NMR Precision Metabolomics: Dynamic Peak Sum Thresholding and Navigators for Highly Standardized and Reproducible Metabolite Profiling of Clinical Urine Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Urine Samples Preparation
2.2. NMR Data Collection and Processing
2.3. Statistical Analysis
3. Results
3.1. Influence of Specific Gravity
3.2. Navigating Sources of Error in Sample Processing
3.3. Navigating Protein Contamination
3.4. Navigating Urine pH
3.5. Using myOLARIS Navigators for Biomarker Discovery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Havelund, J.F.; Heegaard, N.H.H.; Færgeman, N.J.K.; Gramsbergen, J.B. Biomarker Research in Parkinson’s Disease Using Metabolite Profiling. Metabolites 2017, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, L.; O’Day, E.M. Perspective: A Potential Role for NUS in Metabolite-Based In Vitro Diagnostics. Magn. Reson. Chem. 2021, 59, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Varma, V.R.; Oommen, A.M.; Varma, S.; Casanova, R.; An, Y.; Andrews, R.M.; O’Brien, R.; Pletnikova, O.; Troncoso, J.C.; Toledo, J.; et al. Brain and Blood Metabolite Signatures of Pathology and Progression in Alzheimer Disease: A Targeted Metabolomics Study. PLoS Med. 2018, 15, e1002482. [Google Scholar] [CrossRef] [PubMed]
- Bragg, F.; Trichia, E.; Aguilar-Ramirez, D.; Bešević, J.; Lewington, S.; Emberson, J. Predictive Value of Circulating NMR Metabolic Biomarkers for Type 2 Diabetes Risk in the UK Biobank Study. BMC Med. 2022, 20, 159. [Google Scholar] [CrossRef]
