, E.R.; Gibney, M.J.; Roche, H.M.; Brennan, L. Relationship between the lipidome, inflammatory markers and insulin resistance. Mol. Biosyst. 2014, 10, 1586–1595. [Google Scholar] [CrossRef]
- Frigerio, G.; Favero, C.; Savino, D.; Mercadante, R.; Albetti, B.; Dioni, L.; Vigna, L.; Bollati, V.; Pesatori, A.C.; Fustinoni, S. Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study. Metabolites 2021, 11, 194. [Google Scholar] [CrossRef]
- Carayol, M.; Leitzmann, M.F.; Ferrari, P.; Zamora-Ros, R.; Achaintre, D.; Stepien, M.; Schmidt, J.A.; Travis, R.C.; Overvad, K.; Tjonneland, A.; et al. Blood Metabolic Signatures of Body Mass Index: A Targeted Metabolomics Study in the EPIC Cohort. J. Proteome Res. 2017, 16, 3137–3146. [Google Scholar] [CrossRef]
- Bagheri, M.; Farzadfar, F.; Qi, L.; Yekaninejad, M.S.; Chamari, M.; Zeleznik, O.A.; Kalantar, Z.; Ebrahimi, Z.; Sheidaie, A.; Koletzko, B.; et al. Obesity-Related Metabolomic Profiles and Discrimination of Metabolically Unhealthy Obesity. J. Proteome Res. 2018, 17, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Tonks, K.T.; Coster, A.C.; Christopher, M.J.; Chaudhuri, R.; Xu, A.; Gagnon-Bartsch, J.; Chisholm, D.J.; James, D.E.; Meikle, P.J.; Greenfield, J.R.; et al. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity 2016, 24, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Piko, P.; Pal, L.; Szucs, S.; Kosa, Z.; Sandor, J.; Adany, R. Obesity-Related Changes in Human Plasma Lipidome Determined by the Lipidyzer Platform. Biomolecules 2021, 11, 326. [Google Scholar] [CrossRef]
- Wang, S.M.; Yang, R.Y.; Wang, M.; Ji, F.S.; Li, H.X.; Tang, Y.M.; Chen, W.X.; Dong, J. Identification of serum metabolites associated with obesity and traditional risk factors for metabolic disease in Chinese adults. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Heimerl, S.; Fischer, M.; Baessler, A.; Liebisch, G.; Sigruener, A.; Wallner, S.; Schmitz, G. Alterations of plasma lysophosphatidylcholine species in obesity and weight loss. PLoS ONE 2014, 9, e111348. [Google Scholar] [CrossRef]
- Cole, L.K.; Vance, J.E.; Vance, D.E. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim. Biophys. Acta 2012, 1821, 754–761. [Google Scholar] [CrossRef]
- Bellot, P.; Moia, M.N.; Reis, B.Z.; Pedrosa, L.F.C.; Tasic, L.; Barbosa, F., Jr.; Sena-Evangelista, K.C.M. Are Phosphatidylcholine and Lysophosphatidylcholine Body Levels Potentially Reliable Biomarkers in Obesity? A Review of Human Studies. Mol. Nutr. Food Res. 2023, 67, e2200568. [Google Scholar] [CrossRef]
- Schwab, U.; Seppanen-Laakso, T.; Yetukuri, L.; Agren, J.; Kolehmainen, M.; Laaksonen, D.E.; Ruskeepaa, A.L.; Gylling, H.; Uusitupa, M.; Oresic, M.; et al. Triacylglycerol fatty acid composition in diet-induced weight loss in subjects with abnormal glucose metabolism--the GENOBIN study. PLoS ONE 2008, 3, e2630. [Google Scholar] [CrossRef]
- Schwenk, R.W.; Vogel, H.; Schurmann, A. Genetic and epigenetic control of metabolic health. Mol. Metab. 2013, 2, 337–347. [Google Scholar] [CrossRef]
