Metabolomics of Benzene Exposure and Development of Biomarkers for Exposure Hazard Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Structured Questionnaires and Sample Collection
2.3. Untargeted Metabolomics Based on LC-MS
2.4. Targeted Metabolomics
2.5. Constructing a Benzene Exposure Animal Model Using Benzene-Poisoned Mice
2.6. Statistical Analysis
3. Results
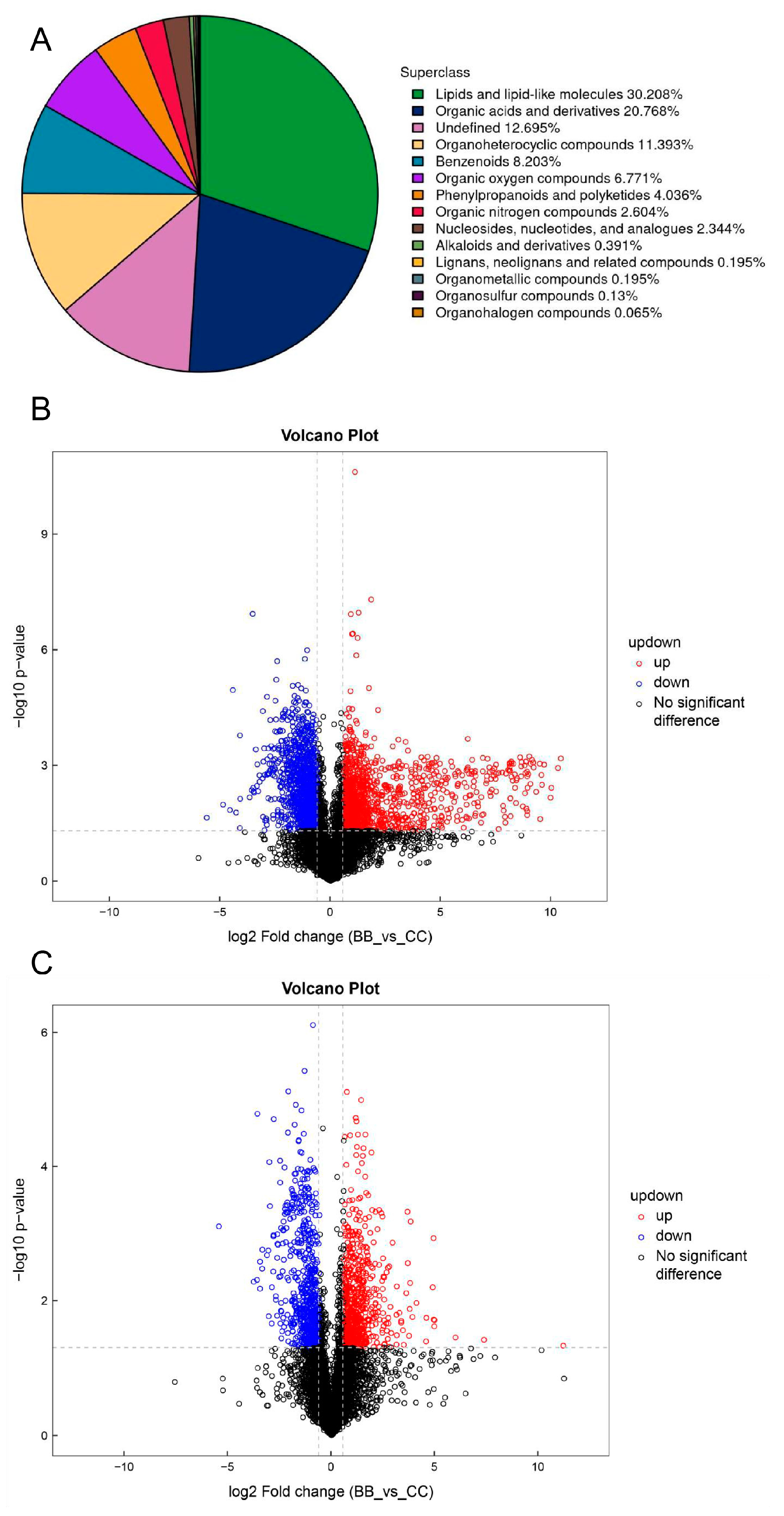
3.1. Analysis of Plasma Untargeted Metabolomics in Benzene-Exposed Workers
3.2. Plasma Differential Metabolite Targeting and Biomarker Analysis in Benzene-Exposed Workers
3.3. Changes in the Metabolic Profile of Benzene-Exposed Mice and the Toxic Effects of Benzene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McHale, C.M.; Zhang, L.; Smith, M.T. Current understanding of the mechanism of benzene-induced leukemia in humans: Implications for risk assessment. Carcinogenesis 2012, 33, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Paustenbach, D. Shanghai Health Study (2001–2009): What was learned about benzene health effects? Crit. Rev. Toxicol. 2018, 48, 217–251. [Google Scholar] [CrossRef]
- Gut, I.; Nedelcheva, V.; Souček, P.; Stopka, P.; Tichavská, B. Cytochromes P450 in benzene metabolism and involvement of their metabolites and reactive oxygen species in toxicity. Environ. Health Perspect. 1996, 104, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Louise, M. Winn, Homologous Recombination Initiated by Benzene Metabolites: A Potential Role of Oxidative Stress. Toxicol. Sci. 2003, 72, 143–149. [Google Scholar] [CrossRef]
- Cordiano, R.; Papa, V.; Cicero, N.; Spatari, G.; Allegra, A.; Gangemi, S. Effects of Benzene: Hematological and Hypersensitivity Manifestations in Resident Living in Oil Refinery Areas. Toxics 2022, 10, 678. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Hirabayashi, Y.; Choi, Y.; Kodama, Y.; Kanno, J.; Han, J.; Inoue, T.; Yoon, B. Benzene activates caspase-4 and -12 at the transcription level, without an association with apoptosis, in mouse bone marrow cells lacking the p53 gene. Arch. Toxicol. 2009, 83, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A. The role of DNA repair in benzene-induced carcinogenesis. Chem.-Biol. Interact. 2010, 184, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Wang, J.; Zhang, W.; Ren, J.; Chen, Y.; Zhang, Y.; Gao, A. Plasma metabolomics study reveals the critical metabolic signatures for benzene-induced hematotoxicity. JCI Insight 2022, 7, e154999. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Umashankar, S.; Swarup, S. Plant metabolomics: From experimental design to knowledge extraction. Methods Mol. Biol. 2013, 1069, 279–312. [Google Scholar] [CrossRef]
- Smolinska, A.; Blanchet, L.; Buydens, L.; Wijmenga, S. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef]
