Identification and Characterization of Cannabichromene’s Major Metabolite Following Incubation with Human Liver Microsomes
Abstract
1. Introduction

2. Materials and Methods
2.1. Enzymes, Reagents, and Chemicals
2.2. Generation of CBC Metabolites with Pooled Human Liver Microsomes
2.3. Derivatization of CBC Metabolite and GC-MS/MS Analysis
2.4. Chemical Synthesis of the Major Metabolite of CBC
2.5. Identification of the Major Metabolite Structure via NMR Analysis
2.6. Molecular Docking
2.7. Competitive Binding Assays
2.8. Data Analysis
3. Results
3.1. Metabolism of CBC by Human Liver Microsomes
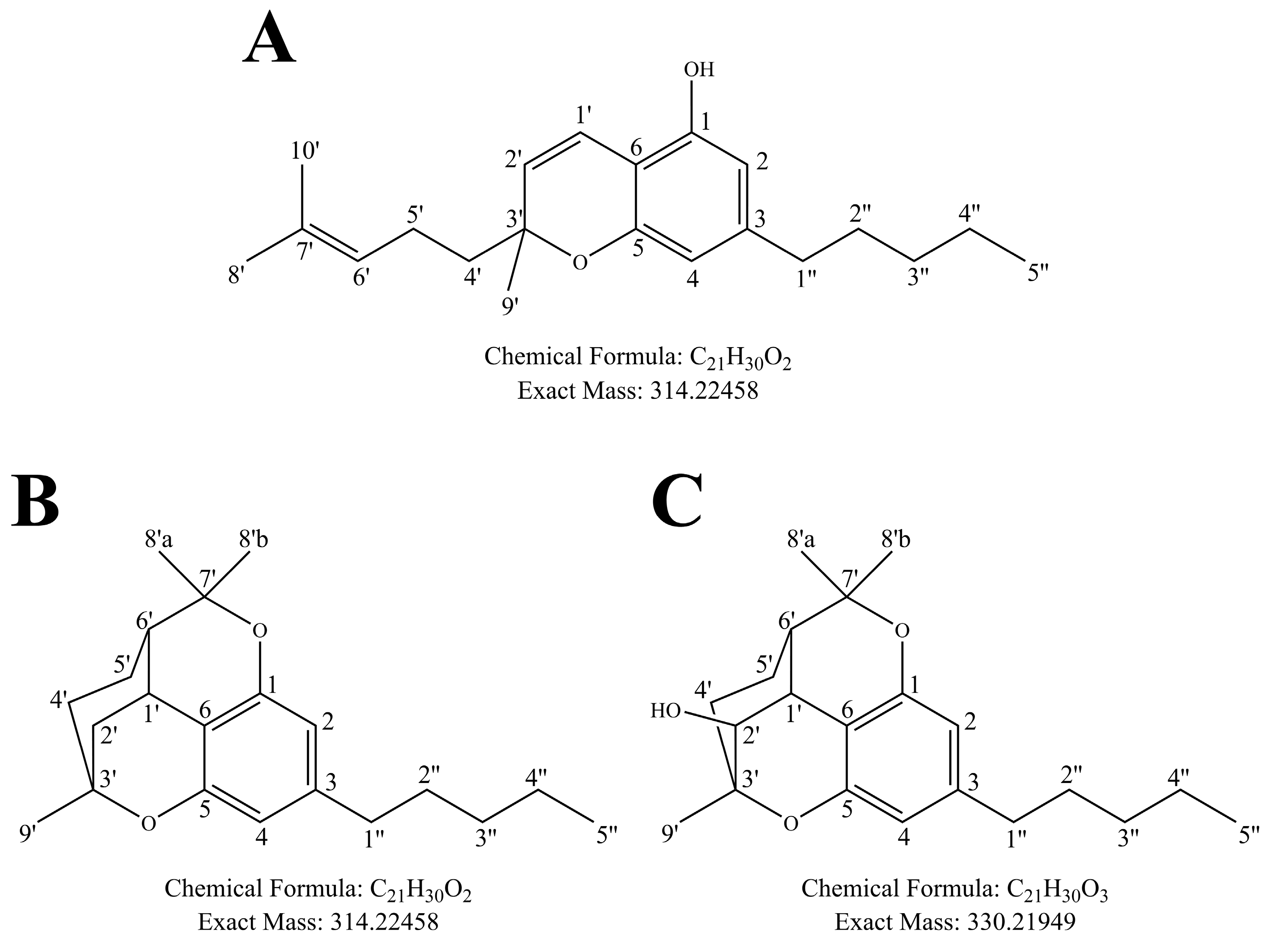
3.2. Identification of the Structure of the Major CBC Metabolite Generated by Human Liver Microsomes Using GC-MS/MS
3.3. Structural Identification of 2′-Hydroxycannabicitran via NMR
3.4. Molecular Docking Prediction of CB1 and CB2 Receptor Binding
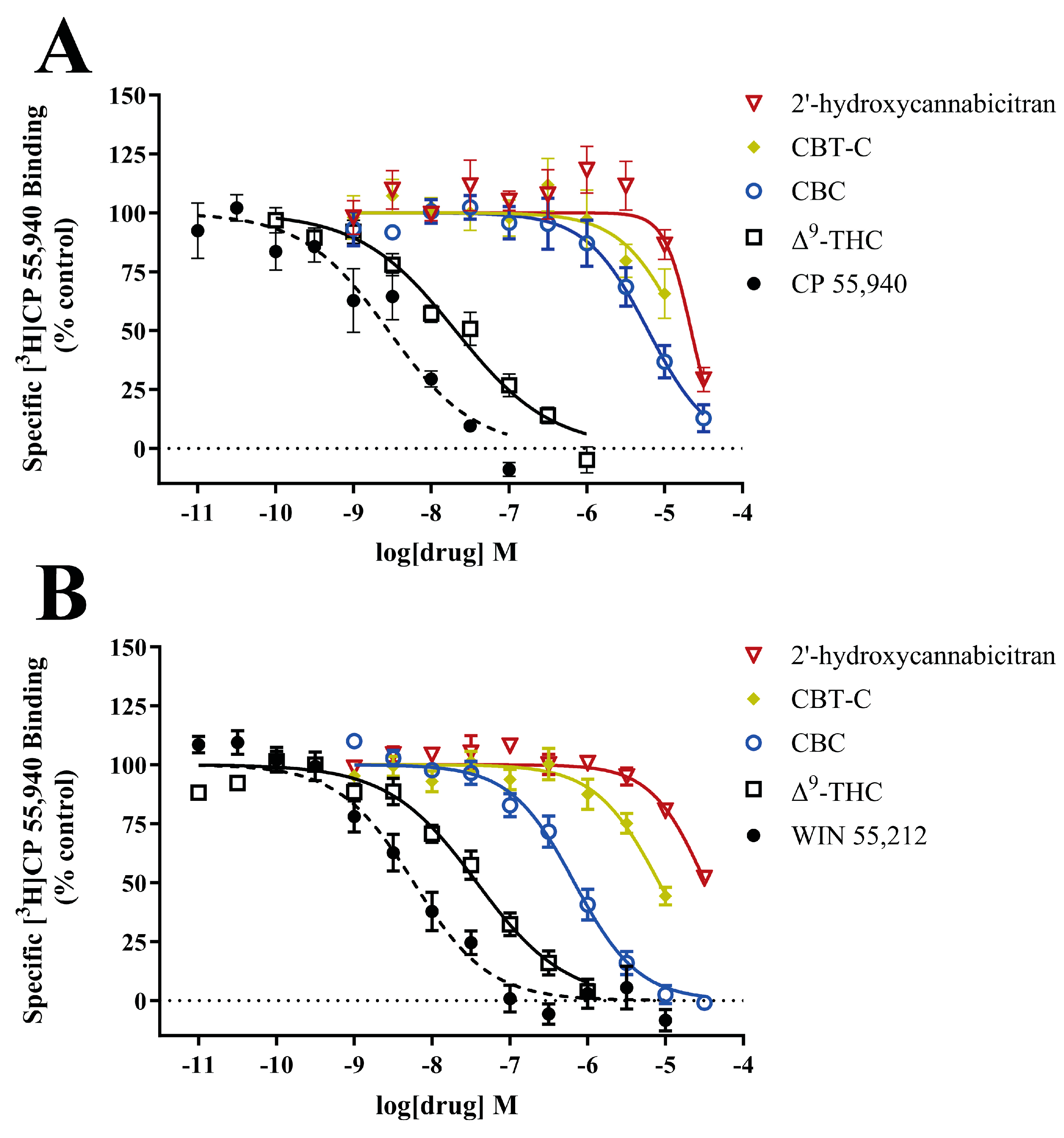
3.5. In Vitro Interactions at the CB1 and CB2 Receptors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pertwee, R.G. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br. J. Pharmacol. 