- Holmes, M.V.; Millwood, I.Y.; Kartsonaki, C.; Hill, M.R.; Bennett, D.A.; Boxall, R.; Guo, Y.; Xu, X.; Bian, Z.; Hu, R.; et al. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Dudka, I.; Lundquist, K.; Wikström, P.; Bergh, A.; Gröbner, G. Metabolomic Profiles of Intact Tissues Reflect Clinically Relevant Prostate Cancer Subtypes. J. Transl. Med. 2023, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jasbi, P.; Shi, X.; Turner, C.; Hrovat, J.; Liu, L.; Rabena, Y.; Porter, P.; Gu, H. Early Breast Cancer Detection Using Untargeted and Targeted Metabolomics. J. Proteome Res. 2021, 20, 3124–3133. [Google Scholar] [CrossRef] [PubMed]
- Perazzoli, G.; García-Valdeavero, O.M.; Peña, M.; Prados, J.; Melguizo, C.; Jiménez-Luna, C. Evaluating Metabolite-Based Biomarkers for Early Diagnosis of Pancreatic Cancer: A Systematic Review. Metabolites 2023, 13, 872. [Google Scholar] [CrossRef]
- Stavarache, C.; Nicolescu, A.; Duduianu, C.; Ailiesei, G.L.; Balan-Porcăraşu, M.; Cristea, M.; Macsim, A.M.; Popa, O.; Stavarache, C.; Hîrtopeanu, A.; et al. A Real-Life Reproducibility Assessment for NMR Metabolomics. Diagnostics 2022, 12, 559. [Google Scholar] [CrossRef]
- Lin, Y.; Caldwell, G.W.; Li, Y.; Lang, W.; Masucci, J. Inter-Laboratory Reproducibility of an Untargeted Metabolomics GC–MS Assay for Analysis of Human Plasma. Sci. Rep. 2020, 10, 10918. [Google Scholar] [CrossRef]
- Roth, H.E.; Powers, R. Meta-Analysis Reveals Both the Promises and the Challenges of Clinical Metabolomics. Cancers 2022, 14, 3992. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The Role of Metabolites and Metabolomics in Clinically Applicable Biomarkers of Disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef]
- Zhang, B.; Powers, R.; O’Day, E.M. Evaluation of Non-Uniform Sampling 2D1H–13C HSQC Spectra for Semi-Quantitative Metabolomics. Metabolites 2020, 10, 203. [Google Scholar] [CrossRef]
