The Exometabolome of Xylella fastidiosa in Contact with Paraburkholderia phytofirmans Supernatant Reveals Changes in Nicotinamide, Amino Acids, Biotin, and Plant Hormones
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance and Cultivation of Bacteria
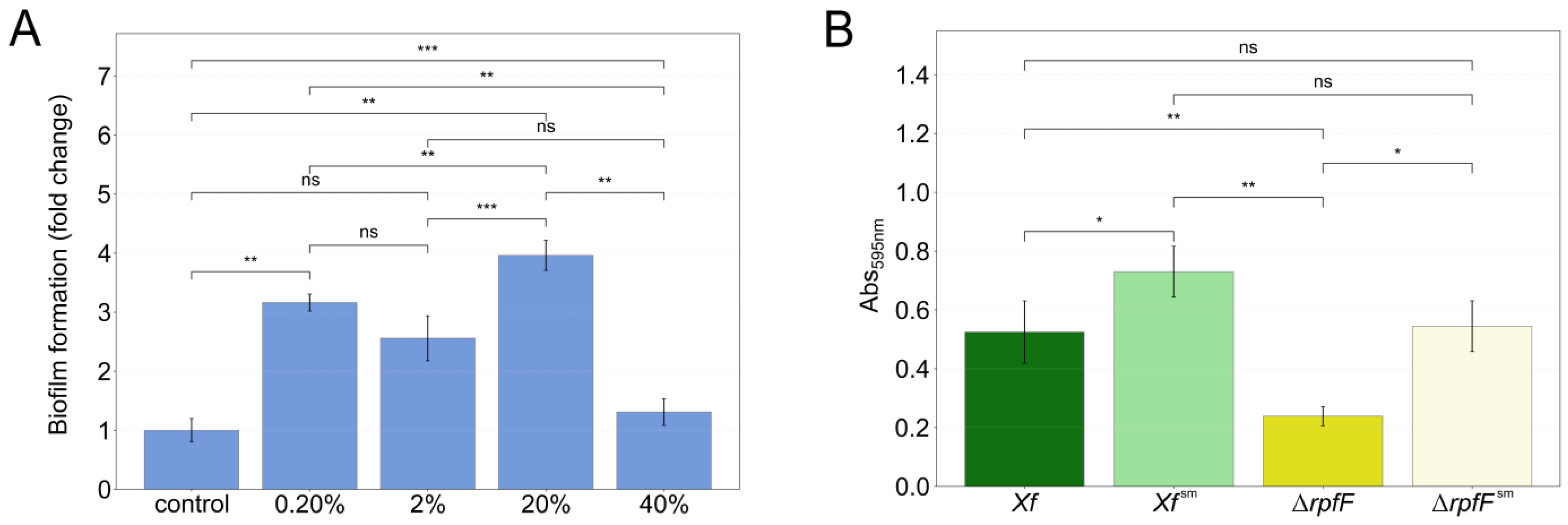
2.2. X. fastidiosa Biofilm Measurement
2.3. Analysis of Metabolites of the Supernatant of X. fastidiosa Cultures by Liquid Chromatography Coupled to Mass Spectrometry (LC-MS)
2.4. LCMS Data Processing and Analysis
2.5. Integration of Metabolites and Gene Set for Functional Annotation of Exometabolomes
2.6. Bioinformatic Analysis
2.7. Cooperative Metabolic Interactions between X. fastidiosa and P. phytofirmans
3. Results
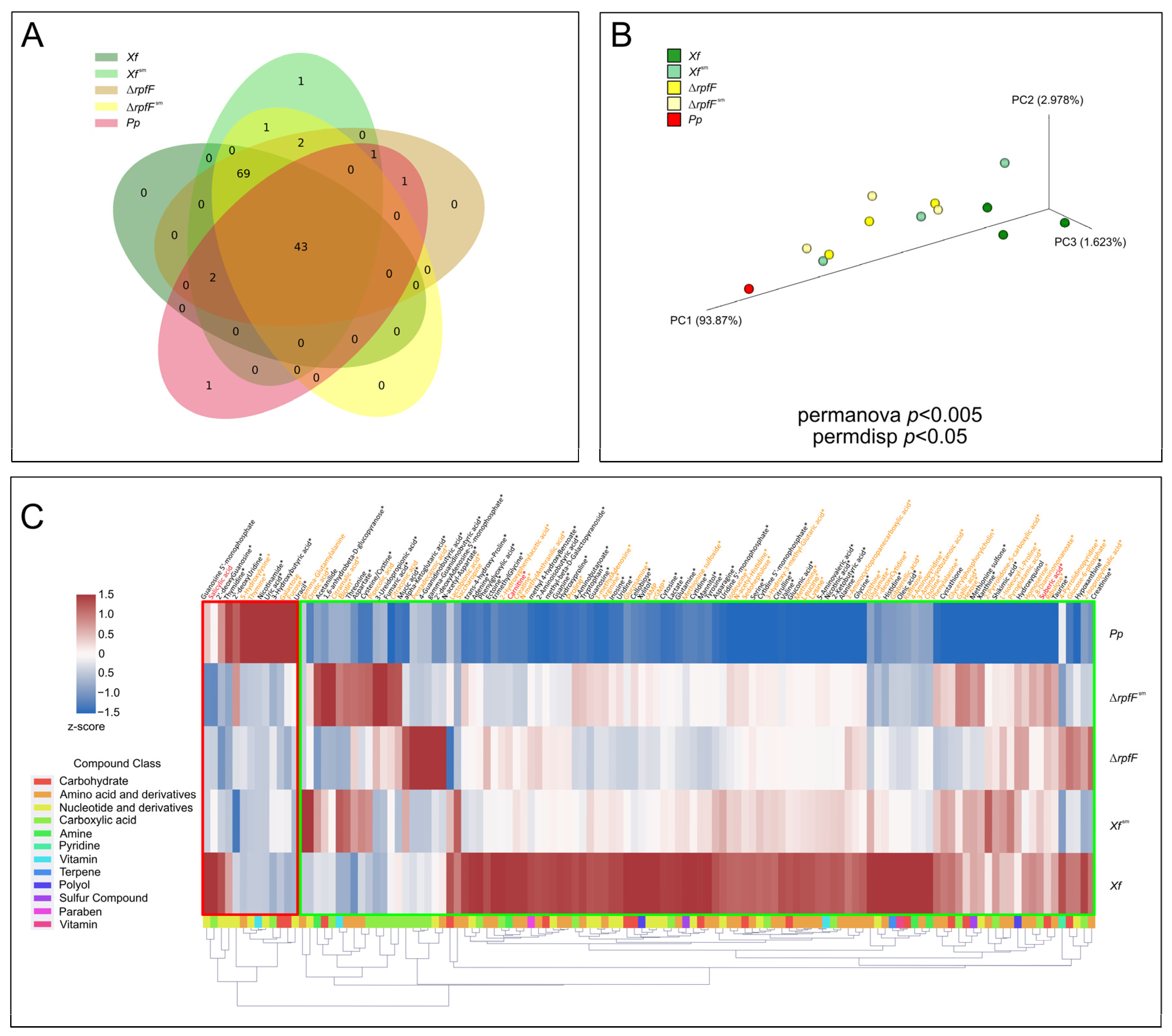
3.1. P. phytofirmans Interacts with X. fastidiosa through Its Exometabolome
3.2. Exometabolome Variation among X. fastidiosa Strains in Response to P. phytofirmans
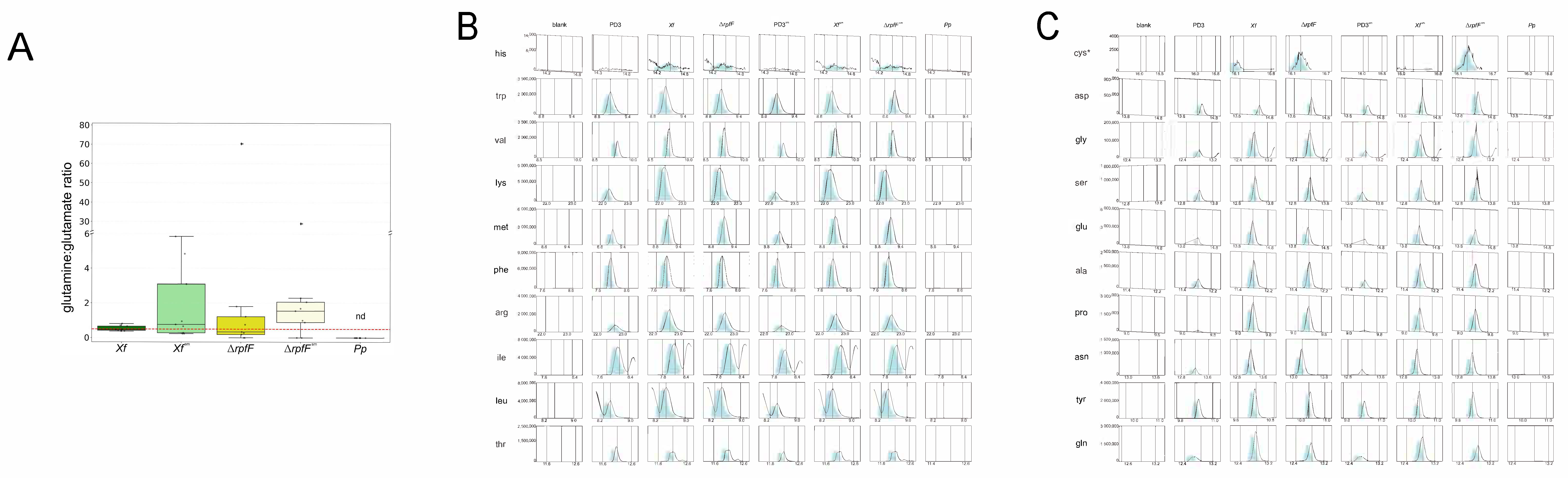
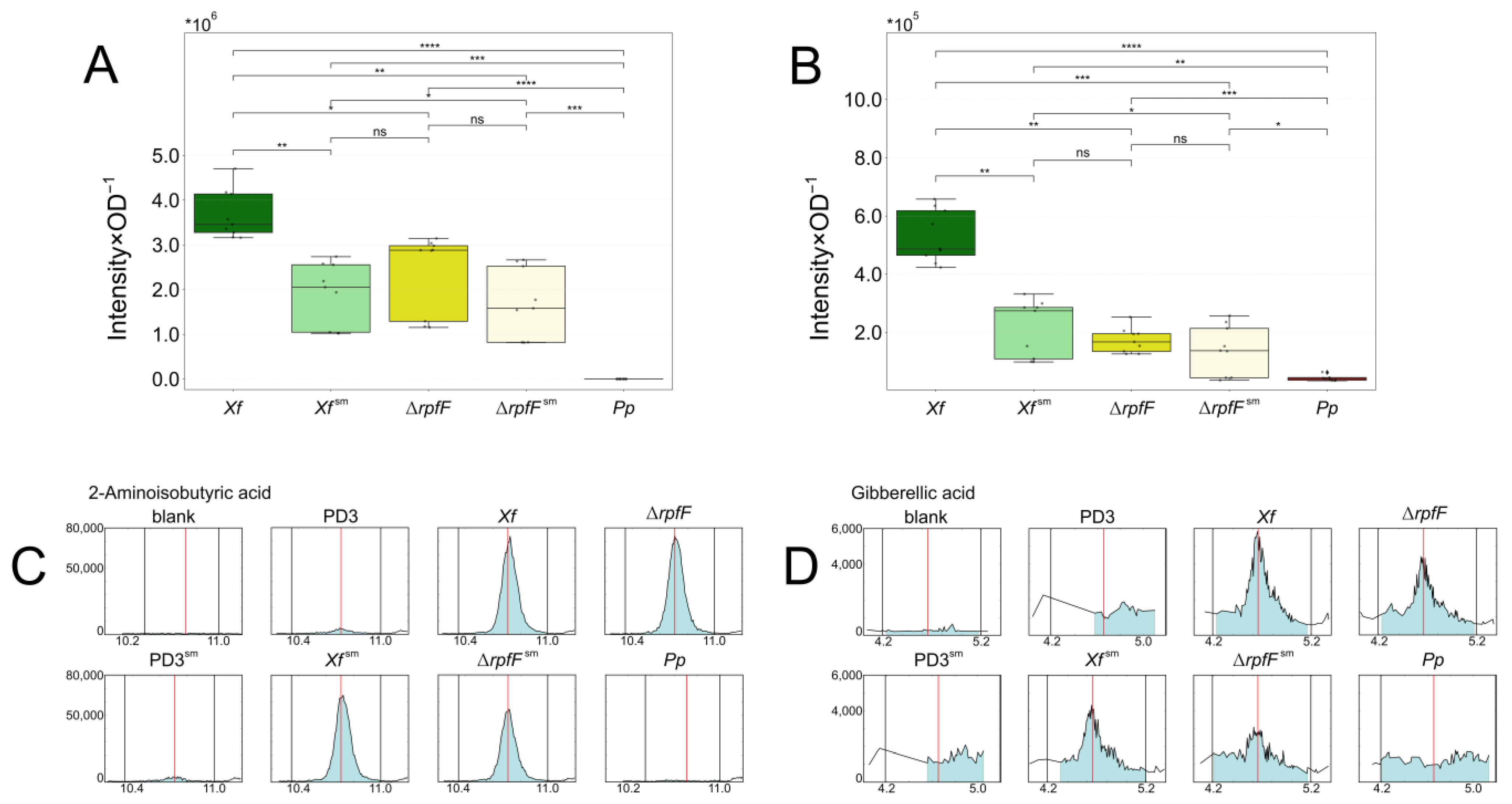
3.3. X. fastidiosa Secretes High Amounts of Amino Acids and Vitamins
3.4. Two Plant Hormones Are Exclusively Secreted by X. fastidiosa
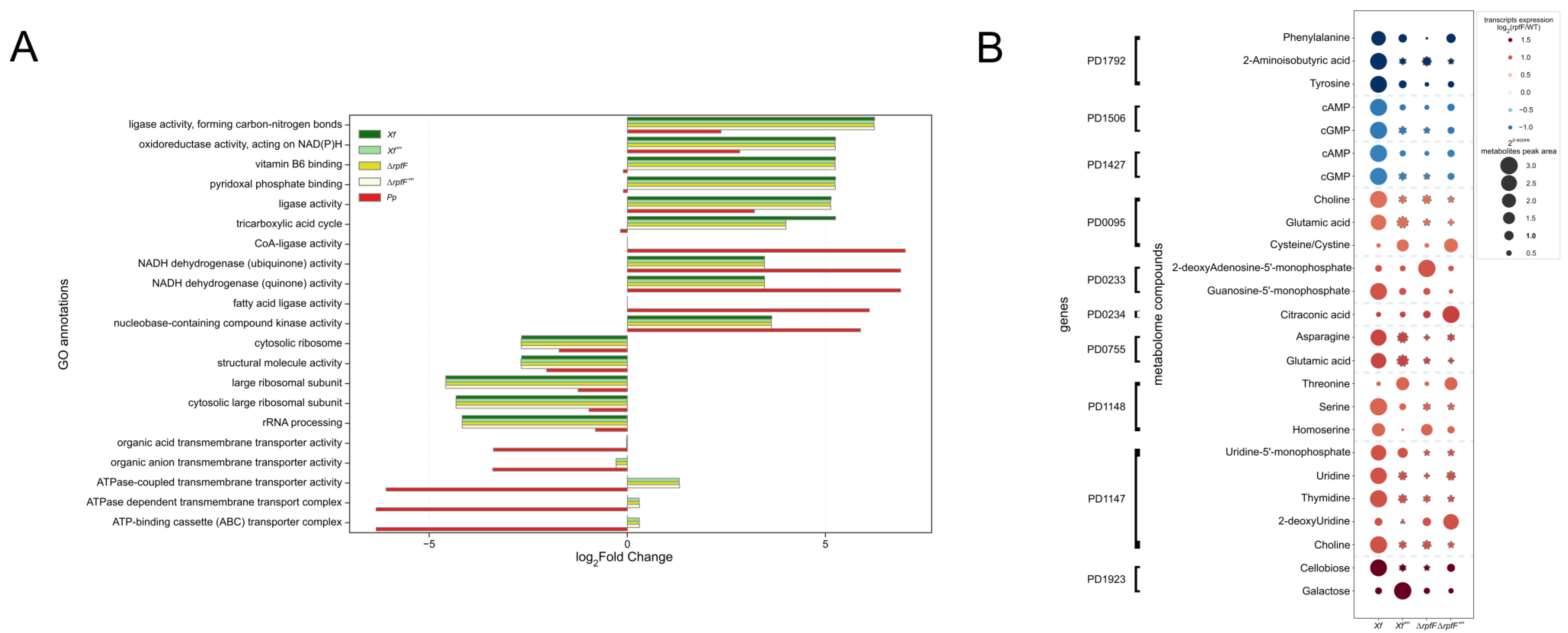
3.5. Exometabolome, Genome, and Transcriptome Integration for X. fastidiosa and P. phytofirmans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sicard, A.; Zeilinger, A.R.; Vanhove, M.; Schartel, T.E.; Beal, D.J.; Daugherty, M.P.; Almeida, R.P.P. Xylella fastidiosa: Insights into an Emerging Plant Pathogen. Annu. Rev. Phytopathol. 2018, 56, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Almeida, R.P.; Lindow, S. Living in two worlds: The plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 2008, 46, 243–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, J.L.; Lindow, S.E. RpfF-dependent regulon of Xylella fastidiosa. Phytopathology 2012, 102, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, M.; Yokota, K.; Antonova, E.; Garcia, A.; Beaulieu, E.; Hayes, T.; Iavarone, A.T.; Lindow, S.E. Promiscuous Diffusible Signal Factor Production and Responsiveness of the Xylella fastidiosa Rpf System. MBio 2016, 7, e01054-16. [Google Scholar] [CrossRef]
- Roper, C.; Castro, C.; Ingel, B. Xylella fastidiosa: Bacterial parasitism with hallmarks of commensalism. Curr. Opin. Plant Biol. 2019, 50, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, J.; Ingel, B.; Blanco-Ulate, B.; Cantu, D.; Roper, C. Xylella fastidiosa: An examination of a re-emerging plant pathogen. Mol. Plant Pathol. 2018, 19, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.E.; Moury, B. Revisiting the Concept of Host Range of Plant Pathogens. Annu. Rev. Phytopathol. 2019, 57, 63–90. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Uptake of bacteria into living plant cells, the unifying and distinct feature of the nitrogen-fixing root nodule symbiosis. Curr. Opin. Plant Biol. 2018, 44, 164–174. [Google Scholar] [CrossRef]