- Nestel, P.J.; Straznicky, N.; Mellett, N.A.; Wong, G.; De Souza, D.P.; Tull, D.L.; Barlow, C.K.; Grima, M.T.; Meikle, P.J. Specific plasma lipid classes and phospholipid fatty acids indicative of dairy food consumption associate with insulin sensitivity. Am. J. Clin. Nutr. 2014, 99, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Shindou, H.; Hishikawa, D.; Harayama, T.; Eto, M.; Shimizu, T. Generation of membrane diversity by lysophospholipid acyltransferases. J. Biochem. 2013, 154, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Agren, J.J.; Kurvinen, J.P.; Kuksis, A. Isolation of very low density lipoprotein phospholipids enriched in ethanolamine phospholipids from rats injected with Triton WR 1339. Biochim. Biophys. Acta 2005, 1734, 34–43. [Google Scholar] [CrossRef]
- Skipski, V.P.; Barclay, M.; Barclay, R.K.; Fetzer, V.A.; Good, J.J.; Archibald, F.M. Lipid composition of human serum lipoproteins. Biochem. J. 1967, 104, 340–352. [Google Scholar] [CrossRef]
- Verkade, H.J.; Fast, D.G.; Rusinol, A.E.; Scraba, D.G.; Vance, D.E. Impaired biosynthesis of phosphatidylcholine causes a decrease in the number of very low density lipoprotein particles in the Golgi but not in the endoplasmic reticulum of rat liver. J. Biol. Chem. 1993, 268, 24990–24996. [Google Scholar]
- Yao, Z.M.; Vance, D.E. Head group specificity in the requirement of phosphatidylcholine biosynthesis for very low density lipoprotein secretion from cultured hepatocytes. J. Biol. Chem. 1989, 264, 11373–11380. [Google Scholar]
- Pol, A.; Gross, S.P.; Parton, R.G. Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 2014, 204, 635–646. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Lingrell, S.; da Silva, R.P.; Jacobs, R.L.; Vance, D.E. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes 2014, 63, 2620–2630. [Google Scholar] [CrossRef]
- Wang, B.; Rong, X.; Duerr, M.A.; Hermanson, D.J.; Hedde, P.N.; Wong, J.S.; Vallim, T.Q.; Cravatt, B.F.; Gratton, E.; Ford, D.A.; et al. Intestinal Phospholipid Remodeling Is Required for Dietary-Lipid Uptake and Survival on a High-Fat Diet. Cell Metab. 2016, 23, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Funai, K.; Lodhi, I.J.; Spears, L.D.; Yin, L.; Song, H.; Klein, S.; Semenkovich, C.F. Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 2016, 65, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Zembowicz, A.; Tang, J.L.; Wu, K.K. Transcriptional induction of endothelial nitric oxide synthase type III by lysophosphatidylcholine. J. Biol. Chem. 1995, 270, 17006–17010. [Google Scholar] [CrossRef]
- Brkic, L.; Riederer, M.; Graier, W.F.; Malli, R.; Frank, S. Acyl chain-dependent effect of lysophosphatidylcholine on cyclooxygenase (COX)-2 expression in endothelial cells. Atherosclerosis 2012, 224, 348–354. [Google Scholar] [CrossRef]
- Zembowicz, A.; Jones, S.L.; Wu, K.K. Induction of cyclooxygenase-2 in human umbilical vein endothelial cells by lysophosphatidylcholine. J. Clin. Investig. 1995, 96, 1688–1692. [Google Scholar] [CrossRef]