- O’Connell, G.C.; Alder, M.; Smothers, C.; Still, C.; Webel, A.; Moore, S. Use of high-sensitivity digital ELISA improves the diagnostic performance of circulating brain-specific proteins for detection of traumatic brain injury during triage. Neurol. Res. 2020, 42, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Deng, T.; Yuan, W.; Deng, H.; Jin, M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Li, L.; Tang, S.; Liu, C.; Fu, X.; Shi, Z.; Mao, H. Metabolomics Reveals that Crossbred Dairy Buffaloes are more Thermotolerant than Holstein cows under Chronic Heat Stress. J. Agric. Food Chem. 2018, 66, 12889–12897. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, R.; Kumar, P.; Sharma, U. Metabolomics techniques: A brief update. Epigenetics Metabolomics 2021, 28, 1–29. [Google Scholar] [CrossRef]
- Goldansaz, S.; Guo, A.; Sajed, T.; Steele, M.; Plastow, G.; Wishart, D. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS ONE 2017, 12, e0177675. [Google Scholar] [CrossRef] [PubMed]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; Angelis, M.; Kronenberg, F.; Meitinger, T.; Mewes, H.; Wichmann, H.; Weinberger, K.; Adamski, J.; et al. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef]
- James, E.; Parkinson, E. Serum metabolomics in animal models and human disease. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 478–483. [Google Scholar] [CrossRef]
- Allwood, J.; Parker, D.; Beckmann, M.; Draper, J.; Goodacre, R. Fourier Transform Ion Cyclotron Resonance mass spectrometry for plant metabolite profiling and metabolite identification. Methods Mol. Biol. 2012, 860, 157–176. [Google Scholar] [CrossRef]
- Liu, M.; Liu, H.; Mei, F.; Yang, N.; Zhao, D.; Ai, G.; Xiang, H.; Zheng, Y. Identification of the Biosynthetic Pathway of Glycine Betaine That Is Responsible for Salinity Tolerance in Halophilic Thioalkalivibrio versutus D301. Front. Microbiol. 2022, 13, 875843. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Rajendiran, T.; Ateeq, B.; Asangani, I.; Athanikar, J.; Yocum, A.; Mehra, R.; Siddiqui, J.; Palapattu, G.; Wei, J.; et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia 2013, 15, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, X.; Ren, J.; Chen, Y.; Wang, J.; Gao, A. Glycine/glycine N-methyltransferase/sarcosine axis mediates benzene-induced hematotoxicity. Toxicol. Appl. Pharmacol. 2021, 428, 115682. [Google Scholar] [CrossRef] [PubMed]
- Kline, P.; Schramm, V. Purine nucleoside phosphorylase. Inosine hydrolysis, tight binding of the hypoxanthine intermediate, and third-the-sites reactivity. Biochemistry 1992, 31, 5964–5973. [Google Scholar] [CrossRef]
- Cohen, A.; Gudas, L.; Ammann, A.; Staal, G.; Martin, D. Deoxyguanosine triphosphate as a possible toxic metabolite in the immunodeficiency associated with purine nucleoside phosphorylase deficiency. J. Clin. Investig. 1978, 61, 1405–1409. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Grunebaum, E.; Arpaia, E.; Roifman, C. Immunodeficiency Caused by Purine Nucleoside Phosphorylase Deficiency. Radiol. Clin. N. Am. 2000, 20, 143–159. [Google Scholar] [CrossRef]
- Rothman, N.; Vermeulen, R.; Zhang, L.; Hu, W.; Yin, S.; Rappaport, S.M.; Smith, M.T.; Jones, D.P.; Rahman, M.; Lan, Q.; et al. Metabolome-wide association study of occupational exposure to benzene. Carcinogenesis 2021, 42, 1326–1336. [Google Scholar] [CrossRef]
- Sun, Y.; Rose, J.; Wang, B.; Hsiao, C. The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: Comparisons with other amino acid binding proteins. J. Mol. Biol. 1998, 278, 219–229. [Google Scholar] [CrossRef]