2008, 153, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.; Di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: Beyond CB(1) and CB(2). Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef] [PubMed]
- Britch, S.C.; Babalonis, S.; Walsh, S.L. Cannabidiol: Pharmacology and therapeutic targets. Psychopharmacology 2021, 238, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, S.; Srebro, D.; Vujovic, K.S.; Vucetic, C.; Prostran, M. Cannabinoids and Pain: New Insights From Old Molecules. Front. Pharmacol. 2018, 9, 1259. [Google Scholar] [CrossRef] [PubMed]
- Zendulka, O.; Dovrtělová, G.; Nosková, K.; Turjap, M.; Šulcová, A.; Hanuš, L.; Juřica, J. Cannabinoids and Cytochrome P450 Interactions. Curr. Drug Metab. 2016, 17, 206–226. [Google Scholar] [CrossRef] [PubMed]
- Huestis, M.A. Pharmacokinetics and Metabolism of the Plant Cannabinoids, Δ9-Tetrahydrocannabinol, Cannabidiol and Cannabinol. Handb. Exp. Pharmacol. 2005, 168, 657–690. [Google Scholar] [CrossRef]
- EPIDIOLEX (Cannabidiol) Oral Solution [Package Insert]; Greenwich Biosciences, Inc.: Carlsbad, CA, USA, 2018. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf (accessed on 14 November 2023).