- Jougasaki, M.; Kugiyama, K.; Saito, Y.; Nakao, K.; Imura, H.; Yasue, H. Suppression of endothelin-1 secretion by lysophosphatidylcholine in oxidized low density lipoprotein in cultured vascular endothelial cells. Circ. Res. 1992, 71, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Wolf, A.; Menon, N.K.; Saeed, M.; Alves, C.; Bing, R.J. Lysolecithins as endothelium-dependent vascular smooth muscle relaxants that differ from endothelium-derived relaxing factor (nitric oxide). Proc. Natl. Acad. Sci. USA 1988, 85, 8246–8250. [Google Scholar] [CrossRef]
- Bing, R.J.; Saeed, M. The role of lysolecithin in the relaxation of vascular smooth muscle. Biosci. Rep. 1987, 7, 783–789. [Google Scholar] [CrossRef]
- Schilcher, I.; Ledinski, G.; Radulovic, S.; Hallstrom, S.; Eichmann, T.; Madl, T.; Zhang, F.; Leitinger, G.; Kolb-Lenz, D.; Darnhofer, B.; et al. Endothelial lipase increases antioxidative capacity of high-density lipoprotein. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1363–1374. [Google Scholar] [CrossRef]
- Wolf, A.; Saito, T.; Dudek, R.; Bing, R.J. The effect of lysophosphatidylcholine on coronary and renal circulation in the rabbit. Lipids 1991, 26, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, B.; Zieseniss, S.; Brand, K.; Page, S.; Lentschat, A.; Ulmer, A.J.; Gerlach, E. Tissue factor expression of human monocytes is suppressed by lysophosphatidylcholine. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Hara, H.; Adachi, T. The expression of extracellular-superoxide dismutase is increased by lysophosphatidylcholine in human monocytic U937 cells. Atherosclerosis 2002, 163, 223–228. [Google Scholar] [CrossRef]
- Chang, M.K.; Hartvigsen, K.; Ryu, J.; Kim, Y.; Han, K.H. The pro-atherogenic effects of macrophages are reduced upon formation of a complex between C-reactive protein and lysophosphatidylcholine. J. Inflamm. 2012, 9, 42. [Google Scholar] [CrossRef]
- Mendonca, M.C.; Rezende, A.; Doi, S.Q.; Sellitti, D.F. Lysophosphatidylcholine increases C-type natriuretic peptide expression in human vascular smooth muscle cells via membrane distortion. Vascul. Pharmacol. 2009, 51, 29–36. [Google Scholar] [CrossRef]
- Leermakers, E.T.; Moreira, E.M.; Kiefte-de Jong, J.C.; Darweesh, S.K.; Visser, T.; Voortman, T.; Bautista, P.K.; Chowdhury, R.; Gorman, D.; Bramer, W.M.; et al. Effects of choline on health across the life course: A systematic review. Nutr. Rev. 2015, 73, 500–522. [Google Scholar] [CrossRef]
- Vance, D.E.; Ridgway, N.D. The methylation of phosphatidylethanolamine. Prog. Lipid Res. 1988, 27, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, J.K.; Tuomainen, T.P.; Voutilainen, S. Dietary intake of choline and phosphatidylcholine and risk of type 2 diabetes in men: The Kuopio Ischaemic Heart Disease Risk Factor Study. Eur. J. Nutr. 2020, 59, 3857–3861. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Karlsson, T. Choline—A scoping review for Nordic Nutrition Recommendations 2023. Food Nutr. Res. 2023, 67. [Google Scholar] [CrossRef]