- Peng, C.T.; Wu, K.H.; Lan, S.J.; Tsai, J.J.; Tsai, F.J.; Tsai, C.H. Amino acid concentrations in cerebrospinal fluid in children with acute lymphoblastic leukemia undergoing chemotherapy. Eur. J. Cancer 2005, 41, 1158–1163. [Google Scholar] [CrossRef]
- Tsuruoka, M.; Hara, J.; Hirayama, A.; Sugimoto, M.; Soga, T.; Shankle, W.R.; Tomita, M. Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis 2013, 34, 2865–2872. [Google Scholar] [CrossRef]
- Wang, Z.; He, Z.; Emara, A.; Gan, X.; Li, H. Effects of malondialdehyde as a byproduct of lipid oxidation on protein oxidation in rabbit meat. Food Chem. 2019, 288, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Całyniuk, B.; Grochowska-Niedworok, E.; Walkiewicz, K.W.; Kawecka, S.; Popiołek, E.; Fatyga, E. Malondialdehyde (MDA)—Product of lipid peroxidation as marker of homeostasis disorders and aging. Ann. Acad. Medicae Silesiensis 2016, 70, 224–228. [Google Scholar] [CrossRef]
- Cao, G.; Song, Z.; Hong, Y.; Yang, Z.; Song, Y.; Chen, Z.; Chen, Z.; Cai, Z. Large-scale targeted metabolomics method for metabolite profiling of human samples. Anal. Chim. Acta 2020, 1125, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Singh, Z.; Karthigesu, I.; Singh, P.; Kaur, R. Use of Malondialdehyde as a Biomarker for Assessing Oxidative Stress in Different Disease Pathologies: A Review. Iran. J. Public Health 2014, 43, 7–16. [Google Scholar]
- Amin, M.; Rafiei, N.; Poursafa, P.; Ebrahimpour, K.; Mozafarian, N.; Shoshtari-Yeganeh, B.; Hashemi, M.; Kelishadi, R. Association of benzene exposure with insulin resistance, SOD, and MDA as markers of oxidative stress in children and adolescents. Environ. Sci. Pollut. Res. 2018, 25, 34046–34052. [Google Scholar] [CrossRef]
- Yu, K.; Yang, K.Y.; Ren, X.Z.; Chen, Y.; Liu, X.H. Amifostine Protects Bone Marrow from Benzene-Induced Hematotoxicity in Mice. Int. J. Toxicol. 2007, 26, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Z.; Park, S.; Reagan, W.J.; Goldstein, R.; Zhong, S.; Lawton, M.; Rajamohan, F.; Qian, K.; Liu, L.; Gong, D.W. Alanine aminotransferase isoenzymes: Molecular cloning and quantitative analysis of tissue expression in rats and serum elevation in liver toxicity. Hepatology 2009, 49, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Jiang, Y.; Pan, F.; Cai, Q. Lipid metabolism after autologous orthotopic liver transplantation in rat models of fatty liver. J. Clin. Rehabil. Tissue Eng. Res. 2018, 22, 5821–5827. [Google Scholar]
- Leung, L.; Kalgutkar, A.; Obach, S. Metabolic activation in drug-induced liver injury. Drug Metab. Rev. 2012, 44, 18–33. [Google Scholar] [CrossRef]
- Abd El-Shakour, A.; El-Ebiarie, A.S.; Ibrahim, Y.H.; Moneim, A.E.; El-Mekawy, A.M. Effect of benzene on oxidative stress and the functions of liver and kidney in rats. J. Environ. Occup. Health 2015, 4, 34–39. [Google Scholar]
