Optimising Extracellular Vesicle Metabolomic Methodology for Prostate Cancer Biomarker Discovery
Abstract
1. Introduction
2. Methods and Experimental Design
2.1. Preparation for lEVs Isolation from Cell Culture Medium (CCM)
2.2. Nanoparticle Tracking Analysis (NTA)
2.3. Transmission Electron Microscopy (TEM)
2.4. Western Blot (WB)
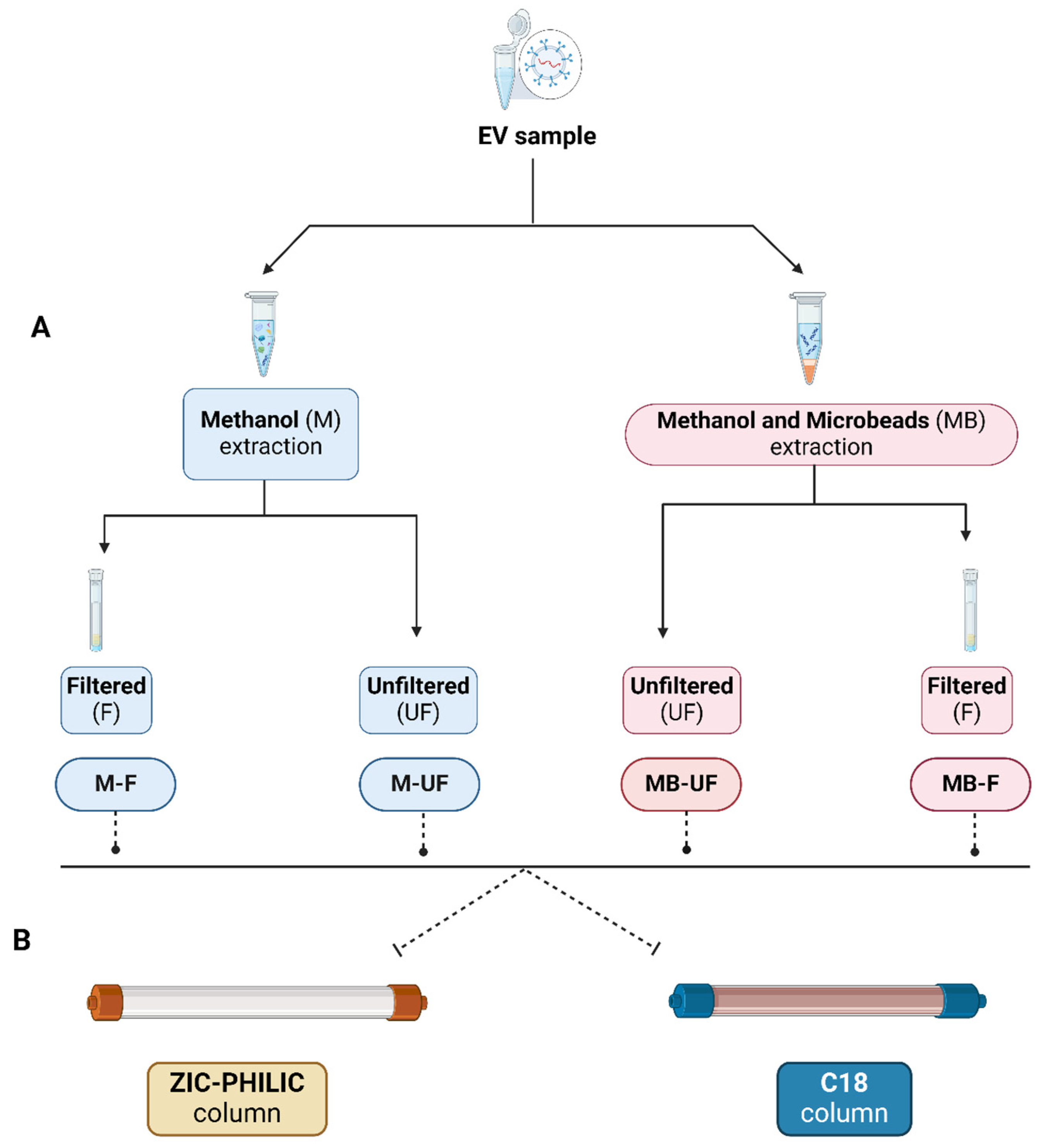
2.5. Metabolite Extraction
- 1
- M-UF approach:
- 2
- MB-UF approach:
- 3
- M-F approach:
- 4
- MB-F approach:
2.6. Global Metabolomics: Using ZIC-pHILIC and C18 Chromatography
2.7. Statistical Analysis
3. Results and Discussion
3.1. Confirmation and Characterisation of EVs Isolated from PC3 Cell Line
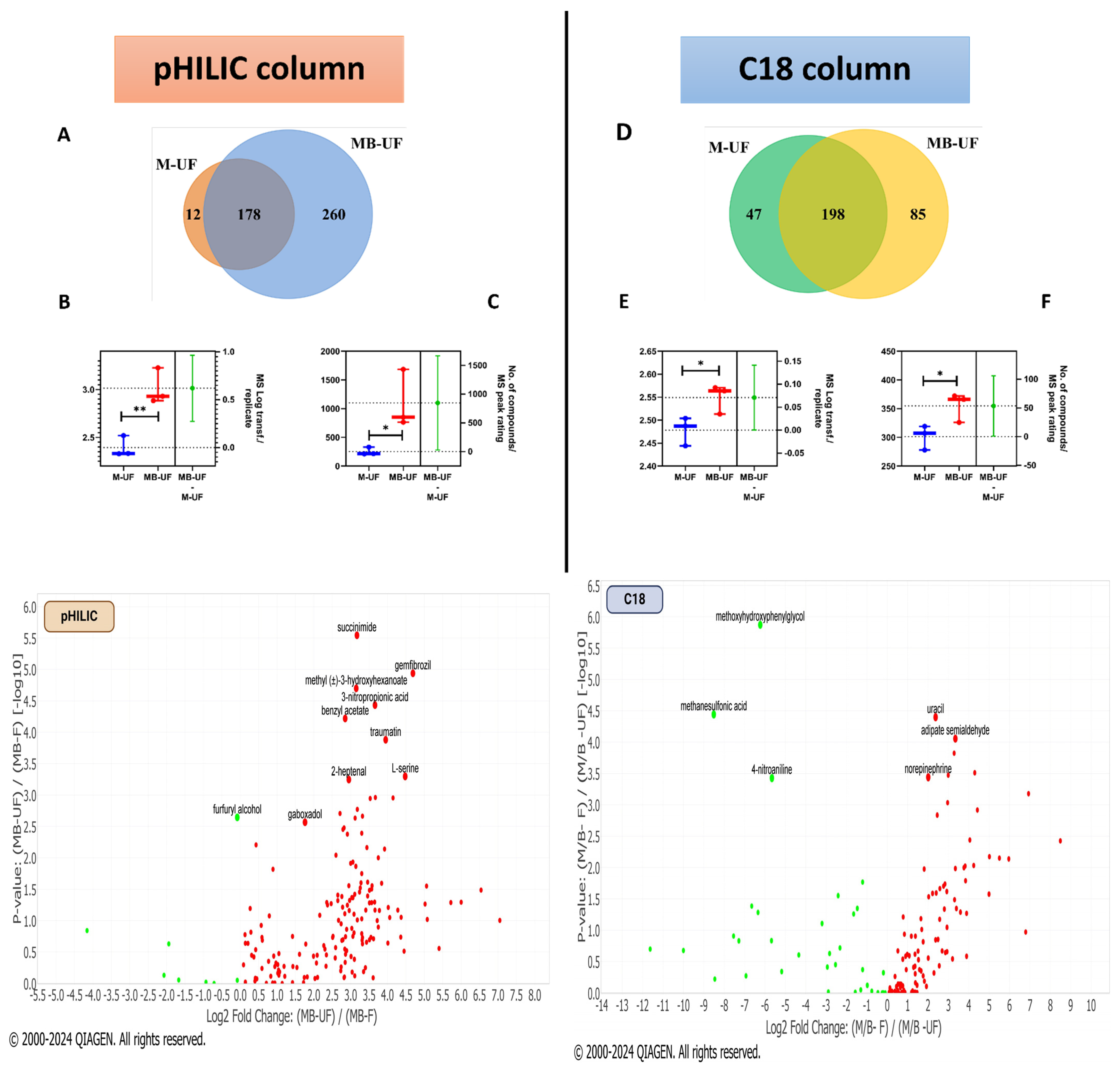
3.2. Analysing PC3 Cell-Derived lEVs Metabolites: Four Extraction Methods Compared Using pHILIC and C18
3.3. Identifying Key Metabolites: Comparison of pHILIC and C18 Chromatography
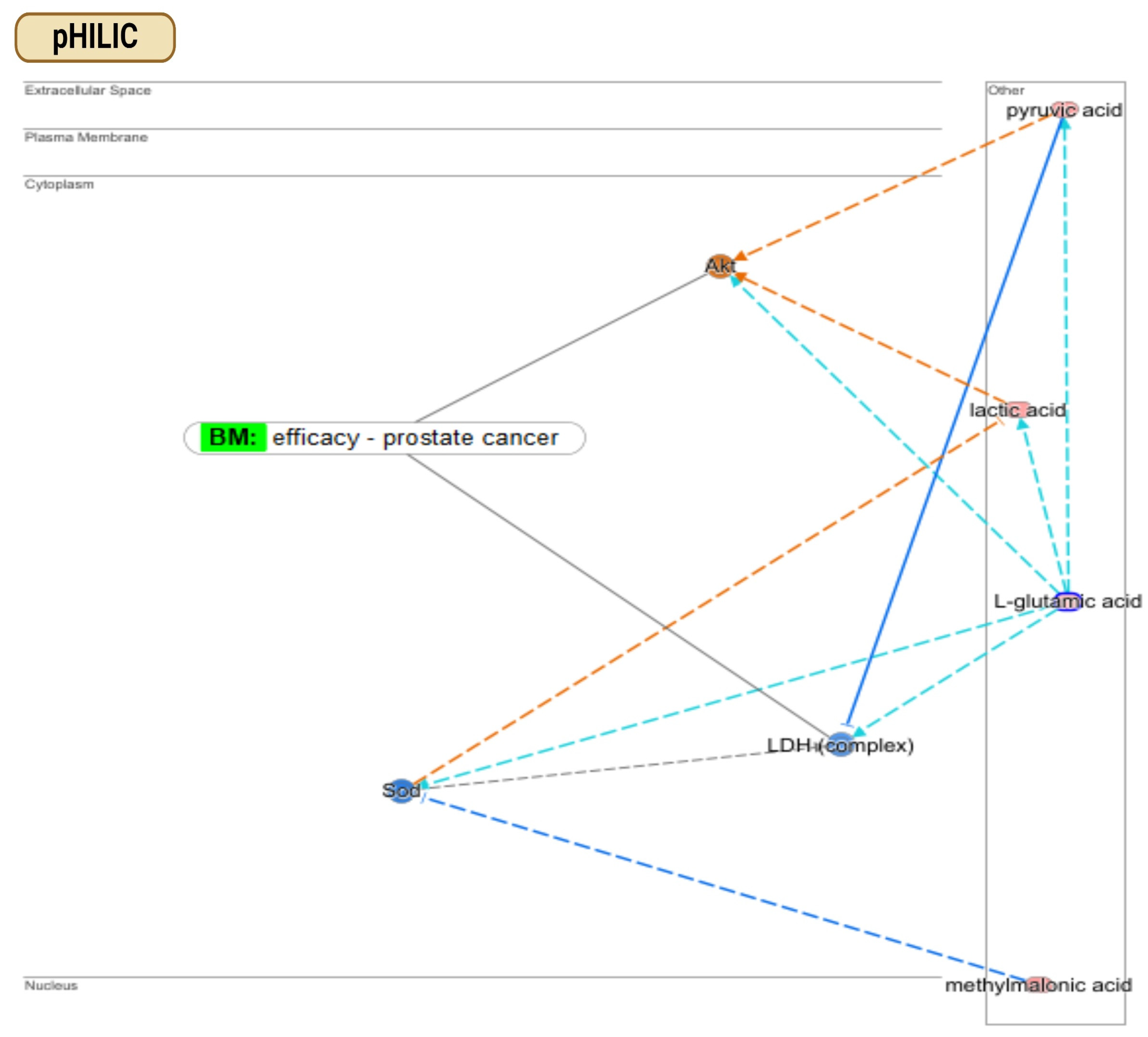
3.4. Finding Metabolic Pathways in PCa: Comparison of pHILIC and C18 Chromatography
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers That Differentiate Benign Prostatic Hyperplasia from Prostate Cancer: A Literature Review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef] [PubMed]
- De Visschere, P.; Oosterlinck, W.; De Meerleer, G.; Villeirs, G. Clinical and imaging tools in the early diagnosis of prostate cancer, a review. J. Belg. Soc. Radiol. 2010, 93, 62. [Google Scholar] [CrossRef] [PubMed]