- Field, B.C.; Gordillo, R.; Scherer, P.E. The Role of Ceramides in Diabetes and Cardiovascular Disease Regulation of Ceramides by Adipokines. Front. Endocrinol. 2020, 11, 569250. [Google Scholar] [CrossRef]
- Mirza, I.; Naquiallah, D.; Hassan, C.; MAsrur, M.; Bianco, F.M.; Ali, M.; Mahmoud, A.M. Abstract 13966: The Effect of Diabetes-Associated Adiposomes in Promoting Caveolar Loss and Endothelial Dysfunction. Circulation 2022, 146, A13966. [Google Scholar] [CrossRef]


| Variable | Lean (n = 10, 5 Females) | Obese (n = 10, 5 Females) | p-Value |
|---|---|---|---|
| Age, y | 36 ± 5 | 35 ± 7 | NS |
| Anthropometric | |||
| Weight, kg | 69.5 ± 3.1 | 152.4 ± 9.7 | <0.0001 |
| BMI, kg/m2 | 22.4 ± 1.4 | 47.8 ± 5.9 | <0.0001 |
| Cardiometabolic | |||
| FPI, µU/mL | 6.4 ± 0.3 | 14.2 ± 1.9 | 0.0007 |
| FPG, mg/dL | 85 ± 7 | 127 ± 10 | 0.0029 |
| HOMA-IR | 2.2 ± 0.3 | 5.2 ± 0.5 | 0.0001 |
| HbA1c, % | 5.4 ± 0.2 | 5.7 ± 0.3 | NS |
| Chol, mg/dL | 147.5 ± 5.5 | 192.0 ± 11 | 0.0020 |
| LDL-C, mg/dL | 85.1 ± 4.6 | 111.3 ± 6.1 | 0.0030 |
| HDL-C, mg/dL | 57.0 ± 3.7 | 32.0 ± 4.7 | 0.0006 |
| Trig, mg/dL | 91.6 ± 4.2 | 127.0 ± 5.2 | <0.0001 |
| HR, bpm | 75 ± 4 | 80 ± 5 | NS |
| SBP, mmHg | 118 ± 2 | 122 ± 4 | NS |
| DBP, mmHg | 75 ± 2 | 84 ± 2 | 0.0052 |
| BMI | Fat % | Lean % | ||||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| LPC 17:0 | −0.552 ** | 0.006 | −0.764 ** | 0.000 | 0.804 ** | 0.000 |
| LPC 18:0 | −0.562 ** | 0.005 | −0.721 ** | 0.000 | 0.777 ** | 0.000 |
| LPC 18:0 | −0.569 ** | 0.004 | −0.728 ** | 0.000 | 0.785 ** | 0.000 |
| LPC 24:0 | −0.604 ** | 0.002 | −0.698 ** | 0.000 | 0.769 ** | 0.000 |
| LPC 18:1 | −0.559 ** | 0.005 | −0.731 ** | 0.000 | 0.760 ** | 0.000 |
| LPC 17:1 | −0.535 ** | 0.008 | −0.740 ** | 0.000 | 0.784 ** | 0.000 |
| LPC 20:2 | −0.552 ** | 0.006 | −0.735 ** | 0.000 | 0.806 ** | 0.000 |
| LPC 18:2 | −0.631 ** | 0.001 | −0.793 ** | 0.000 | 0.830 ** | 0.000 |
| PC 18:2/22:6 | −0.614 ** | 0.002 | −0.731 ** | 0.000 | 0.747 ** | 0.000 |
| PC 18:2/20:4 | −0.543 ** | 0.007 | −0.709 ** | 0.000 | 0.769 ** | 0.000 |
| LPC 20:0 | −0.587 ** | 0.003 | −0.729 ** | 0.000 | 0.785 ** | 0.000 |
| LPC 19:0 | −0.550 ** | 0.006 | −0.702 ** | 0.000 | 0.791 ** | 0.000 |
| PC 36:3 | −0.576 ** | 0.004 | −0.696 ** | 0.000 | 0.768 ** | 0.000 |
| PC 18:1/18:3 | −0.552 ** | 0.006 | −0.683 ** | 0.000 | 0.760 ** | 0.000 |
| PC 18:2/18:2 | −0.544 ** | 0.007 | −0.670 ** | 0.001 | 0.749 ** | 0.000 |
| PC 16:1/18:2 | −0.590 ** | 0.003 | −0.580 ** | 0.004 | 0.660 ** | 0.001 |
| LPC 24:1 | −0.491 * | 0.014 | −0.624 ** | 0.002 | 0.724 ** | 0.000 |
| PC 18:1/18:2 | −0.564 ** | 0.005 | −0.606 ** | 0.002 | 0.700 ** | 0.000 |
| PC 18:1/22:5 | −0.525 ** | 0.009 | −0.522 ** | 0.009 | 0.596 ** | 0.003 |
| LPC 22:0 | −0.329 | 0.078 | −0.652 ** | 0.001 | 0.713 ** | 0.000 |
| PC 17:0/18:2 | −0.559 ** | 0.005 | −0.578 ** | 0.004 | 0.656 ** | 0.001 |