- Nagel, R.; Turrini, P.C.; Nett, R.S.; Leach, J.E.; Verdier, V.; Van Sluys, M.A.; Peters, R.J. An operon for production of bioactive gibberellin A(4) phytohormone with wide distribution in the bacterial rice leaf streak pathogen Xanthomonas oryzae pv. oryzicola. New Phytol. 2017, 214, 1260–1266. [Google Scholar] [CrossRef]
- Ankrah, N.Y.D.; Wilkes, R.A.; Zhang, F.Q.; Aristilde, L.; Douglas, A.E. The Metabolome of Associations between Xylem-Feeding Insects and their Bacterial Symbionts. J. Chem. Ecol. 2020, 46, 735–744. [Google Scholar] [CrossRef]
- Yang, J.; Masoudi, A.; Li, H.; Gu, Y.; Wang, C.; Wang, M.; Yu, Z.; Liu, J. Microbial community structure and niche differentiation under different health statuses of Pinus bungeana in the Xiong’an New Area in China. Front. Microbiol. 2022, 13, 913349. [Google Scholar] [CrossRef]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Saldanha, L.L.; Allard, P.M.; Dilarri, G.; Codesido, S.; Gonzalez-Ruiz, V.; Queiroz, E.F.; Ferreira, H.; Wolfender, J.L. Metabolomic-and Molecular Networking-Based Exploration of the Chemical Responses Induced in Citrus sinensis Leaves Inoculated with Xanthomonas citri. J. Agric. Food Chem. 2022, 70, 14693–14705. [Google Scholar] [CrossRef]
- Ryffel, F.; Helfrich, E.J.N.; Kiefer, P.; Peyriga, L.; Portais, J.C.; Piel, J.; Vorholt, J.A. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 2016, 10, 632–643. [Google Scholar] [CrossRef]
- Chen, X.L.; Sun, M.C.; Chong, S.L.; Si, J.P.; Wu, L.S. Transcriptomic and Metabolomic Approaches Deepen Our Knowledge of Plant-Endophyte Interactions. Front. Plant Sci. 2022, 12, 700200. [Google Scholar] [CrossRef]
- Zaini, P.A.; Nascimento, R.; Gouran, H.; Cantu, D.; Chakraborty, S.; Phu, M.; Goulart, L.R.; Dandekar, A.M. Molecular Profiling of Pierce’s Disease Outlines the Response Circuitry of Vitis vinifera to Xylella fastidiosa Infection. Front. Plant Sci. 2018, 9, 771. [Google Scholar] [CrossRef]
- Cariddi, C.; Saponari, M.; Boscia, D.; De Stradis, A.; Loconsole, G.; Nigro, F.; Porcelli, F.; Potere, O.; Martelli, G.P. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J. Plant Pathol. 2014, 96, 425–429. [Google Scholar]
- Desprez-Loustau, M.-L.; Balci, Y.; Cornara, D.; Gonthier, P.; Robin, C.; Jacques, M.-A. Is Xylella fastidiosa a serious threat to European forests? For. Int. J. For. Res. 2020, 94, 1–17. [Google Scholar]
- Krugner, R.; Sisterson, M.S.; Backus, E.A.; Burbank, L.P.; Redak, R.A. Sharpshooters: A review of what moves Xylella fastidiosa. Austral. Entomol. 2019, 58, 248–267. [Google Scholar] [CrossRef]
- Saponari, M.; Loconsole, G.; Cornara, D.; Yokomi, R.K.; De Stradis, A.; Boscia, D.; Bosco, D.; Martelli, G.P.; Krugner, R.; Porcelli, F. Infectivity and transmission of Xylellua fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. J. Econ. Entomol. 2014, 107, 1316–1319. [Google Scholar] [CrossRef]
- Huang, W.J.; Reyes-Caldas, P.; Mann, M.; Seifbarghi, S.; Kahn, A.; Almeida, R.P.P.; Béven, L.; Heck, M.; Hogenhout, S.A.; Coaker, G. Bacterial Vector-Borne Plant Diseases: Unanswered Questions and Future Directions. Molecular. Plant 2020, 13, 1379–1393. [Google Scholar] [CrossRef]