- Andriole, G.L.; Crawford, E.D.; Grubb, R.L., III; Buys, S.S.; Chia, D.; Church, T.R.; Fouad, M.N.; Isaacs, C.; Kvale, P.A.; Reding, D.J.; et al. Prostate Cancer Screening in the Randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: Mortality Results after 13 Years of Follow-up. JNCI J. Natl. Cancer Inst. 2012, 104, 125–132. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Zijlstra, A.; Rubin, M.A.; Di Vizio, D. Extracellular vesicles for liquid biopsy in prostate cancer: Where are we and where are we headed? Prostate Cancer Prostatic Dis. 2017, 20, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Oey, O.; Ghaffari, M.; Li, J.J.; Hosseini-Beheshti, E. Application of extracellular vesicles in the diagnosis and treatment of prostate cancer: Implications for clinical practice. Crit. Rev. Oncol. Hematol. 2021, 167, 103495. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.L.; Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer 2004, 4, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Trock, B.J. Application of metabolomics to prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Loizaga-Iriarte, A.; Zuñiga-Garcia, P.; Sánchez-Mosquera, P.; Rosa Cortazar, A.; González, E.; Torrano, V.; Alonso, C.; Pérez-Cormenzana, M.; Ugalde-Olano, A.; et al. Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J. Extracell. Vesicles 2018, 7, 1470442. [Google Scholar] [CrossRef]