- Beers, J.L.; Fu, D.; Jackson, K.D. Cytochrome P450-Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol. Drug Metab. Dispos. 2021, 49, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Sempio, C.; Almaraz-Quinones, N.; Jackson, M.; Zhao, W.; Wang, G.S.; Liu, Y.; Leehey, M.; Knupp, K.; Klawitter, J.; Christians, U.; et al. Simultaneous Quantification of 17 Cannabinoids by LC-MS-MS in Human Plasma. J. Anal. Toxicol. 2022, 46, 383–392. [Google Scholar] [CrossRef] [PubMed]
- ElSohly, M.A.; Mehmedic, Z.; Foster, S.; Gon, C.; Chandra, S.; Church, J.C. Changes in Cannabis Potency Over the Last 2 Decades (1995-2014): Analysis of Current Data in the United States. Biol. Psychiatry 2016, 79, 613–619. [Google Scholar] [CrossRef]
- Swift, W.; Wong, A.; Li, K.M.; Arnold, J.C.; McGregor, I.S. Analysis of cannabis seizures in NSW, Australia: Cannabis potency and cannabinoid profile. PLoS ONE 2013, 8, e70052. [Google Scholar] [CrossRef]
- Potter, D.J.; Clark, P.; Brown, M.B. Potency of delta 9-THC and other cannabinoids in cannabis in England in 2005: Implications for psychoactivity and pharmacology. J. Forensic Sci. 2008, 53, 90–94. [Google Scholar] [CrossRef]
- Mehmedic, Z.; Chandra, S.; Slade, D.; Denham, H.; Foster, S.; Patel, A.S.; Ross, S.A.; Khan, I.A.; ElSohly, M.A. Potency trends of Delta9-THC and other cannabinoids in confiscated cannabis preparations from 1993 to 2008. J. Forensic Sci. 2010, 55, 1209–1217. [Google Scholar] [CrossRef]
- Agricultural Improvement Act of 2018. H.R.2—115th Congress (2017–2018) ed.; United States of America. 2018. Available online: https://www.congress.gov/bill/115th-congress/house-bill/2 (accessed on 20 October 2023).
- U.S. Minor Cannabinoids Market Size, Share & Trends Analysis Report by Product (Cannabigerol (CBG), Tetrahydrocannabivarin (THCV), Cannabichromene (CBC)), By Application (Cancer, Inflammation, Neurological Disorders), and Segment Forecasts, 2024–2030; Grand View Research: San Francisco, CA, USA, 2023; pp. 1–80. Available online: https://www.grandviewresearch.com/industry-analysis/us-minor-cannabinoids-market-report# (accessed on 16 February 2024).
- Udoh, M.; Santiago, M.; Devenish, S.; McGregor, I.S.; Connor, M. Cannabichromene is a cannabinoid CB2 receptor agonist. Br. J. Pharmacol. 2019, 176, 4537–4547. [Google Scholar] [CrossRef]
- Zagzoog, A.; Mohamed, K.A.; Kim, H.J.J.; Kim, E.D.; Frank, C.S.; Black, T.; Jadhav, P.D.; Holbrook, L.A.; Laprairie, R.B. In vitro and in vivo pharmacological activity of minor cannabinoids isolated from Cannabis sativa. Sci. Rep. 2020, 10, 20405. [Google Scholar] [CrossRef]
- Turner, C.E.; ElSohly, M.A.; Boeren, E.G. Constituents of Cannabis sativa L. XVII. A Review of the Natural Constituents. J. Nat. Prod. 1980, 43, 169–234. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Shani, A.; Edery, H.; Grunfeld, Y. Chemical Basis of Hashish Activity. Science 1970, 169, 611–612. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Vellani, V.; Schiano-Moriello, A.; Marini, P.; Magherini, P.C.; Orlando, P.; Di Marzo, V. Plant-derived cannabinoids modulate the activity of transient receptor potential channels of ankyrin type-1 and melastatin type-8. J. Pharmacol. Exp. Ther. 2008, 325, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- De Petrocellis, L.; Ligresti, A.; Moriello, A.S.; Allara, M.; Bisogno, T.; Petrosino, S.; Stott, C.G.; Di Marzo, V. Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br. J. Pharmacol. 2011, 163, 1479–1494. [Google Scholar] [CrossRef] [PubMed]
- Romano, B.; Borrelli, F.; Fasolino, I.; Capasso, R.; Piscitelli, F.; Cascio, M.; Pertwee, R.; Coppola, D.; Vassallo, L.; Orlando, P.; et al. The cannabinoid TRPA1 agonist cannabichromene inhibits nitric oxide production in macrophages and ameliorates murine colitis. Br. J. Pharmacol. 2013, 169, 213–229. [Google Scholar] [CrossRef]