- Saude, E.J.; Adamko, D.J.; Rowe, B.H.; Marrie, T.; Sykes, B.D. Variation of Metabolites in Normal Human Urine. Metabolomics 2007, 3, 439–451. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The Human Urine Metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Schreier, C.; Kremer, W.; Huber, F.; Neumann, S.; Pagel, P.; Lienemann, K.; Pestel, S. Reproducibility of NMR Analysis of Urine Samples: Impact of Sample Preparation, Storage Conditions, and Animal Health Status. Biomed. Res. Int. 2013, 2013, 878374. [Google Scholar] [CrossRef]
- Xiao, C.; Hao, F.; Qin, X.; Wang, Y.; Tang, H. An Optimized Buffer System for NMR-Based Urinary Metabonomics with Effective PH Control, Chemical Shift Consistency and Dilution Minimization. Analyst 2009, 134, 916–925. [Google Scholar] [CrossRef]
- Warrack, B.M.; Hnatyshyn, S.; Ott, K.H.; Reily, M.D.; Sanders, M.; Zhang, H.; Drexler, D.M. Normalization Strategies for Metabonomic Analysis of Urine Samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 547–552. [Google Scholar] [CrossRef]
- Button, K.S.; Ioannidis, J.P.A.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.J.; Munafò, M.R. Power Failure: Why Small Sample Size Undermines the Reliability of Neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef]
- Dong, C.; Honrao, C.; Rodrigues, L.O.; Wolf, J.; Sheehan, K.B.; Surface, M.; Alcalay, R.N.; O’day, E.M. Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients. Metabolites 2022, 12, 149. [Google Scholar] [CrossRef]
- Honrao, C.; Teissier, N.; Zhang, B.; Powers, R.; O’Day, E.M. Gadolinium-Based Paramagnetic Relaxation Enhancement Agent Enhances Sensitivity for NUS Multidimensional NMR-Based Metabolomics. Molecules 2021, 26, 5115. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise; AAAI Press: Washington, DA, USA, 1996. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. Available online: https://ggplot2.tidyverse.org (accessed on 13 December 2021).