- Vallabhaneni, K.C.; Penfornis, P.; Dhule, S.; Guillonneau, F.; Adams, K.V.; Mo, Y.Y.; Xu, R.; Liu, Y.; Watabe, K.; Vemuri, M.C.; et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 2015, 6, 4953–4967. [Google Scholar] [CrossRef]
- Luo, X.; An, M.; Cuneo, K.C.; Lubman, D.M.; Li, L. High-Performance Chemical Isotope Labeling Liquid Chromatography Mass Spectrometry for Exosome Metabolomics. Anal. Chem. 2018, 90, 8314–8319. [Google Scholar] [CrossRef]
- Hamed, M.A.; Wasinger, V.; Wang, Q.; Graham, P.; Malouf, D.; Bucci, J.; Li, Y. Prostate cancer-derived extracellular vesicles metabolic biomarkers: Emerging roles for diagnosis and prognosis. J. Control. Release 2024, 371, 126–145. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Puhka, M.; Takatalo, M.; Nordberg, M.E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T.; et al. Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics 2017, 7, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T.; Campoy, I.; Lanau, L.; Gill, K.; Rigau, M.; Gil-Moreno, A.; Reventos, J.; Byers, S.; Colas, E.; Cheema, A.K. Enabling Metabolomics Based Biomarker Discovery Studies Using Molecular Phenotyping of Exosome-Like Vesicles. PLoS ONE 2016, 11, e0151339. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Beheshti, E.; Pham, S.; Adomat, H.; Li, N.; Tomlinson Guns, E. Exosomes as Biomarker Enriched Microvesicles: Characterization of Exosomal Proteins Derived from a Panel of Prostate Cell Lines with Distinct AR Phenotypes. Mol. Cell. Proteom. MCP 2012, 11, 863–885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and valuable tools towards diagnosis, prognosis and treatment of COVID-19 and other diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef]
- Waddell, O.; Frizelle, F.A.; Keenan, J.I. The role of biomarkers to increase the detection of early-onset colorectal cancer. Med. Res. Arch. 2023, 11. [Google Scholar] [CrossRef]
- Mejía-García, T.A.; Portugal, C.C.; Encarnação, T.G.; Prado, M.A.; Paes-de-Carvalho, R. Nitric oxide regulates AKT phosphorylation and nuclear translocation in cultured retinal cells. Cell Signal 2013, 25, 2424–2439. [Google Scholar] [CrossRef]
- A Pharmacodynamic Study of Pre-Prostatectomy Rapamycin in Men with Advanced Localized Prostate Cancer. Available online: https://clinicaltrials.gov/ (accessed on 13 April 2024).
- A Phase II Study of the Efficacy and Safety of AP23573 in Patients with Taxane-Resistant Androgen-Independent Prostate Cancer (AIPC). Available online: https://clinicaltrials.gov/ (accessed on 14 April 2024).