- Dadiotis, E.; Cui, M.; Gerasi, M.; Mitsis, V.; Melliou, E.; Makriyannis, A.; Logothetis, D.E.; Magiatis, P. A Simple Chiral (1)H NMR Method for the Discrimination of (R)- and (S)-Cannabichromene in Complex Natural Mixtures and Their Effects on TRPA1 Activity. J. Nat. Prod. 2023, 87, 77–84. [Google Scholar] [CrossRef]
- Izzo, A.A.; Capasso, R.; Aviello, G.; Borrelli, F.; Romano, B.; Piscitelli, F.; Gallo, L.; Capasso, F.; Orlando, P.; Di Marzo, V. Inhibitory effect of cannabichromene, a major non-psychotropic cannabinoid extracted from Cannabis sativa, on inflammation-induced hypermotility in mice. Br. J. Pharmacol. 2012, 166, 1444–1460. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi, H.; Salles, E.L.; Shin, E.; Jarrahi, A.; Costigliola, V.; Kumar, P.; Yu, J.C.; Morgan, J.C.; Hess, D.C.; Vaibhav, K.; et al. A potential role for cannabichromene in modulating TRP channels during acute respiratory distress syndrome. J. Cannabis Res. 2021, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- DeLong, G.T.; Wolf, C.E.; Poklis, A.; Lichtman, A.H. Pharmacological evaluation of the natural constituent of Cannabis sativa, cannabichromene and its modulation by Delta(9)-tetrahydrocannabinol. Drug Alcohol. Depend. 2010, 112, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Kim, J.H.; Han, J.H.; Ryu, B.R.; Lim, Y.S.; Lim, J.D.; Park, S.H.; Kim, C.H.; Lee, S.U.; Kwon, T.H. In Vitro and In Vivo Anti-Inflammatory Potential of Cannabichromene Isolated from Hemp. Plants 2023, 12, 3966. [Google Scholar] [CrossRef] [PubMed]
- Anis, O.; Vinayaka, A.C.; Shalev, N.; Namdar, D.; Nadarajan, S.; Anil, S.M.; Cohen, O.; Belausov, E.; Ramon, J.; Mayzlish Gati, E.; et al. Cannabis-Derived Compounds Cannabichromene and Delta9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization. Molecules 2021, 26, 465. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, A.; Moriello, A.S.; Starowicz, K.; Matias, I.; Pisanti, S.; De Petrocellis, L.; Laezza, C.; Portella, G.; Bifulco, M.; Di Marzo, V. Antitumor activity of plant cannabinoids with emphasis on the effect of cannabidiol on human breast carcinoma. J. Pharmacol. Exp. Ther. 2006, 318, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Rosenthaler, S.; Pohn, B.; Kolmanz, C.; Huu, C.N.; Krewenka, C.; Huber, A.; Kranner, B.; Rausch, W.D.; Moldzio, R. Differences in receptor binding affinity of several phytocannabinoids do not explain their effects on neural cell cultures. Neurotoxicol. Teratol. 2014, 46, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Whynot, E.G.; Tomko, A.M.; Dupre, D.J. Anticancer properties of cannabidiol and Delta(9)-tetrahydrocannabinol and synergistic effects with gemcitabine and cisplatin in bladder cancer cell lines. J. Cannabis Res. 2023, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J.; Brown, N.K. A Method for the Structural Determination of Cannabichromene Metabolites by Mass Spectrometry. Rapid Commun. Mass Spectrom. 1990, 4, 135–136. [Google Scholar] [CrossRef]
- Harvey, D.J.; Brown, N.K. Identification of Cannabichromene Metabolites by Mass Spectrometry: Identification of Eight New Dihydroxy Metabolites in the Rabbit. Biol. Mass Spectrom. 1991, 20, 275–285. [Google Scholar] [CrossRef]
- Harvey, D.J.; Brown, N.K. Comparative In Vitro Metabolism of the Cannabinoids. Pharmacol. Biochem. Behav. 1991, 40, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Maturano, J.; Hasdemir, H.; Lopez, A.; Xu, F.; Hellman, J.; Tajkhorshid, E.; Sarlah, D.; Das, A. Elucidating the Mechanism of Metabolism of Cannabichromene by Human Cytochrome P450s. J. Nat. Prod. 2024, 87, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Hanus, L.O.; Meyer, S.M.; Munoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [PubMed]
- Elsohly, M.A.; Slade, D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, J.; Westland, J. Quantitation of Cannabinoids in Hemp Flower by Derivatization GC/MS. Available online: https://www.agilent.com/cs/library/applications/application-hemp-cannabis-thc-gcms-5994-2757en-agilent.pdf (accessed on 11 November 2023).