- Minton, D.M.; O’Neal, E.K.; Torres-McGehee, T.M. Agreement of Urine Specific Gravity Measurements between Manual and Digital Refractometers. J. Athl. Train. 2015, 50, 59–64. [Google Scholar] [CrossRef]
- Lindon, J.C.; Nicholson, J.K.; Holmes, E.; Everett, J.R. Metabonomics: Metabolic Processes Studied by NMR Spectroscopy of Biofluids. Concepts Magn. Reson. 2000, 12, 289–320. [Google Scholar] [CrossRef]
- Tredwell, G.D.; Bundy, J.G.; De Iorio, M.; Ebbels, T.M.D. Modelling the Acid/Base 1H NMR Chemical Shift Limits of Metabolites in Human Urine. Metabolomics 2016, 12, 152. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H. A Simple Method for the Isolation of Total Lipids from Animal Tissues. J. Biol. Chem. 1956, 55, 497–509. [Google Scholar]
- Vachek, J.; Zakiyanov, O.; Tesar, V. Proteinuria. Interni Med. Pro Praxi 2018, 20, 96–98. [Google Scholar]
- Nagana Gowda, G.A.; Raftery, D. Can NMR Solve Some Significant Challenges in Metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef]
- Khaniani, Y.; Lipfert, M.; Bhattacharyya, D.; Pineiro, R.P.; Zheng, J.; Wishart, D.S. A Simple and Convenient Synthesis of Unlabeled And13 C-Labeled 3-(3-Hydroxyphenyl)-3-Hydroxypropionic Acid and Its Quantification in Human Urine Samples. Metabolites 2018, 8, 80. [Google Scholar] [CrossRef]