- Roper, M.C.; Greve, L.C.; Warren, J.G.; Labavitch, J.M.; Kirkpatrick, B.C. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol. Plant-Microbe Interact. 2007, 20, 411–419. [Google Scholar] [CrossRef]
- Nascimento, R.; Gouran, H.; Chakraborty, S.; Gillespie, H.W.; Almeida-Souza, H.O.; Tu, A.; Rao, B.J.; Feldstein, P.A.; Bruening, G.; Goulart, L.R.; et al. The Type II Secreted Lipase/Esterase LesA is a Key Virulence Factor Required for Xylella fastidiosa Pathogenesis in Grapevines. Sci. Rep. 2016, 6, 18598. [Google Scholar] [CrossRef]
- Feitosa, O.R.; Stefanello, E.; Zaini, P.A.; Nascimento, R.; Pierry, P.M.; Dandekar, A.M.; Lindow, S.E.; da Silva, A.M. Proteomic and Metabolomic Analyses of Xylella fastidiosa OMV-Enriched Fractions Reveal Association with Virulence Factors and Signaling Molecules of the DSF Family. Phytopathology 2019, 109, 1344–1353. [Google Scholar] [CrossRef]
- Block, A.; Li, G.Y.; Fu, Z.Q.; Alfano, J.R. Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 2008, 11, 396–403. [Google Scholar] [CrossRef]
- Kvitko, B.H.; Collmer, A. Discovery of the Hrp Type III Secretion System in Phytopathogenic Bacteria: How Investigation of Hypersensitive Cell Death in Plants Led to a Novel Protein Injector System and a World of Inter-Organismal Molecular Interactions Within Plant Cells. Phytopathology 2023, 113, 626–636. [Google Scholar] [CrossRef]
- Van Sluys, M.A.; Monteiro-Vitorello, C.B.; Camargo, L.E.; Menck, C.F.; Da Silva, A.C.; Ferro, J.A.; Oliveira, M.C.; Setubal, J.C.; Kitajima, J.P.; Simpson, A.J. Comparative genomic analysis of plant-associated bacteria. Annu. Rev. Phytopathol. 2002, 40, 169–189. [Google Scholar] [CrossRef]
- De La Fuente, L.; Merfa, M.V.; Cobine, P.A.; Coleman, J.J. Pathogen Adaptation to the Xylem Environment. Annu. Rev. Phytopathol. 2022, 60, 163–186. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef]
- Sessitsch, A.; Coenye, T.; Sturz, A.V.; Vandamme, P.; Barka, E.A.; Salles, J.F.; Van Elsas, J.D.; Faure, D.; Reiter, B.; Glick, B.R.; et al. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef]
- Miotto-Vilanova, L.; Jacquard, C.; Courteaux, B.; Wortham, L.; Michel, J.; Clement, C.; Barka, E.A.; Sanchez, L. Burkholderia phytofirmans PsJN Confers Grapevine Resistance against Botrytis cinerea via a Direct Antimicrobial Effect Combined with a Better Resource Mobilization. Front. Plant Sci. 2016, 7, 1236. [Google Scholar] [CrossRef]
- Baccari, C.; Antonova, E.; Lindow, S. Biological Control of Pierce’s Disease of Grape by an Endophytic Bacterium. Phytopathology 2019, 109, 248–256. [Google Scholar] [CrossRef]
- Lindow, S.; Koutsoukis, R.; Meyer, K.M.; Baccari, C. Control of Pierce’s disease of grape with Paraburkholderia phytofirmans PsJN in the field. Phytopathology 2023. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Sue, T.; Obolonkin, V.; Griffiths, H.; Villas-Boas, S.G. An Exometabolomics Approach to Monitoring Microbial Contamination in Microalgal Fermentation Processes by Using Metabolic Footprint Analysis. Appl. Environ. Microb. 2011, 77, 7605–7610. [Google Scholar] [CrossRef]