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.; Vemuri, K.; Nikas, S.P.; Laprairie, R.B.; Wu, Y.; Qu, L.; Pu, M.; Korde, A.; Jiang, S.; Ho, J.H.; et al. Crystal structures of agonist-bound human cannabinoid receptor CB(1). Nature 2017, 547, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal Structure of the Human Cannabinoid Receptor CB2. Cell 2019, 176, 459–467.e413. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Torralva, R.; Eshleman, A.J.; Swanson, T.L.; Schmachtenberg, J.L.; Schutzer, W.E.; Bloom, S.H.; Wolfrum, K.M.; Reed, J.F.; Janowsky, A. Fentanyl but not Morphine Interacts with Nonopioid Recombinant Human Neurotransmitter Receptors and Transporters. J. Pharmacol. Exp. Ther. 2020, 374, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuki, T.; Friesen, J.B.; Chen, S.; McAlpine, J.B.; Pauli, G.F. Selective Preparation and High Dynamic-Range Analysis of Cannabinoids in “CBD Oil” and Other Cannabis sativa Preparations. J. Nat. Prod. 2022, 85, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.S.; Gordon, W.H.; Morgan, J.B.; Williamson, R.T. Calculated and experimental (1) H and (13) C NMR assignments for cannabicitran. Magn. Reson. Chem. 2022, 60, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Agua, A.R.; Barr, P.J.; Marlowe, C.K.; Pirrung, M.C. Cannabichromene Racemization and Absolute Stereochemistry Based on a Cannabicyclol Analog. J. Org. Chem. 2021, 86, 8036–8040. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, J.M.; Umstead, W.J. Chiral Separation of Cannabichromene, Cannabicyclol, and Their Acidic Analogs on Polysaccharide Chiral Stationary Phases. Molecules 2023, 28, 1164. [Google Scholar] [CrossRef] [PubMed]
- Calcaterra, A.; Cianfoni, G.; Tortora, C.; Manetto, S.; Grassi, G.; Botta, B.; Gasparrini, F.; Mazzoccanti, G.; Appendino, G. Natural Cannabichromene (CBC) Shows Distinct Scalemicity Grades and Enantiomeric Dominance in Cannabis sativa Strains. J. Nat. Prod. 2023, 86, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, F.; Grandi, V.; Banerjee, A.; Trant, J.F. Cannabinoids and Cannabinoid Receptors: The Story so Far. iScience 2020, 23, 101301. [Google Scholar] [CrossRef]
- Pertwee, R.G. Ligands that target cannabinoid receptors in the brain: From THC to anandamide and beyond. Addict. Biol. 2008, 13, 147–159. [Google Scholar] [CrossRef]
- Havlasek, J.; Vrba, J.; Zatloukalova, M.; Papouskova, B.; Modriansky, M.; Storch, J.; Vacek, J. Hepatic biotransformation of non-psychotropic phytocannabinoids and activity screening on cytochromes P450 and UDP-glucuronosyltransferases. Toxicol. Appl. Pharmacol. 2023, 476, 116654. [Google Scholar] [CrossRef]