| PC 17:0/18:1 | −0.551 ** | 0.006 | −0.606 ** | 0.002 | 0.623 ** | 0.002 |
| PC 35:3 | −0.490 * | 0.014 | −0.609 ** | 0.002 | 0.692 ** | 0.000 |
| PC 16:0/18:0 | −0.578 ** | 0.004 | −0.659 ** | 0.001 | 0.722 ** | 0.000 |
| PC 18:0/20:2 | −0.574 ** | 0.004 | −0.608 ** | 0.002 | 0.687 ** | 0.000 |
| LPC 26:0 | −0.712 ** | 0.000 | −0.684 ** | 0.000 | 0.720 ** | 0.000 |
| PC 14:0/18:3 | −0.515 * | 0.010 | −0.491 * | 0.014 | 0.520 ** | 0.009 |
| PC 18:0/18:1 | −0.650 ** | 0.001 | −0.623 ** | 0.002 | 0.649 ** | 0.001 |
| PC 19:0/18:2 | −0.537 ** | 0.007 | −0.582 ** | 0.004 | 0.691 ** | 0.000 |
| LPC 20:3 | −0.521 ** | 0.009 | −0.660 ** | 0.001 | 0.688 ** | 0.000 |
| LPC 15:0 | −0.535 ** | 0.008 | −0.604 ** | 0.002 | 0.651 ** | 0.001 |
| SUM LPC | −0.635 ** | 0.001 | −0.715 ** | 0.000 | 0.794 ** | 0.000 |
| SBP | NO | FMD | FID | |||||
|---|---|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| LPC 17:0 | −0.306 | 0.095 | 0.352 | 0.064 | 0.289 | 0.108 | 0.687 ** | 0.000 |
| LPC 18:0 | −0.281 | 0.115 | 0.515 * | 0.010 | 0.341 | 0.070 | 0.684 ** | 0.000 |
| LPC 18:0 | −0.289 | 0.108 | 0.525 ** | 0.009 | 0.355 | 0.062 | 0.692 ** | 0.000 |
| LPC 24:0 | −0.223 | 0.172 | 0.523 ** | 0.009 | 0.388 * | 0.046 | 0.722 ** | 0.000 |
| LPC 18:1 | −0.314 | 0.089 | 0.459 * | 0.021 | 0.254 | 0.140 | 0.691 ** | 0.000 |
| LPC 17:1 | −0.347 | 0.067 | 0.350 | 0.065 | 0.254 | 0.140 | 0.684 ** | 0.000 |
| LPC 20:2 | −0.334 | 0.075 | 0.351 | 0.065 | 0.381 * | 0.049 | 0.657 ** | 0.001 |
| LPC 18:2 | −0.332 | 0.077 | 0.397 * | 0.042 | 0.475 * | 0.017 | 0.659 ** | 0.001 |
| PC 18:2/22:6 | −0.357 | 0.061 | 0.355 | 0.062 | 0.233 | 0.161 | 0.707 ** | 0.000 |
| PC 18:2/20:4 | −0.367 | 0.056 | 0.309 | 0.093 | 0.359 | 0.060 | 0.656 ** | 0.001 |
| LPC 20:0 | −0.273 | 0.122 | 0.479 * | 0.016 | 0.419 * | 0.033 | 0.704 ** | 0.000 |
| LPC 19:0 | −0.360 | 0.059 | 0.378 * | 0.050 | 0.381 * | 0.049 | 0.711 ** | 0.000 |
| PC 36:3 | −0.421 * | 0.032 | 0.368 | 0.055 | 0.599 ** | 0.003 | 0.684 ** | 0.000 |
| PC 18:1/18:3 | −0.411 * | 0.036 | 0.350 | 0.065 | 0.602 ** | 0.002 | 0.650 ** | 0.001 |
| PC 18:2/18:2 | −0.400 * | 0.040 | 0.332 | 0.076 | 0.588 ** | 0.003 | 0.644 ** | 0.001 |
| PC 16:1/18:2 | −0.465 * | 0.019 | 0.314 | 0.088 | 0.259 | 0.135 | 0.707 ** | 0.000 |
| LPC 24:1 | −0.280 | 0.116 | 0.531 ** | 0.008 | 0.406 * | 0.038 | 0.695 ** | 0.000 |
| PC 18:1/18:2 | −0.371 | 0.053 | 0.353 | 0.064 | 0.531 ** | 0.008 | 0.662 ** | 0.001 |
| PC 18:1/22:5 | −0.368 | 0.055 | 0.281 | 0.115 | 0.081 | 0.367 | 0.622 ** | 0.002 |
| LPC 22:0 | −0.363 | 0.058 | 0.327 | 0.080 | 0.435 * | 0.028 | 0.588 ** | 0.003 |
| PC 17:0/18:2 | −0.376 | 0.051 | 0.215 | 0.181 | 0.413 * | 0.035 | 0.620 ** | 0.002 |
| PC 17:0/18:1 | −0.263 | 0.131 | 0.398 * | 0.041 | 0.120 | 0.307 | 0.670 ** | 0.001 |
| PC 35:3 | −0.414 * | 0.035 | 0.277 | 0.118 | 0.428 * | 0.030 | 0.636 ** | 0.001 |
| PC 16:0/18:0 | −0.272 | 0.123 | 0.571 ** | 0.004 | 0.292 | 0.106 | 0.689 ** | 0.000 |