- Reily, M.D.; Robosky, L.C.; Manning, M.L.; Butler, A.; Baker, J.D.; Winters, R.T. DFTMP, an NMR Reagent for Assessing the near-Neutral PH of Biological Samples. J. Am. Chem. Soc. 2006, 128, 12360–12361. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; Bosque-Plata, L. Del The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Peters, S.R.; Overmyer, K.A.; Paulson, B.R.; Westphall, M.S.; Coon, J.J. Real-Time Health Monitoring through Urine Metabolomics. npj Digit. Med. 2019, 2, 109. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, L. Sample Normalization Methods in Quantitative Metabolomics. J. Chromatogr. A 2016, 1430, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.L.; Paulina de la Mata, A.; Dias, R.P.; Harynuk, J.J. Towards Standardization of Data Normalization Strategies to Improve Urinary Metabolomics Studies by Gc×gc-Tofms. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Edmands, W.M.B.; Ferrari, P.; Scalbert, A. Normalization to Specific Gravity Prior to Analysis Improves Information Recovery from High Resolution Mass Spectrometry Metabolomic Profiles of Human Urine. Anal. Chem. 2014, 86, 10925–10931. [Google Scholar] [CrossRef] [PubMed]
- Meister, I.; Zhang, P.; Sinha, A.; Sköld, C.M.; Wheelock, Å.M.; Izumi, T.; Chaleckis, R.; Wheelock, C.E. High-Precision Automated Workflow for Urinary Untargeted Metabolomic Epidemiology. Anal. Chem. 2021, 93, 5248–5258. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Boudreau, N.; Lévesque, A. Internal Standards for Quantitative LC-MS Bioanalysis. In LC-MS in Drug Bioanalysis; Springer: Boston, MA, USA, 2012; ISBN 978-1-4614-3828-1. [Google Scholar]
- Voinescu, G.C.; Shoemaker, M.; Moore, H.; Khanna, R.; Nolph, K.D. The Relationship between Urine Osmolality and Specific Gravity. Am. J. Med. Sci. 2002, 323, 39–42. [Google Scholar] [CrossRef]