- Shokati, T.; Drake, S.H.; Zhao, W.; Klawitter, J.; Klawitter, J.; Christians, U. Structural Identification of Zotarolimus (ABT-578) Metabolites Generated by Human Liver Microsomes Using Ion-Trap and High-Resolution Time-of-Flight Mass Spectrometry in Combination with the Analysis of Fragmentation Patterns. Metabolites 2023, 13, 1093. [Google Scholar] [CrossRef]
- Shokati, T.; Hartmann, M.; Davari, B.; Klawitter, J.; Klawitter, J.; Christians, U. Temsirolimus metabolic pathways revisited. Xenobiotica 2020, 50, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Dadiotis, E.; Mitsis, V.; Melliou, E.; Magiatis, P. Direct Quantitation of Phytocannabinoids by One-Dimensional (1)H qNMR and Two-Dimensional (1)H-(1)H COSY qNMR in Complex Natural Mixtures. Molecules 2022, 27, 2965. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J.; Brown, N.K. In vitro Metabolism of Cannabigerol in Several Mammalian Species. Biomed. Environ. Mass. Spectrom. 1990, 19, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Dennis, D.G.; Eschbach, M.D.; Anand, S.D.; Xu, F.; Maturano, J.; Hellman, J.; Sarlah, D.; Das, A. Metabolites of Cannabigerol Generated by Human Cytochrome P450s Are Bioactive. Biochemistry 2022, 61, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.; Vemuri, K.; Pu, M.; Qu, L.; Han, G.W.; Wu, Y.; Zhao, S.; Shui, W.; Li, S.; Korde, A.; et al. Crystal Structure of the Human Cannabinoid Receptor CB(1). Cell 2016, 167, 750–762.e714. [Google Scholar] [CrossRef]
- Jakowiecki, J.; Filipek, S. Hydrophobic Ligand Entry and Exit Pathways of the CB1 Cannabinoid Receptor. J. Chem. Inf. Model. 2016, 56, 2457–2466. [Google Scholar] [CrossRef]

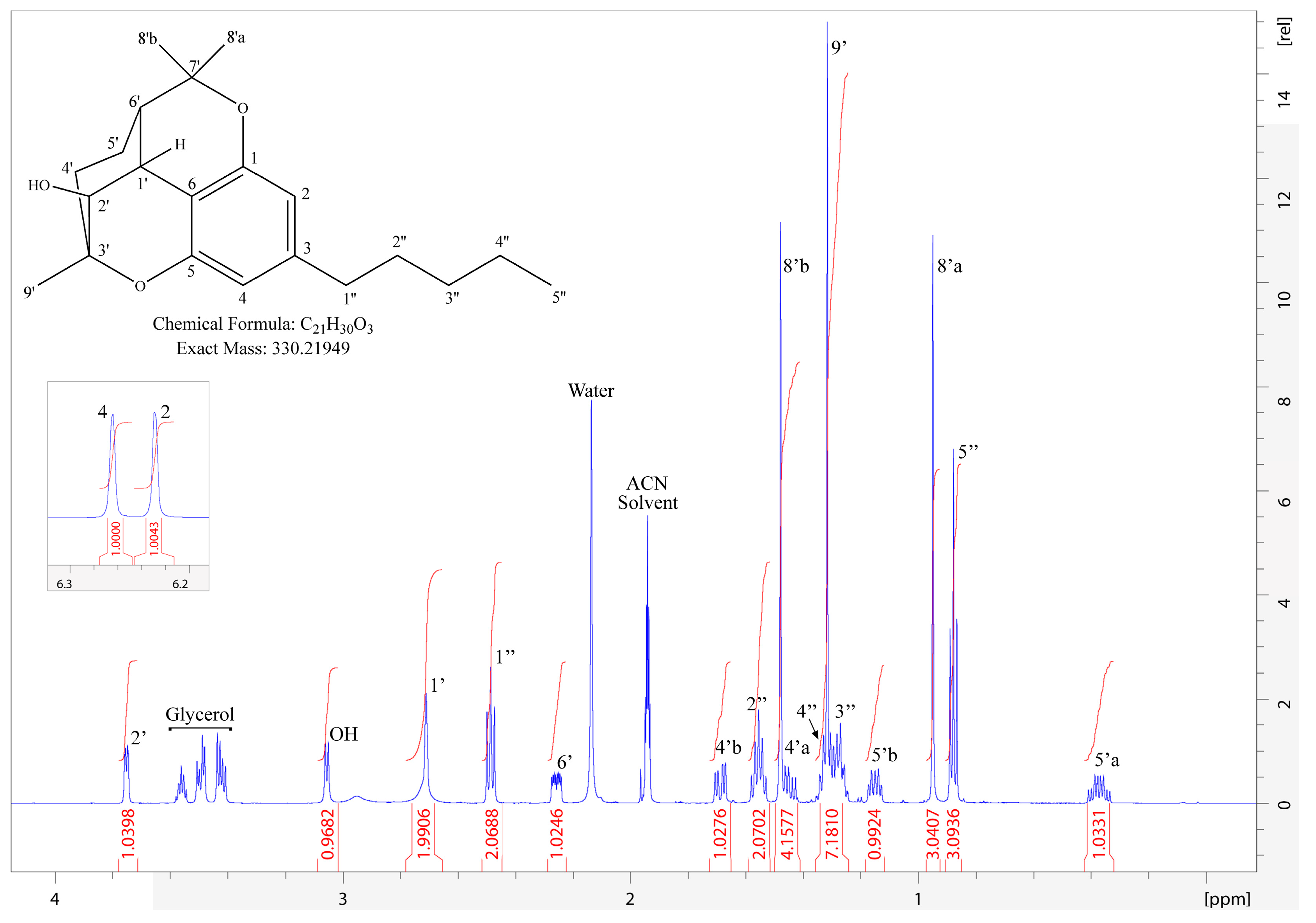
| 1H Chemical Shift (δ ppm) | 1H Multiplicity J (Hz) | Assignment | Integration | 13C Chemical Shift (δ ppm) | HMBC Correlations |
|---|---|---|---|---|---|
| 6.814 | s | OH | 1H | - | 5, 6 |
| 6.580 | dd (0.6, 10.1) | 1′ | 1H | 116.919 | 1, 5, 6, 2′, 3′, 4′, 9′ |
| 6.176 | s | 4 | 1H | 107.543 | 1, 2, 5, 6, 1′, 1″ |
| 6.130 | m | 2 | 1H | 107.962 | 1, 4, 5, 6, 1′, 1″ |
| 5.520 | d (10.0) | 2′ | 1H | 126.980 | 1, 6, 3′, 4′, 9′ |
| 5.104 | tq (1.4, 7.3) | 6′ | 1H | 124.150 | 4′, 5′, 8′, 10′ |
| 2.424 | dd (7.0, 7.8) | 1″ | 2H | 35.443 | 2, 3, 4, 2″ |
| 2.068 | q (8.0) | 5′ | 2H | 22.381 | 3′, 4′, 6′, 7′ |
| 1.641 | s | 8′ | 5H | 24.787 | 6′, 7′, 10′ |
| 1.625 | m | 4′ | 40.543 | 2′, 3′, 4′, 9′ | |
| 1.555 | s | 10′ | 5H | 16.658 | 6′, 7′, 8′ |
| 1.544 | m | 2″ | 30.620 | 3, 1″, 3″, 4″ | |
| 1.329 | m | 4″ | 7H | 22.232 | 3″, 5″ |
| 1.319 | s | 9′ | 25.452 | 1′, 2′, 3′, 4′ | |
| 1.278 | m | 3″ | 31.205 | 1″, 2″, 4″, 5″ | |
| 0.887 | t (7.0) | 5″ | 3H | 13.340 | 3″, 4″ |