- Curi, R.; Newsholme, P.; Procopio, J.; Lagranha, C.; Gorjão, R.; Pithon-Curi, T.C. Glutamine, gene expression, and cell function. Front. Biosci. 2007, 12, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Jacque, N.; Ronchetti, A.M.; Larrue, C.; Meunier, G.; Birsen, R.; Willems, L.; Saland, E.; Decroocq, J.; Maciel, T.T.; Lambert, M.; et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015, 126, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.; Jacque, N.; Jacquel, A.; Neveux, N.; Trovati Maciel, T.; Lambert, M.; Schmitt, A.; Poulain, L.; Green, A.S.; Uzunov, M.; et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 2013, 122, 3521–3532. [Google Scholar] [CrossRef] [PubMed]






| Characteristics | Control | Benzene Exposure | p-Value |
|---|---|---|---|
| Sex | 0.700 | ||
| Male | 14 | 13 | |
| Female | 4 | 5 | |
| Age | 32.06 ± 2.35 | 34.50 ± 2.29 | 0.214 |
| Educational level | 0.054 | ||
| High school/Junior college | 2 | 7 | |
| College/University | 16 | 11 | |
| Smoking | 0.480 | ||
| Yes | 11 | 13 | |
| No | 7 | 5 | |
| Drinks alcohol | 0.457 | ||
| Yes | 12 | 12 | |
| No | 6 | 6 | |
| Length of service | 9.56 ± 2.55 | 13.67 ± 2.46 | 0.037 * |
| Benzene exposure concentration | 0.03 ± 0.003 | 1.66 ± 0.130 | <0.000 * |
| Total | 18 | 18 |
| Characteristics | Control | Benzene Exposure | p-Value |
|---|---|---|---|
| Sex | 0.969 | ||
| Male | 138 | 102 | |
| Female | 20 | 16 | |
| Age | 36.35 ± 10.68 | 34.57 ± 10.58 | 0.170 |
| Educational level | 0.168 | ||
| High school/Junior College | 33 | 34 | |
| College/University | 125 | 84 | |
| Smoking | 0.028 * | ||
| Yes | 67 | 34 | |
| No | 91 | 84 | |
| Drinks alcohol | 0.569 | ||
| Yes | 46 | 12 | |
| No | 112 | 6 | |
| Length of service | 14.03 ± 11.93 | 13.95 ± 11.58 | 0.954 |
| Benzene exposure Concentration | 0.03 ± 0.01 | 1.37 ± 0.63 | <0.000 * |
| Total | 158 | 118 |
| Poisoning Time | Indicator | Control | Benzene 150 mg (kg·d) |
|---|---|---|---|
| 15 days | White blood cells (109/L) | 5.07 ± 0.93 | 1.17 ± 0.47 * |
| Red blood cells (1012/L) | 9.65 ± 0.35 | 8.65 ± 0.37 | |
| Hemoglobin (g/L) | 154.67 ± 9.24 | 141.33 ± 6.66 | |
| Platelets (109/L) | 1094.67 ± 59.72 | 1133.33 ± 130.09 * | |
| 60 days | White blood cells (109/L) | 6.30 ± 0.78 | 4.20 ± 0.87 * |
| Red blood cells (1012/L) | 9.21 ± 0.41 | 7.80 ± 0.49 * | |
| Hemoglobin (g/L) | 140.33 ± 5.13 | 127.00 ± 7.21 | |
| Platelets (109/L) | 914.67 ± 104.74 | 607.33 ± 234.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Sun, Q.; Li, F.; Wang, B.; Zhu, B. Metabolomics of Benzene Exposure and Development of Biomarkers for Exposure Hazard Assessment. Metabolites 2024, 14, 377. https://doi.org/10.3390/metabo14070377
Li H, Sun Q, Li F, Wang B, Zhu B. Metabolomics of Benzene Exposure and Development of Biomarkers for Exposure Hazard Assessment. Metabolites. 2024; 14(7):377. https://doi.org/10.3390/metabo14070377
Chicago/Turabian StyleLi, Hao, Qianyu Sun, Fei Li, Boshen Wang, and Baoli Zhu. 2024. "Metabolomics of Benzene Exposure and Development of Biomarkers for Exposure Hazard Assessment" Metabolites 14, no. 7: 377. https://doi.org/10.3390/metabo14070377
APA StyleLi, H., Sun, Q., Li, F., Wang, B., & Zhu, B. (2024). Metabolomics of Benzene Exposure and Development of Biomarkers for Exposure Hazard Assessment. Metabolites, 14(7), 377. https://doi.org/10.3390/metabo14070377