- Akarsu, E.; Buyukhatipoglu, H.; Aktaran, S.; Geyik, R. The Value of Urine Specific Gravity in Detecting Diabetes Insipidus in a Patient with Uncontrolled Diabetes Mellitus: Urine Specific Gravity in Differential Diagnosis. J. Gen. Intern. Med. 2006, 21, C1–C2. [Google Scholar] [CrossRef]
- Morash, B.; Sarker, M.; Rainey, J.K. Concentration-Dependent Changes to Diffusion and Chemical Shift of Internal Standard Molecules in Aqueous and Micellar Solutions. J. Biomol. NMR 2018, 71, 79–89. [Google Scholar] [CrossRef]
- Alum, M.F.; Shaw, P.A.; Sweatman, B.C.; Ubhi, B.K.; Haselden, J.N.; Connor, S.C. 4,4-Dimethyl-4-Silapentane-1-Ammonium Trifluoroacetate (DSA), a Promising Universal Internal Standard for NMR-Based Metabolic Profiling Studies of Biofluids, Including Blood Plasma and Serum. Metabolomics 2008, 4, 122–127. [Google Scholar] [CrossRef]
- Bhinderwala, F.; Roth, H.E.; Noel, H.; Feng, D.; Powers, R. Chemical Shift Variations in Common Metabolites. J. Magn. Reson. 2022, 345, 107335. [Google Scholar] [CrossRef]
- Ye, L.; De Iorio, M.; Ebbels, T.M.D. Bayesian Estimation of the Number of Protonation Sites for Urinary Metabolites from NMR Spectroscopic Data. Metabolomics 2018, 14, 56. [Google Scholar] [CrossRef]
- Quattrini, S.; Pampaloni, B.; Brandi, M.L. Natural Mineral Waters: Chemical Characteristics and Health Effects. Clin. Cases Miner. Bone Metab. 2016, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Anton, S.D.; Lu, X.; Bank, G.; Heekin, K.; Saha, D.; Dubyak, P.J.; Hausenblas, H.A. nn Effect of a Novel Dietary Supplement on PH Levels of Healthy Volunteers: A Pilot Study. J. Integr. Med. 2013, 11, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Yıldırım, İ.; Koçan, H. The PH of Drinking Water and Its Effect on the PH of Urine. Cureus 2023, 15, e47437. [Google Scholar] [CrossRef] [PubMed]