| - | 1 | - | 153.947 | ||
| - | 3 | - | 144.776 | ||
| - | 5 | - | 152.295 | ||
| - | 6 | - | 106.905 | ||
| - | 3′ | - | 77.815 | ||
| - | 7′ | - | 131.386 |
| 1H Chemical Shift (δ ppm) | 1H Multiplicity J (Hz) | Assignment | Integration | 13C Chemical Shift (δ ppm) | 13C Multiplicity | HMBC Correlations |
|---|---|---|---|---|---|---|
| 6.266 | s | 4 | 1H | 109.261 | CH | 2, 5, 6, 1″ |
| 6.228 | s | 2 | 1H | 110.699 | CH | 1, 4, 6, 1″ |
| 3.749 | dd (1.9, 5.8) | 2′ | 1H | 71.161 | CH | 6, 1′, 3′, 6′ |
| 3.060 | d (6.1) | OH | 1H | - | - | 1′, 2′, 3′ |
| 2.719 | t (2.3) | 1′ | 1H* | 37.036 | CH | 1, 5, 6, 2′, 3′, 5′, 6′, 7′ |
| 2.485 | dd (7.4) | 1″ | 2H | 36.583 | CH2 | 2, 3, 4, 2″ |
| 2.258 | ddd (2.8, 5.3, 11.5) | 6′ | 1H | 48.335 | CH | 6, 1′, 2′, 8′b |
| 1.690 | ddd (0.9, 6.1, 15.4) | 4′b | 1H | 37.398 | CH2 | 2′, 3′, 5′, 6′, 7′ |
| 1.554 | m | 2″ | 2H | 31.918 | CH2 | 3, 1″, 3″, 4″ |
| 1.478 | s | 8′b | 4H | 29.767 | CH3 | 1, 6′, 7′, 8′a |
| 1.455 | td (7.1, 15.3) | 4′a | 37.398 | CH2 | 5′, 9′ | |
| 1.321 | m | 4″ | 7H | 23.166 | CH2 | 3″, 5″ |
| 1.315 | s | 9′ | 24.796 | CH3 | 5, 2′, 3′, 4′, 5′ | |
| 1.270 | m | 3″ | 32.144 | CH2 | 1″, 2″, 4″, 5″ | |
| 1.152 | dt (5.9, 12.8) | 5′b | 1H | 22.418 | CH2 | 1′, 3′, 6′ |
| 0.948 | s | 8′a | 3H | 23.936 | CH3 | 2′, 5′, 6′, 7′, 8′b |
| 0.880 | t (7.1) | 5″ | 3H | 14.300 | CH3 | 3″, 4″ |
| 0.373 | tdd (6.2, 11.9, 13.4) | 5′a | 1H | 22.418 | CH2 | 4′, 6′, 7′ |
| - | 3′ | - | 78.238 | C | ||
| - | 7′ | - | 84.205 | C | ||
| - | 6 | - | 113.632 | C | ||
| - | 3 | - | 143.421 | C | ||
| - | 5 | - | 156.996 | C | ||
| - | 1 | - | 158.480 | C |
| Computer Ranking | Ligand | Docking Score | Highlighted Residue-Ligand Interactions | Distance of Interaction (Å) | Type of Interaction |
|---|---|---|---|---|---|
| 1 | (+)-CBC | −10.600 | Ser505-OH | 1.69 | H-bond |
| Phe170-AR | 3.70 | π–π stacking | |||
| Phe170-P | 3.70 | π–π stacking | |||
| Phe268-AR | 3.78 | π–π stacking | |||
| Phe268-P | 3.78 | π-π stacking | |||
| 2 | (−)-Δ9-THC | −10.569 | Ser505-OH | 1.84 | H-bond |
| Phe268-AR | 3.75 | π–π stacking | |||
| Phe170-AR | 3.84 | π–π stacking | |||
| 3 | (−)-CBC | −9.668 | Ser505-OH | 1.81 | H-bond |
| Phe174-AR | 3.33 | π–π stacking | |||
| Phe268-P | 3.46 | π–π stacking | |||
| Phe268-AR | 3.62 | π–π stacking | |||
| Phe170-AR | 3.70 | π–π stacking | |||
| Phe170-P | 3.70 | π–π stacking | |||
| 4 | (R)-2′-OH-(+)-cannabicitran | −9.254 | Ile267-OH | 2.49 | H-bond |
| 5 | (+)-CBT-C | −8.345 | Phe170-AR | 3.38 | π–π stacking |
| 6 | (−)-CBT-C | −7.967 | Phe170-AR | 3.55 | π–π stacking |
| Phe268-AR | 3.55 | π–π stacking | |||