| PC 18:0/20:2 | −0.245 | 0.149 | 0.343 | 0.069 | 0.462 * | 0.020 | 0.651 ** | 0.001 |
| LPC 26:0 | −0.329 | 0.078 | 0.575 ** | 0.004 | 0.243 | 0.151 | 0.751 ** | 0.000 |
| PC 14:0/18:3 | −0.389 * | 0.045 | 0.459 * | 0.021 | 0.514 * | 0.010 | 0.564 ** | 0.005 |
| PC 18:0/18:1 | −0.216 | 0.181 | 0.509 * | 0.011 | 0.314 | 0.089 | 0.693 ** | 0.000 |
| PC 19:0/18:2 | −0.302 | 0.098 | 0.394 * | 0.043 | 0.549 ** | 0.006 | 0.609 ** | 0.002 |
| LPC 20:3 | −0.216 | 0.180 | 0.287 | 0.110 | 0.290 | 0.107 | 0.577 ** | 0.004 |
| LPC 15:0 | −0.348 | 0.066 | 0.257 | 0.137 | 0.242 | 0.152 | 0.605 ** | 0.002 |
| SUM LPC | −0.395 * | 0.042 | 0.407 * | 0.037 | 0.528 ** | 0.008 | 0.733 ** | 0.000 |
| FPI | FPG | HOMA_IR | HDL-C | IL6 | CRP | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| LPC 17:0 | −0.406 * | 0.038 | 0.114 | 0.317 | −0.252 | 0.142 | 0.270 | 0.125 | −0.478 * | 0.017 | −0.520 ** | 0.009 |
| LPC 18:0 | −0.453 * | 0.022 | 0.023 | 0.462 | −0.310 | 0.092 | 0.216 | 0.181 | −0.490 * | 0.014 | −0.518 ** | 0.010 |
| LPC 18:0 | −0.458 * | 0.021 | 0.024 | 0.460 | −0.313 | 0.090 | 0.220 | 0.175 | −0.494 * | 0.013 | −0.523 ** | 0.009 |
| LPC 24:0 | −0.556 ** | 0.005 | −0.144 | 0.272 | −0.443 * | 0.025 | 0.270 | 0.125 | −561 ** | 0.005 | −0.507 * | 0.011 |
| LPC 18:1 | −0.398 * | 0.041 | 0.082 | 0.366 | −0.238 | 0.156 | 0.208 | 0.189 | −0.473 * | 0.017 | −0.512 * | 0.010 |
| LPC 17:1 | −0.396 * | 0.042 | 0.098 | 0.341 | −0.243 | 0.151 | 0.260 | 0.134 | −0.494 * | 0.013 | −0.494 * | 0.013 |
| LPC 20:2 | −0.455 * | 0.022 | 0.015 | 0.474 | −0.325 | 0.081 | 0.275 | 0.120 | −0.522 ** | 0.009 | −0.484 * | 0.015 |
| LPC 18:2 | −0.476 * | 0.017 | −0.020 | 0.466 | −0.351 | 0.065 | 0.383 * | 0.048 | −0.469 * | 0.019 | −0.558 ** | 0.005 |
| PC 18:2/22:6 | −0.525 ** | 0.009 | 0.058 | 0.404 | −0.357 | 0.061 | 0.321 | 0.084 | −0.379 * | 0.050 | −0.649 ** | 0.001 |
| PC 18:2/20:4 | −0.538 ** | 0.007 | −0.064 | 0.395 | −0.412 * | 0.036 | 0.253 | 0.141 | −0.523 ** | 0.009 | −0.533 ** | 0.008 |
| LPC 20:0 | −0.501 * | 0.012 | −0.029 | 0.451 | −0.362 | 0.058 | 0.188 | 0.213 | −0.539 ** | 0.007 | −0.568 ** | 0.004 |
| LPC 19:0 | −0.509 * | 0.011 | −0.011 | 0.482 | −0.369 | 0.055 | 0.305 | 0.095 | −0.604 ** | 0.002 | −0.504 * | 0.012 |
| PC 36:3 | −0.515 * | 0.010 | −0.060 | 0.401 | −0.402 * | 0.039 | 0.250 | 0.143 | −0.538 ** | 0.007 | −0.539 ** | 0.007 |
| PC 18:1/18:3 | −0.507 * | 0.011 | −0.093 | 0.348 | −0.406 * | 0.038 | 0.222 | 0.174 | −0.565 ** | 0.005 | −0.501 * | 0.012 |
| PC 18:2/18:2 | −0.505 * | 0.012 | −0.105 | 0.329 | −0.410 * | 0.036 | 0.204 | 0.194 | −0.572 ** | 0.004 | −0.489 * | 0.014 |
| PC 16:1/18:2 | −0.524 ** | 0.009 | −0.074 | 0.379 | −0.410 * | 0.036 | 0.411 * | 0.036 | −0.562 ** | 0.005 | −0.466 * | 0.019 |
| LPC 24:1 | −0.411 * | 0.036 | 0.016 | 0.473 | −0.263 | 0.131 | 0.207 | 0.190 | −0.635 ** | 0.001 | −0.398 * | 0.041 |
| PC 18:1/18:2 | −0.558 ** | 0.005 | −0.191 | 0.210 | −0.480 * | 0.016 | 0.296 | 0.102 | −0.625 ** | 0.002 | −0.472 * | 0.018 |