- Amin, N.; Pearce, B. Glutamate toxicity in neuron-enriched and neuron-astrocyte co-cultures: Effect of the glutamate uptake inhibitor L-trans-pyrrolidine-2,4-dicarboxylate. Neurochem. Int. 1997, 30, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Bu, K.; An, H.; Tao, N. A retrospective study: Analysis of the relationship between lactate dehydrogenase and castration-resistant prostate cancer based on restricted cubic spline model. PeerJ 2023, 11, e16158. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Eisenberger, M.A.; Halabi, S.; Oudard, S.; Nanus, D.M.; Petrylak, D.P.; Sartor, A.O.; Scher, H.I. Biomarkers in the management and treatment of men with metastatic castration-resistant prostate cancer. Eur. Urol. 2012, 61, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saari, H.; Kärkkäinen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J. Extracell. Vesicles 2019, 8, 1596669. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Li, X.; Gong, S.; Ge, X.; Zhu, T.; Ge, X.; Weng, L.; Tao, Q.; Guo, J. Dual Roles of Lactate in EGFR-TKI-Resistant Lung Cancer by Targeting GPR81 and MCT1. J. Oncol. 2022, 2022, 3425841. [Google Scholar] [CrossRef]
- Ryou, M.G.; Liu, R.; Ren, M.; Sun, J.; Mallet, R.T.; Yang, S.H. Pyruvate protects the brain against ischemia-reperfusion injury by activating the erythropoietin signaling pathway. Stroke 2012, 43, 1101–1107. [Google Scholar] [CrossRef]
- Xu, X.Y.; Nie, Y.; Wang, F.F.; Bai, Y.; Lv, Z.Z.; Zhang, Y.Y.; Li, Z.J.; Gao, W. Growth differentiation factor (GDF)-15 blocks norepinephrine-induced myocardial hypertrophy via a novel pathway involving inhibition of epidermal growth factor receptor transactivation. J. Biol. Chem. 2014, 289, 10084–10094. [Google Scholar] [CrossRef]
- Zhuang, H.; Jiang, W.; Zhang, X.; Qiu, F.; Gan, Z.; Cheng, W.; Zhang, J.; Guan, S.; Tang, B.; Huang, Q.; et al. Suppression of HSP70 expression sensitizes NSCLC cell lines to TRAIL-induced apoptosis by upregulating DR4 and DR5 and downregulating c-FLIP-L expressions. J. Mol. Med. 2012, 91, 219–235. [Google Scholar] [CrossRef]
- Wang, J.; Xu, L.F.; Liu, C.; Huang, T.; Liang, C.Z.; Fan, Y.D. Identifying the role of apolipoprotein A-I in prostate cancer. Asian J. Androl. 2021, 23, 400–408. [Google Scholar] [CrossRef]
- An Open Label Phase II Study of Oral Treatment with Sunitinib (SUTENT) in Patients Suffering from Hormone Refractory Prostate Cancer after Progression with Docetaxel Based Regimen. Available online: https://clinicaltrials.gov/ (accessed on 15 April 2024).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamed, M.A.; Wasinger, V.; Wang, Q.; Biazik, J.; Graham, P.; Malouf, D.; Bucci, J.; Li, Y. Optimising Extracellular Vesicle Metabolomic Methodology for Prostate Cancer Biomarker Discovery. Metabolites 2024, 14, 367. https://doi.org/10.3390/metabo14070367
Hamed MA, Wasinger V, Wang Q, Biazik J, Graham P, Malouf D, Bucci J, Li Y. Optimising Extracellular Vesicle Metabolomic Methodology for Prostate Cancer Biomarker Discovery. Metabolites. 2024; 14(7):367. https://doi.org/10.3390/metabo14070367
Chicago/Turabian StyleHamed, Mahmoud Assem, Valerie Wasinger, Qi Wang, Joanna Biazik, Peter Graham, David Malouf, Joseph Bucci, and Yong Li. 2024. "Optimising Extracellular Vesicle Metabolomic Methodology for Prostate Cancer Biomarker Discovery" Metabolites 14, no. 7: 367. https://doi.org/10.3390/metabo14070367
APA StyleHamed, M. A., Wasinger, V., Wang, Q., Biazik, J., Graham, P., Malouf, D., Bucci, J., & Li, Y. (2024). Optimising Extracellular Vesicle Metabolomic Methodology for Prostate Cancer Biomarker Discovery. Metabolites, 14(7), 367. https://doi.org/10.3390/metabo14070367