- Drenos, F. Mechanistic insights from combining genomics with metabolomics. Curr. Opin. Lipidol. 2017, 28, 99–103. [Google Scholar] [CrossRef]
- Liu, R.; Bao, Z.X.; Zhao, P.J.; Li, G.H. Advances in the Study of Metabolomics and Metabolites in Some Species Interactions. Molecules 2021, 26, 3311. [Google Scholar] [CrossRef]
- Villas-Boas, S.G.; Noel, S.; Lane, G.A.; Attwood, G.; Cookson, A. Extracellular metabolomics: A metabolic footprinting approach to assess fiber degradation in complex media. Anal. Biochem. 2006, 349, 297–305. [Google Scholar] [CrossRef]
- Villas-Boas, S.G.; Mas, S.; Akesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass. Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.; Purcell, A.H.; Lindow, S.E. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol. 2003, 69, 7319–7327. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.P.; Purcell, A.H.; Lindow, S.E. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 2004, 101, 1737–1742. [Google Scholar] [CrossRef]
- Frommel, M.I.; Nowak, J.; Lazarovits, G. Growth Enhancement and Developmental Modifications of in Vitro Grown Potato (Solanum tuberosum spp. tuberosum) as Affected by a Nonfluorescent Pseudomonas sp. Plant Physiol. 1991, 96, 928–936. [Google Scholar] [CrossRef]
- Weilharter, A.; Mitter, B.; Shin, M.V.; Chain, P.S.G.; Nowak, J.; Sessitsch, A. Complete Genome Sequence of the Plant Growth-Promoting Endophyte Burkholderia phytofirmans Strain PsJN. J. Bacteriol. 2011, 193, 3383–3384. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Moll, L.; Badosa, E.; Planas, M.; Feliu, L.; Montesinos, E.; Bonaterra, A. Antimicrobial Peptides With Antibiofilm Activity Against. Front. Microbiol. 2021, 12, 753874. [Google Scholar] [CrossRef]
- Kosina, S.M.; Danielewicz, M.A.; Mohammed, M.; Ray, J.; Suh, Y.; Yilmaz, S.; Singh, A.K.; Arkin, A.P.; Deutschbauer, A.M.; Northen, T.R. Exometabolomics Assisted Design and Validation of Synthetic Obligate Mutualism. ACS Synth. Biol. 2016, 5, 569–576. [Google Scholar] [CrossRef]
- Bowen, B.P.; Northen, T.R. Dealing with the unknown: Metabolomics and metabolite atlases. J. Am. Soc. Mass. Spectrom. 2010, 21, 1471–1476. [Google Scholar] [CrossRef]
- Yao, Y.; Sun, T.; Wang, T.; Ruebel, O.; Northen, T.; Bowen, B.P. Analysis of Metabolomics Datasets with High-Performance Computing and Metabolite Atlases. Metabolites 2015, 5, 431–442. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Erbilgin, O.; Rubel, O.; Louie, K.B.; Trinh, M.; Raad, M.; Wildish, T.; Udwary, D.; Hoover, C.; Deutsch, S.; Northen, T.R.; et al. MAGI: A Method for Metabolite Annotation and Gene Integration. ACS Chem. Biol. 2019, 14, 704–714. [Google Scholar] [CrossRef]
- Merkel, D. Docker: Lightweight linux containers for consistent development and deployment. Linux J. 2014, 2014, 2. [Google Scholar]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nordberg, H.; Shabalov, I.; Aerts, A.; Cantor, M.; Goodstein, D.; Kuo, A.; Minovitsky, S.; Nikitin, R.; Ohm, R.A.; et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012, 40, D26–D32. [Google Scholar] [CrossRef]