| PC 18:1/22:5 | −0.475 * | 0.017 | −0.041 | 0.433 | −0.350 | 0.065 | 0.625 ** | 0.002 | −0.540 ** | 0.007 | −0.459 * | 0.021 |
| LPC 22:0 | −0.205 | 0.193 | 0.276 | 0.120 | −0.004 | 0.493 | 0.017 | 0.471 | −0.669 ** | 0.001 | −0.339 | 0.072 |
| PC 17:0/18:2 | −0.514 * | 0.010 | −0.161 | 0.249 | −0.454 * | 0.022 | 0.354 | 0.063 | −0.513 * | 0.010 | −0.451 * | 0.023 |
| PC 17:0/18:1 | −0.429 * | 0.030 | −0.017 | 0.471 | −0.316 | 0.087 | 0.275 | 0.121 | −0.384 * | 0.047 | −0.483 * | 0.016 |
| PC 35:3 | −0.480 * | 0.016 | −0.108 | 0.325 | −0.397 * | 0.042 | 0.288 | 0.109 | −0.553 ** | 0.006 | −0.406 * | 0.038 |
| PC 16:0/18:0 | −0.576 ** | 0.004 | −0.122 | 0.304 | −0.460 * | 0.021 | 0.159 | 0.251 | −0.376 | 0.051 | −0.546 ** | 0.006 |
| PC 18:0/20:2 | −0.477 * | 0.017 | −0.118 | 0.310 | −0.421 * | 0.032 | 0.256 | 0.138 | −0.468 * | 0.019 | −0.438 * | 0.027 |
| LPC 26:0 | −0.497 * | 0.013 | −0.052 | 0.413 | −0.339 | 0.072 | 0.517 ** | 0.010 | −0.557 ** | 0.005 | −0.616 ** | 0.002 |
| PC 14:0/18:3 | −0.465 * | 0.019 | −0.115 | 0.315 | −0.367 | 0.056 | 0.230 | 0.165 | −0.387 * | 0.046 | −0.468 * | 0.019 |
| PC 18:0/18:1 | −0.501 * | 0.012 | −0.129 | 0.293 | −0.418 * | 0.033 | 0.231 | 0.163 | −0.404 * | 0.038 | −0.556 ** | 0.005 |
| PC 19:0/18:2 | −0.566 ** | 0.005 | −0.207 | 0.190 | −0.510 * | 0.011 | 0.187 | 0.215 | −0.487 * | 0.015 | −0.457 * | 0.021 |
| LPC 20:3 | −0.380 * | 0.049 | −0.007 | 0.489 | −0.290 | 0.108 | 0.175 | 0.230 | −0.396 * | 0.042 | −0.426 * | 0.031 |
| LPC 15:0 | −0.429 * | 0.029 | 0.041 | 0.431 | −0.318 | 0.086 | 0.386 * | 0.047 | −0.365 | 0.057 | −0.479 * | 0.016 |
| SUM LPC | −0.574 ** | 0.004 | −0.130 | 0.292 | −0.469 * | 0.019 | 0.301 | 0.098 | −0.609 ** | 0.002 | −0.552 ** | 0.006 |
| BMI | Fat % | Lean % | ||||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| Cer d37:0 | 0.410 * | 0.036 | 0.445 * | 0.025 | −0.516 ** | 0.010 |
| Cer d39:0 | 0.394 * | 0.043 | 0.564 ** | 0.005 | −0.639 ** | 0.001 |
| Cer d41:0 | 0.421 * | 0.032 | 0.563 ** | 0.005 | −0.628 ** | 0.002 |
| Cer d18:0/23:0 | 0.421 * | 0.032 | 0.563 ** | 0.005 | −0.628 ** | 0.002 |
| Cer d40:0 | 0.429 * | 0.030 | 0.613 ** | 0.002 | −0.668 ** | 0.001 |
| Cer d18:0/22:0 | 0.429 * | 0.030 | 0.613 ** | 0.002 | −0.668 ** | 0.001 |
| Cer t42:0 | 0.428 * | 0.030 | 0.606 ** | 0.002 | −0.662 ** | 0.001 |
| Cer d18:0/24:0 | 0.395 * | 0.042 | 0.600 ** | 0.003 | −0.652 ** | 0.001 |
| Cer d42:0 | 0.363 | 0.058 | 0.608 ** | 0.002 | −0.654 ** | 0.001 |
| Cer d18:0/18:0 | 0.410 * | 0.036 | 0.421 * | 0.032 | −0.497 * | 0.013 |
| Cer d38:0 | 0.385 * | 0.047 | 0.542 ** | 0.007 | −0.612 ** | 0.002 |
| Cer t40:0 | 0.370 | 0.054 | 0.537 ** | 0.007 | −0.612 ** | 0.002 |
| Cer t43:0 | 0.394 * | 0.043 | 0.540 ** | 0.007 | −0.605 ** | 0.002 |
| Cer d43:0 | 0.353 | 0.063 | 0.556 ** | 0.005 | −0.590 ** | 0.003 |
| Cer d18:1/25:0 | 0.498 * | 0.013 | 0.527 ** | 0.009 | −0.542 ** | 0.007 |
| Cer d19:1/24:0 | 0.502 * | 0.012 | 0.523 ** | 0.009 | −0.537 ** | 0.007 |
| Cer d18:2/24:0 | 0.475 * | 0.017 | 0.441 * | 0.026 | −0.496 * | 0.013 |