| 7 | (S)-2′-OH-(−)-cannabicitran | −6.796 | Phe170-AR | 3.23 | π–π stacking |
| Computer Ranking | Ligand | Docking Score | Highlighted Residue-Ligand Interactions | Distance of Interaction (Å) | Type of Interaction |
|---|---|---|---|---|---|
| 1 | (+)-CBC | −8.545 | Phe87-P | 3.81 | π–π stacking |
| Phe183-P | 3.99 | π–π stacking | |||
| 2 | (+)-CBT-C | −8.472 | - | - | - |
| 3 | (−)-Δ9-THC | −8.312 | Phe87-AR | 3.68 | π–π stacking |
| Phe183-AR | 3.99 | π–π stacking | |||
| 4 | (S)-2′-OH-(−)-cannabicitran | −8.042 | - | - | - |
| 5 | (−)-CBT-C | −7.949 | Phe183-AR | 3.28 | π–π stacking |
| 6 | (−)-CBC | −7.892 | Phe87-P | 3.57 | π–π stacking |
| Phe87-AR | 3.57 | π–π stacking | |||
| Phe183-P | 4.17 | π–π stacking | |||
| 7 | (R)-2′-OH-(+)-cannabicitran | −7.810 | Ser90-OH | 2.67 | H-bond |
| Phe183-AR | 3.63 | π–π stacking | |||
| Phe87-AR | 3.91 | π–π stacking |
| CB1R | CB2R | |||
|---|---|---|---|---|
| Ligand | Ki (nM) | Hill Slope | Ki (nM) | Hill Slope |
| CBC | 3500 ± 1200 | −0.27 ± 0.51 | 301 ± 72 | −1.32 ± 0.36 |
| 2′-hydroxycannabicitran | >10,000 | - | >10,000 | - |
| CBT-C | >10,000 | - | 4200 ± 410 | −1.15 ± 0.14 |
| Δ9-THC | 13.2 ± 1.9 | −0.73 ± 0.09 | 22.6 ± 5.0 | −0.96 ± 0.16 |
| CP 55,940 | 1.77 ± 0.32 | −0.87 ± 0.18 | - | - |
| WIN 55,212 | - | - | 3.9 ± 1.6 | −1.07 ± 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ward, A.M.; Shokati, T.; Klawitter, J.; Klawitter, J.; Nguyen, V.; Kozell, L.; Abbas, A.I.; Jones, D.; Christians, U. Identification and Characterization of Cannabichromene’s Major Metabolite Following Incubation with Human Liver Microsomes. Metabolites 2024, 14, 329. https://doi.org/10.3390/metabo14060329
Ward AM, Shokati T, Klawitter J, Klawitter J, Nguyen V, Kozell L, Abbas AI, Jones D, Christians U. Identification and Characterization of Cannabichromene’s Major Metabolite Following Incubation with Human Liver Microsomes. Metabolites. 2024; 14(6):329. https://doi.org/10.3390/metabo14060329
Chicago/Turabian StyleWard, Alexandra M., Touraj Shokati, Jost Klawitter, Jelena Klawitter, Vu Nguyen, Laura Kozell, Atheir I. Abbas, David Jones, and Uwe Christians. 2024. "Identification and Characterization of Cannabichromene’s Major Metabolite Following Incubation with Human Liver Microsomes" Metabolites 14, no. 6: 329. https://doi.org/10.3390/metabo14060329
APA StyleWard, A. M., Shokati, T., Klawitter, J., Klawitter, J., Nguyen, V., Kozell, L., Abbas, A. I., Jones, D., & Christians, U. (2024). Identification and Characterization of Cannabichromene’s Major Metabolite Following Incubation with Human Liver Microsomes. Metabolites, 14(6), 329. https://doi.org/10.3390/metabo14060329