- Delcher, A.L.; Phillippy, A.; Carlton, J.; Salzberg, S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002, 30, 2478–2483. [Google Scholar] [CrossRef]
- Michelini, S.; Balakrishnan, B.; Parolo, S.; Matone, A.; Mullaney, J.A.; Young, W.; Gasser, O.; Wall, C.; Priami, C.; Lombardo, R.; et al. A reverse metabolic approach to weaning: In silico identification of immune-beneficial infant gut bacteria, mining their metabolism for prebiotic feeds and sourcing these feeds in the natural product space. Microbiome 2018, 6, 171. [Google Scholar] [CrossRef]
- Levy, R.; Borenstein, E. Reverse Ecology: From systems to environments and back. Adv. Exp. Med. Biol. 2012, 751, 329–345. [Google Scholar]
- Carr, R.; Borenstein, E. NetSeed: A network-based reverse-ecology tool for calculating the metabolic interface of an organism with its environment. Bioinformatics 2012, 28, 734–735. [Google Scholar] [CrossRef]
- Levy, R.; Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 2013, 110, 12804–12809. [Google Scholar] [CrossRef]
- Kreimer, A.; Doron-Faigenboim, A.; Borenstein, E.; Freilich, S. NetCmpt: A network-based tool for calculating the metabolic competition between bacterial species. Bioinformatics 2012, 28, 2195–2197. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef]
- Hibi, M.; Fukuda, D.; Kenchu, C.; Nojiri, M.; Hara, R.; Takeuchi, M.; Aburaya, S.; Aoki, W.; Mizutani, K.; Yasohara, Y.; et al. A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes. Commun. Biol. 2021, 4, 16. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Martinez-Montiel, N.; Garcia-Sanchez, J.; Perez, Y.T.R.; Martinez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 208, 85–98. [Google Scholar] [CrossRef]
- Bansal, P.; Morgat, A.; Axelsen, K.B.; Muthukrishnan, V.; Coudert, E.; Aimo, L.; Hyka-Nouspikel, N.; Gasteiger, E.; Kerhornou, A.; Neto, T.B.; et al. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 2022, 50, D693–D700. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes-a 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Ionescu, M.; Zaini, P.A.; Baccari, C.; Tran, S.; da Silva, A.M.; Lindow, S.E. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. USA 2014, 111, E3910–E3918. [Google Scholar] [CrossRef]
- Smolka, M.B.; Martins, D.; Winck, F.V.; Santoro, C.E.; Castellari, R.R.; Ferrari, F.; Brum, I.J.; Galembeck, E.; Coletta, H.D.; Machado, M.A.; et al. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics 2003, 3, 224–237. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Martyn, A.; Kopriva, S. Exometabolomic Profiling of Bacterial Strains as Cultivated Using Arabidopsis Root Extract as the Sole Carbon Source. Mol. Plant-Microbe Interact. 2018, 31, 803–813. [Google Scholar] [CrossRef]
- van Hoogstraten, S.W.G.; Kuik, C.; Arts, J.J.C.; Cillero-Pastor, B. Molecular imaging of bacterial biofilms-a systematic review. Crit. Rev. Microbiol. 2023. [Google Scholar] [CrossRef]