| SUM Cer | 0.273 | 0.123 | 0.427* | 0.030 | −0.470 * | 0.018 |
| SBP | FMD | FID | IL6 | |||||
|---|---|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| Cer d37:0 | 0.224 | 0.171 | −0.328 | 0.079 | −0.483 * | 0.016 | 0.483 * | 0.015 |
| Cer d39:0 | 0.386 * | 0.046 | −0.383 * | 0.048 | −0.593 ** | 0.003 | 0.730 ** | 0.000 |
| Cer d41:0 | 0.393 * | 0.043 | −0.334 | 0.075 | −0.595 ** | 0.003 | 0.670 ** | 0.001 |
| Cer d18:0/23:0 | 0.393 * | 0.043 | −0.334 | 0.075 | −0.595 ** | 0.003 | 0.670 ** | 0.001 |
| Cer d40:0 | 0.429 * | 0.030 | −0.362 | 0.059 | −0.641 ** | 0.001 | 0.768 ** | 0.000 |
| Cer d18:0/22:0 | 0.429 * | 0.030 | −0.362 | 0.059 | −0.641 ** | 0.001 | 0.768 ** | 0.000 |
| Cer t42:0 | 0.421 * | 0.032 | −0.365 | 0.057 | −0.636 ** | 0.001 | 0.767 ** | 0.000 |
| Cer d18:0/24:0 | 0.503 * | 0.012 | −0.348 | 0.066 | −0.623 ** | 0.002 | 0.771 ** | 0.000 |
| Cer d42:0 | 0.596 ** | 0.003 | −0.339 | 0.072 | −0.633 ** | 0.001 | 0.803 ** | 0.000 |
| Cer d18:0/18:0 | 0.204 | 0.194 | −0.320 | 0.084 | −0.458 * | 0.021 | 0.427 * | 0.030 |
| Cer d38:0 | 0.365 | 0.057 | −0.394 * | 0.043 | −0.561 ** | 0.005 | 0.711 ** | 0.000 |
| Cer t40:0 | 0.384 * | 0.047 | −0.404 * | 0.039 | −0.553 ** | 0.006 | 0.712 ** | 0.000 |
| Cer t43:0 | 0.382 * | 0.048 | −0.314 | 0.089 | −0.569 ** | 0.004 | 0.647 ** | 0.001 |
| Cer d43:0 | 0.485 * | 0.015 | −0.290 | 0.107 | −0.573 ** | 0.004 | 0.758 ** | 0.000 |
| Cer d18:1/25:0 | 0.443 * | 0.025 | −0.514 * | 0.010 | −0.516 ** | 0.010 | 0.314 | 0.089 |
| Cer d19:1/24:0 | 0.434 * | 0.028 | −0.507 * | 0.011 | −0.513 * | 0.010 | 0.309 | 0.093 |
| Cer d18:2/24:0 | 0.455 * | 0.022 | −0.560 ** | 0.005 | −0.533 ** | 0.008 | 0.339 | 0.072 |
| SUM Cer | 0.453 * | 0.022 | −0.294 | 0.104 | −0.470 * | 0.018 | 0.704 ** | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.; Mirza, I.; Morsy, M.; Mostafa, A.; Hassan, C.; Masrur, M.; Bianco, F.M.; Papasani, S.; Levitan, I.; Mahmoud, A.M. Comparison of Adiposomal Lipids between Obese and Non-Obese Individuals. Metabolites 2024, 14, 464. https://doi.org/10.3390/metabo14080464
Hussein M, Mirza I, Morsy M, Mostafa A, Hassan C, Masrur M, Bianco FM, Papasani S, Levitan I, Mahmoud AM. Comparison of Adiposomal Lipids between Obese and Non-Obese Individuals. Metabolites. 2024; 14(8):464. https://doi.org/10.3390/metabo14080464
Chicago/Turabian StyleHussein, Mohamed, Imaduddin Mirza, Mohammed Morsy, Amro Mostafa, Chandra Hassan, Mario Masrur, Francesco M. Bianco, Subbaiah Papasani, Irena Levitan, and Abeer M. Mahmoud. 2024. "Comparison of Adiposomal Lipids between Obese and Non-Obese Individuals" Metabolites 14, no. 8: 464. https://doi.org/10.3390/metabo14080464
APA StyleHussein, M., Mirza, I., Morsy, M., Mostafa, A., Hassan, C., Masrur, M., Bianco, F. M., Papasani, S., Levitan, I., & Mahmoud, A. M. (2024). Comparison of Adiposomal Lipids between Obese and Non-Obese Individuals. Metabolites, 14(8), 464. https://doi.org/10.3390/metabo14080464