- Pigoli, G.; Dorizzi, R.M.; Ferrari, F. Variations of the Urinary PH Values in a Population of 13.000 Patients Addressing to the National Health System. Minerva Ginecol. 2010, 62, 85–90. [Google Scholar] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics Enables Precision Medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef]
- Spitzenberger, F.; Patel, J.; Gebuhr, I.; Kruttwig, K.; Safi, A.; Meisel, C. Laboratory-Developed Tests: Design of a Regulatory Strategy in Compliance with the International State-of-the-Art and the Regulation (EU) 2017/746 (EU IVDR [In Vitro Diagnostic Medical Device Regulation]). Ther. Innov. Regul. Sci. 2022, 56, 47–64. [Google Scholar] [CrossRef]
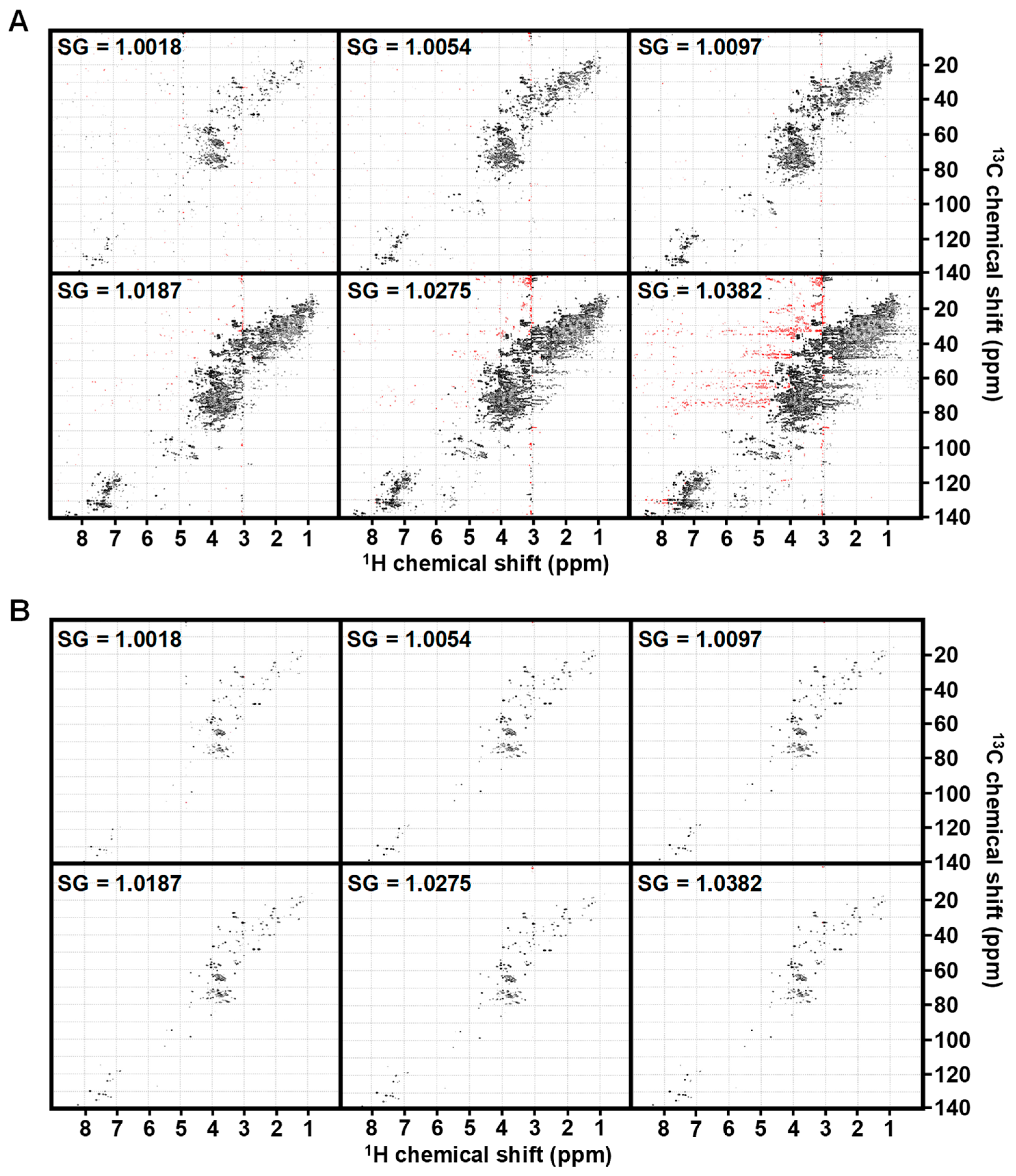


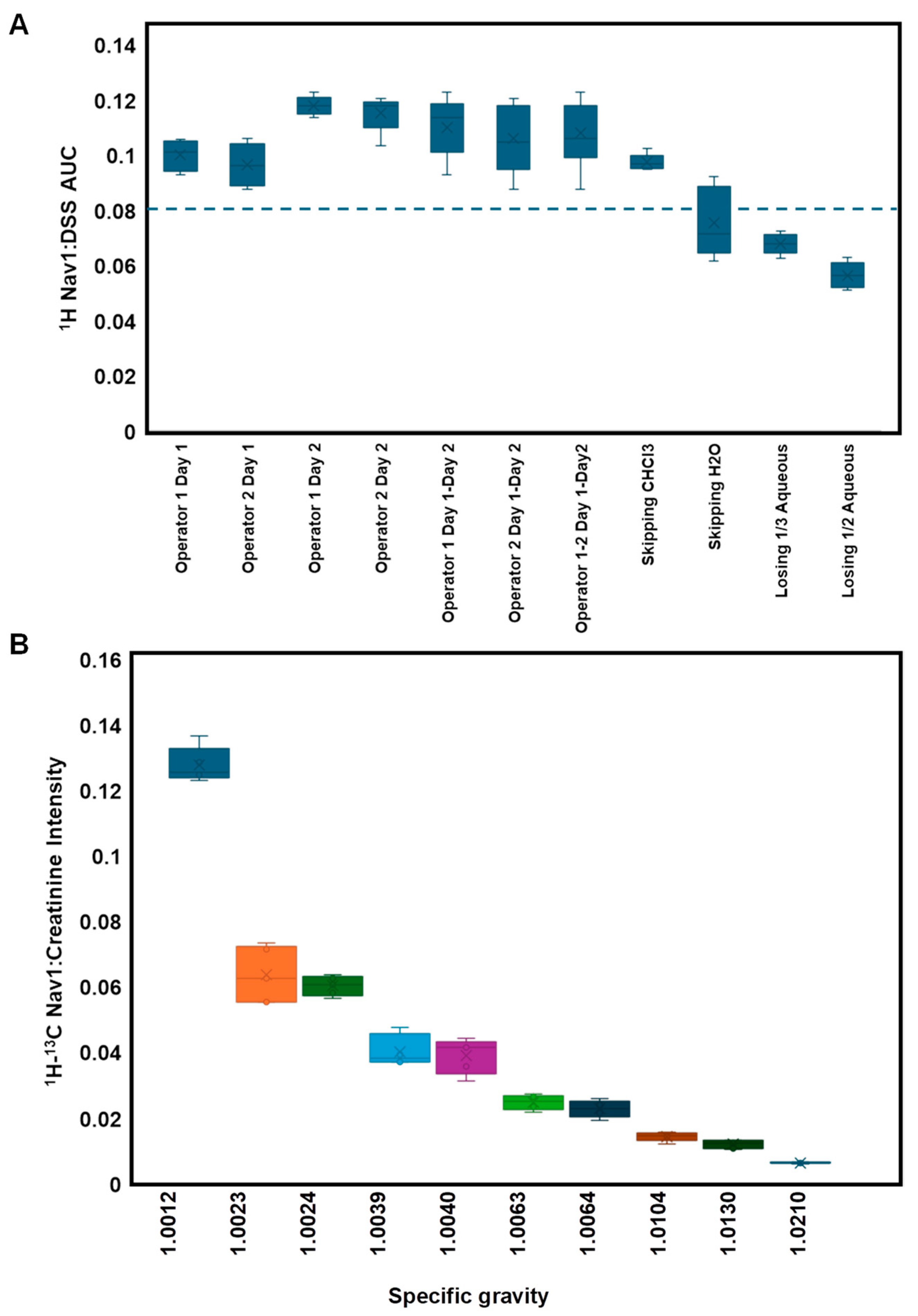
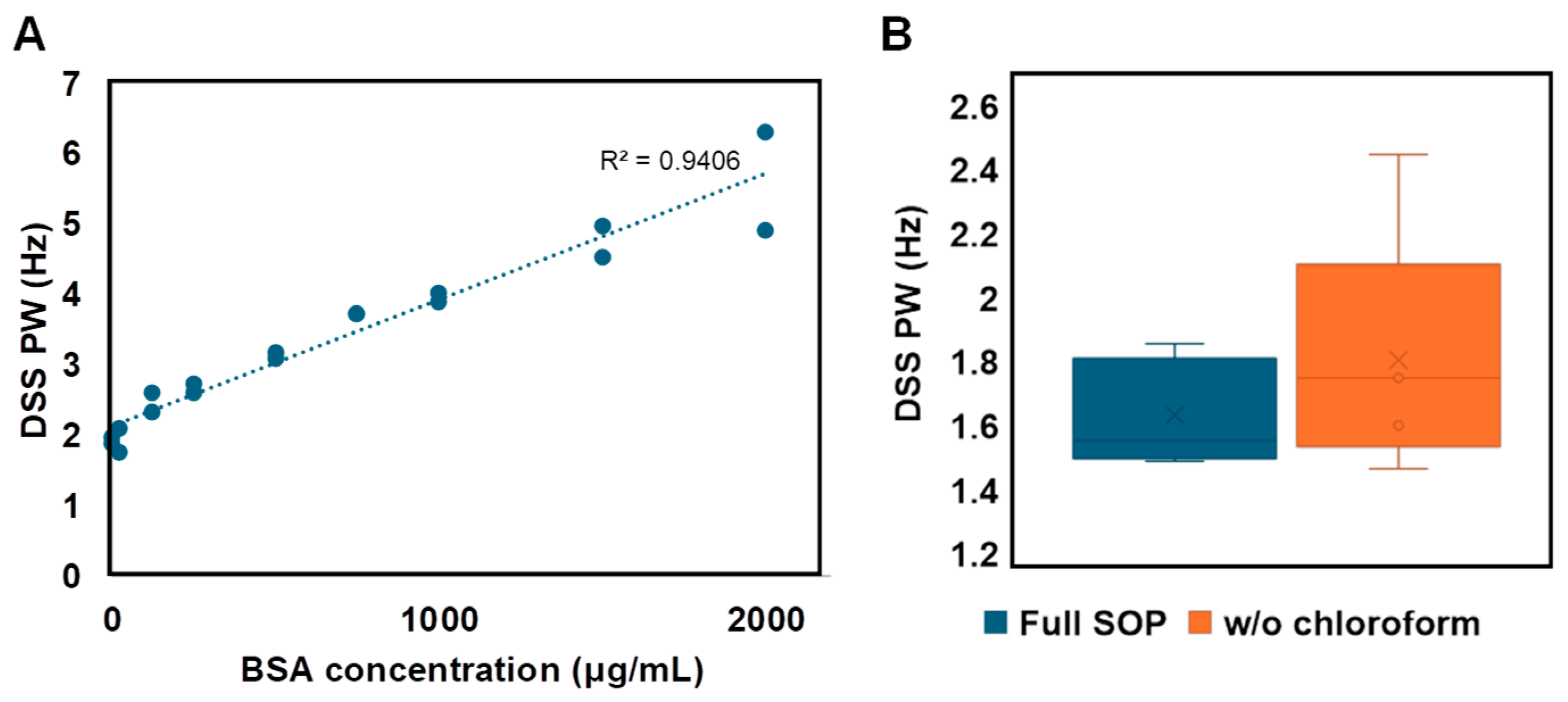

| Analysis | FP Noise-Based | FP Sum-Based |
|---|---|---|
| 1.0054 vs. 1.0097 | 0 | 0 |
| 1.0054 vs. 1.0187 | 79 | 3 |
| 1.0054 vs. 1.0275 | 128 | 8 |
| 1.0097 vs. 1.0187 | 20 | 0 |
| 1.0097 vs. 1.0275 | 80 | 9 |
| 1.0187 vs. 1.0275 | 0 | 0 |
| Analysis | N. Features | Passing by Chance | Passing KW | Passing KW + FC | Passing FDR | Passing FDR + FC |
|---|---|---|---|---|---|---|
| pH 4.99 vs. pH 6.08 | 168 | 8.4 | 22 | 2 | 0 | 0 |
| pH 4.99 vs. pH 8.08 | 167 | 8.35 | 36 | 6 | 0 | 0 |
| pH 6.08 vs. pH 8.08 | 169 | 8.45 | 16 | 3 | 0 | 0 |
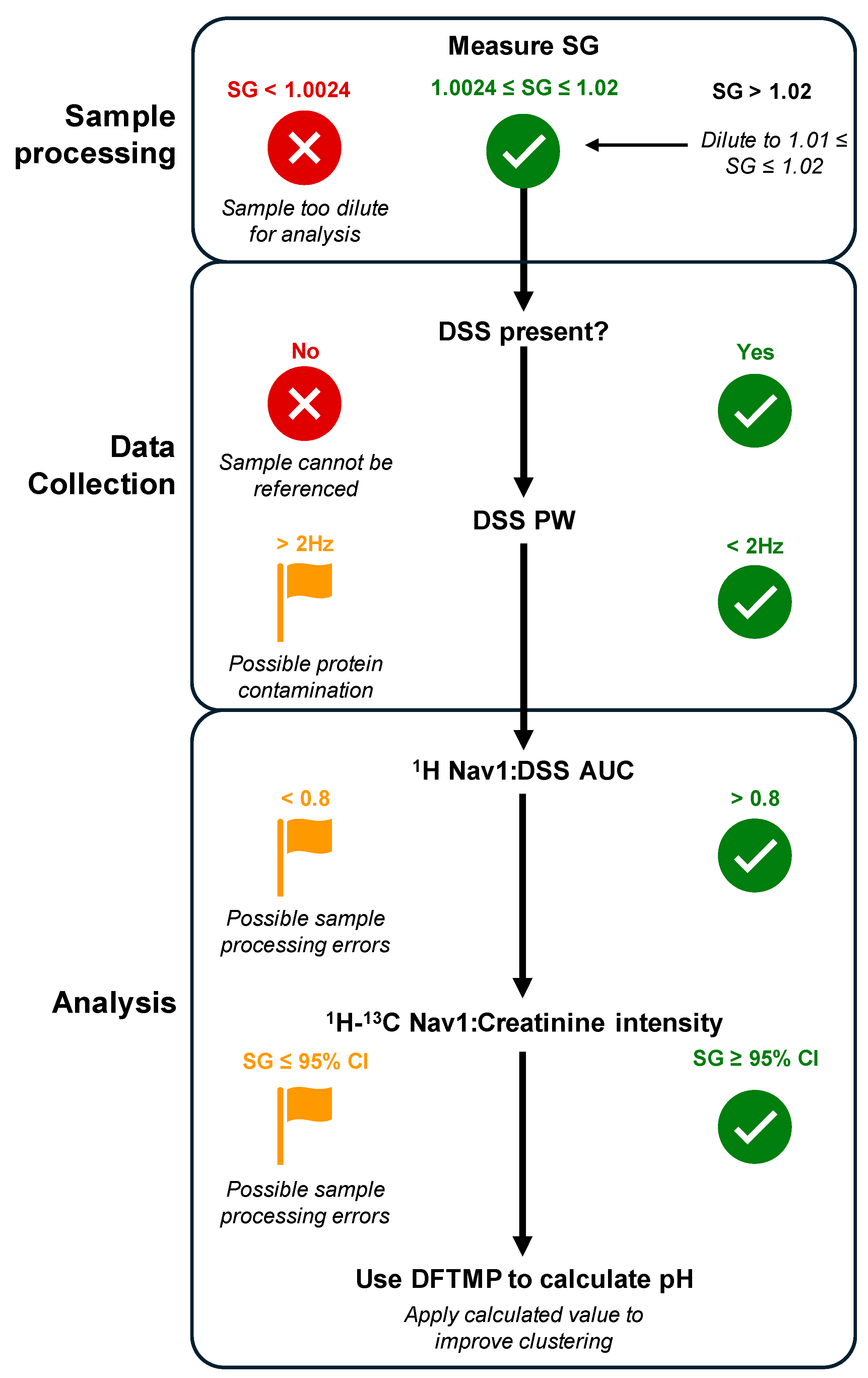
| Navigator | Sources of Error | Description | Automatable? |
|---|---|---|---|
| 1D 1H DSS | Chemical shift differences | Reference DSS peak to 0 ppm | Y |
| 1D 1H DSS PW | Protein contamination; poor shimming | Indicator for lack of protein contamination and proper shimming if PW > 2 Hz | Y |