- Lawson, C.E.; Harcombe, W.R.; Hatzenpichler, R.; Lindemann, S.R.; Löffler, F.E.; O’Malley, M.A.; Martín, H.G.; Pfleger, B.F.; Raskin, L.; Venturelli, O.S.; et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 2019, 17, 725–741. [Google Scholar] [CrossRef]
- Verbeeck, N.; Caprioli, R.M.; Van de Plas, R. Unsupervised machine learning for exploratory data analysis in imaging mass spectrometry. Mass. Spectrom. Rev. 2020, 39, 245–291. [Google Scholar] [CrossRef]
- Flynn, K.J.; Dickson, D.M.J.; Al-Amoudi, O.A. The ratio of glutamine:glutamate in microalgae: A biomarker for N-status suitable for use at natural cell densities. J. Plankton Res. 1989, 11, 165–170. [Google Scholar] [CrossRef]
- Daugherty, M.P.; Rashed, A.; Almeida, R.P.P.; Perring, T.M. Vector preference for hosts differing in infection status: Sharpshooter movement and Xylella fastidiosa transmission. Ecol. Entomol. 2011, 36, 654–662. [Google Scholar] [CrossRef]
- Douglas, A.E. The B vitamin nutrition of insects: The contributions of diet, microbiome and horizontally acquired genes. Curr. Opin. Insect. Sci. 2017, 23, 65–69. [Google Scholar] [CrossRef]
- Sasek, V.; Nováková, M.; Dobrev, P.I.; Valentová, O.; Burketová, L. β-aminobutyric acid protects Brassica napus plants from infection by Leptosphaeria maculans. Resistance induction or a direct antifungal effect? Eur. J. Plant Pathol. 2012, 133, 279–289. [Google Scholar] [CrossRef]
- Pajot, E.; Le Corre, D.; Silué, D. Phytogard® and DL-β-amino Butyric Acid (BABA) Induce Resistance to Downy Mildew (Bremia Lactucae) in Lettuce (Lactuca sativa L). Eur. J. Plant Pathol. 2001, 107, 861–869. [Google Scholar] [CrossRef]
- Obata, K. Synaptic inhibition and γ-aminobutyric acid in the mammalian central nervous system. Proc. Jpn. Acad. B-Phys. 2013, 89, 139–156. [Google Scholar] [CrossRef]
- Wang, H.; Zhi, W.; Qu, H.; Lin, H.; Jiang, Y. Application of alpha-aminoisobutyric acid and beta-aminoisobutyric acid inhibits pericarp browning of harvested longan fruit. Chem. Cent. J. 2015, 9, 54. [Google Scholar] [CrossRef]
- Phinney, B.O. Growth Response of Single-Gene Dwarf Mutants in Maize to Gibberellic Acid. Proc. Natl. Acad. Sci. USA 1956, 42, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Hershey, D.M.; Wang, L.; Bogdanove, A.J.; Peters, R.J. An ent-kaurene-derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. New Phytol. 2015, 206, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, T.; Tarkowská, D.; Strnad, M.; Hedden, P. Gibberellins-Terpenoid Plant Hormones: Biological Importance and Chemical Analysis. Collect Czech. Chem. C 2011, 76, 1669–1686. [Google Scholar] [CrossRef]
- Tudzynski, B. Gibberellin biosynthesis in fungi: Genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 2005, 66, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Nett, R.S.; Montanares, M.; Marcassa, A.; Lul, X.; Nagel, R.; Charles, T.C.; Hedden, P.; Rojas, M.C.; Peters, R.J. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 2017, 13, 69–74. [Google Scholar] [CrossRef]
- Joo, G.J.; Kang, S.M.; Hamayun, M.; Kim, S.K.; Na, C.I.; Shin, D.H.; Lee, I.J. Burkholderia sp KCTC 11096BP as a newly isolated gibberellin producing bacterium. J. Microbiol. 2009, 47, 167–171. [Google Scholar] [CrossRef]
- Vergine, M.; Nicolì, F.; Sabella, E.; Aprile, A.; De Bellis, L.; Luvisi, A. Secondary Metabolites in Xylella fastidiosa–Plant Interaction. Pathogens 2020, 9, 675. [Google Scholar] [CrossRef]
- Novelli, S.; Gismondi, A.; Di Marco, G.; Canuti, L.; Nanni, V.; Canini, A. Plant defense factors involved in Olea europaea resistance against Xylella fastidiosa infection. J. Plant Res. 2019, 132, 439–455. [Google Scholar] [CrossRef]
- Azevedo, J.L.; Araujo, W.L.; Lacava, P.T. The diversity of citrus endophytic bacteria and their interactions with Xylella fastidiosa and host plants. Genet. Mol. Biol. 2016, 39, 476–491. [Google Scholar] [CrossRef]
- Caserta, R.; Souza-Neto, R.R.; Takita, M.A.; Lindow, S.; Souza, A. Ectopic expression of Xylella fastidiosa rpfF conferring production of diffusible signal factor in transgenic tobacco and citrus alters pathogen behavior and reduces disease severity. Mol. Plant-Microbe Interact. MPMI 2017, 30, 866–875. [Google Scholar] [CrossRef] [PubMed]

| Bacteria | Strain | Original Host | Origin | Reference |
|---|---|---|---|---|
| Xylella fastidiosa subsp. fastidiosa | Temecula1 Wild-Type (WT) | Vitis vinifera | Temecula, California, EUA | [27,40] |
| ΔrpfF | – | – | [41] | |
| Paraburkholderia phytofirmans | PsJN | Allium cepa | Ontario, Canada | [42,43] |
| Compound Name | Gene ID | Reciprocal Score | e-Score Reaction-to-Gene | Database ID Reaction-to-Gene | e-Score Gene-to-Reaction | Database ID Gene-to-Reaction | MAGI Score |
|---|---|---|---|---|---|---|---|
| 2-Aminoisobutyric acid | PD0094 | 2.00 | 200.00 | ALANINE--TRNA-LIGASE-RXN | 200.00 | ALANINE--TRNA-LIGASE-RXN | 1.58 |
| PD1696 | 2.00 | 200.00 | RXN-16659 | 200.00 | RXN-16659 | 1.58 | |
| 2.00 | 200.00 | RXN-16649 | 200.00 | RXN-16649 | 1.58 | ||
| PD1823 | 2.00 | 200.00 | RHEA:20249 | 200.00 | RHEA:20249 | 1.58 | |
| PD1864 | 2.00 | 200.00 | RHEA:11224 | 200.00 | RHEA:11224 | 1.58 | |
| PD1865 | 2.00 | 200.00 | RHEA:23374 | 200.00 | RHEA:23374 | 1.58 | |
| Alanine | PD1864 | 2.00 | 200.00 | RHEA:11224 | 200.00 | RHEA:11224 | 6.33 |
| PD1823 | 2.00 | 200.00 | RHEA:20249 | 200.00 | RHEA:20249 | 6.33 | |
| PD0094 | 2.00 | 200.00 | ALANINE--TRNA-LIGASE-RXN | 200.00 | ALANINE--TRNA-LIGASE-RXN | 6.33 | |
| Arginine | PD0116 | 2.00 | 34.19 | ARGININE--TRNA-LIGASE-RXN | 32.31 | ARGININE--TRNA-LIGASE-RXN | 4.01 |
| Asparagine | PD1947 | 2.00 | 200.00 | ASPARAGINE--TRNA-LIGASE-RXN | 200.00 | ASPARAGINE--TRNA-LIGASE-RXN | 6.33 |
| Aspartate | PD0089 | 2.00 | 200.00 | RHEA:12228 | 200.00 | RHEA:12228 | 6.33 |
| 2.00 | 200.00 | RHEA:11375 | 200.00 | RHEA:11375 | 6.33 | ||
| PD0166 | 2.00 | 200.00 | RHEA:22630 | 200.00 | RHEA:22630 | 6.33 | |
| PD0291 | 2.00 | 200.00 | RHEA:10932 | 200.00 | RHEA:10932 | 6.33 | |