| 1D Nav1:DSS AUC ratio | Incomplete or inconsistent sample preparation | Monitor sample processing quality with ratio < 0.8 flagging potential sample processing error(s) | Y |
| 2D 1H-13C HSQC Nav1:creatinine | Inconsistent sample preparation of disease confounder | Ratio can aid in predicting SG of original sample | Y |
| 1D 1H DFTMP | Improper clustering across samples | Determines sample pH and clusters pH-sensitive peaks | Y |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trimigno, A.; Holderman, N.R.; Dong, C.; Boardman, K.D.; Zhao, J.; O’Day, E.M. NMR Precision Metabolomics: Dynamic Peak Sum Thresholding and Navigators for Highly Standardized and Reproducible Metabolite Profiling of Clinical Urine Samples. Metabolites 2024, 14, 275. https://doi.org/10.3390/metabo14050275
Trimigno A, Holderman NR, Dong C, Boardman KD, Zhao J, O’Day EM. NMR Precision Metabolomics: Dynamic Peak Sum Thresholding and Navigators for Highly Standardized and Reproducible Metabolite Profiling of Clinical Urine Samples. Metabolites. 2024; 14(5):275. https://doi.org/10.3390/metabo14050275
Chicago/Turabian StyleTrimigno, Alessia, Nicole R. Holderman, Chen Dong, Kari D. Boardman, Jifang Zhao, and Elizabeth M. O’Day. 2024. "NMR Precision Metabolomics: Dynamic Peak Sum Thresholding and Navigators for Highly Standardized and Reproducible Metabolite Profiling of Clinical Urine Samples" Metabolites 14, no. 5: 275. https://doi.org/10.3390/metabo14050275
APA StyleTrimigno, A., Holderman, N. R., Dong, C., Boardman, K. D., Zhao, J., & O’Day, E. M. (2024). NMR Precision Metabolomics: Dynamic Peak Sum Thresholding and Navigators for Highly Standardized and Reproducible Metabolite Profiling of Clinical Urine Samples. Metabolites, 14(5), 275. https://doi.org/10.3390/metabo14050275