| PD0868 | 2.00 | 200.00 | RHEA:25877 | 200.00 | RHEA:25877 | 6.33 | |
| PD0946 | 2.00 | 200.00 | ASPARTATE--TRNA-LIGASE-RXN | 200.00 | ASPARTATE--TRNA-LIGASE-RXN | 6.33 | |
| PD1273 | 2.00 | 200.00 | RHEA:23777 | 200.00 | RHEA:23777 | 6.33 | |
| PD1274 | 2.00 | 200.00 | RHEA:20015 | 200.00 | RHEA:20015 | 6.33 | |
| PD1627 | 2.00 | 200.00 | RHEA:15753 | 200.00 | RHEA:15753 | 6.33 | |
| Biotin | PD0043 | 2.00 | 140.61 | 2.8.1.6-RXN | 145.10 | 2.8.1.6-RXN | 5.80 |
| PD1071 | 2.00 | 58.42 | RHEA:31118 | 62.94 | RHEA:31118 | 4.66 | |
| 2.00 | 58.42 | BIOTINLIG-RXN | 62.94 | BIOTINLIG-RXN | 4.66 | ||
| PD1494 | 2.00 | 167.61 | DETHIOBIOTIN-SYN-RXN | 165.73 | DETHIOBIOTIN-SYN-RXN | 1.51 | |
| Cysteine | PD0655 | 2.00 | 200.00 | RHEA:13285 | 200.00 | RHEA:13285 | 6.33 |
| PD1812 | 2.00 | 156.15 | RHEA:20400 | 154.24 | RHEA:20400 | 5.93 | |
| PD1841 | 2.00 | 143.27 | ACSERLY-RXN | 137.99 | ACSERLY-RXN | 5.77 | |
| PD0118 | 2.00 | 134.13 | RHEA:19398 | 132.24 | RHEA:19398 | 5.71 | |
| 2.00 | 134.13 | RHEA:25159 | 132.24 | RHEA:25159 | 5.71 | ||
| PD0690 | 2.00 | 133.98 | RXN0-308 | 132.09 | RXN0-308 | 5.71 | |
| PD0287 | 2.00 | 126.84 | CYSTEINE--TRNA-LIGASE-RXN | 124.08 | CYSTEINE--TRNA-LIGASE-RXN | 5.62 | |
| Gibberellic acid | PD0286 | 0.01 | 0.89 | RHEA:36115 | 7.41 | RHEA:25891 | 0.43 |
| PD0716 | 0.01 | 0.54 | RHEA:36115 | 159.33 | RHEA:13804 | 0.38 | |
| Nicotinic acid | PD0393 | 2.00 | 200.00 | RHEA:36166 | 200.00 | RHEA:36166 | 6.33 |
| PD1310 | 2.00 | 42.72 | RHEA:14545 | 40.84 | RHEA:14545 | 4.26 | |
| Glutamic acid | PD0650 | 2.00 | 200.00 | RHEA:17130 | 200.00 | RHEA:17130 | 6.33 |
| PD0654 | 2.00 | 200.00 | RHEA:18052 | 200.00 | RHEA:18052 | 6.33 | |
| PD0399 | 2.00 | 200.00 | RHEA:18633 | 200.00 | RHEA:18633 | 6.33 | |
| PD2062 | 2.00 | 200.00 | RHEA:11613 | 200.00 | RHEA:11613 | 6.33 | |
| 2.00 | 200.00 | RHEA:15504 | 200.00 | RHEA:15504 | 6.33 | ||
| PD1358 | 2.00 | 200.00 | RHEA:16574 | 200.00 | RHEA:16574 | 6.33 | |
| 2.00 | 200.00 | RHEA:14329 | 200.00 | RHEA:14329 | 6.33 | ||
| PD1266 | 2.00 | 200.00 | RHEA:23746 | 200.00 | RHEA:23746 | 6.33 | |
| PD0089 | 2.00 | 200.00 | RHEA:12228 | 200.00 | RHEA:12228 | 6.33 | |
| PD0839 | 2.00 | 200.00 | RHEA:24385 | 200.00 | RHEA:24385 | 6.33 | |
| PD1447 | 2.00 | 200.00 | GMP-SYN-GLUT-RXN | 200.00 | GMP-SYN-GLUT-RXN | 6.33 | |
| PD1848 | 2.00 | 200.00 | GLURS-RXN | 200.00 | GLURS-RXN | 6.33 | |
| PD1026 | 2.00 | 200.00 | RHEA:45804 | 200.00 | RHEA:45804 | 6.33 | |
| 2.00 | 200.00 | RHEA:16170 | 200.00 | RHEA:16170 | 6.33 | ||
| PD0851 | 2.00 | 200.00 | RHEA:14908 | 200.00 | RHEA:14908 | 6.33 | |
| PD0785 | 2.00 | 200.00 | RHEA:15136 | 200.00 | RHEA:15136 | 6.33 | |
| PD2063 | 2.00 | 200.00 | RHEA:12131 | 200.00 | RHEA:12131 | 6.33 | |
| 2.00 | 200.00 | RHEA:32192 | 200.00 | RHEA:32192 | 6.33 | ||
| 2.00 | 200.00 | GLUTAMATE-SYNTHASE-FERREDOXIN-RXN | 200.00 | GLUTAMATE-SYNTHASE-FERREDOXIN-RXN | 6.33 | ||
| PD0398 | 2.00 | 200.00 | RHEA:18633 | 200.00 | RHEA:18633 | 6.33 | |
| PD0170 | 2.00 | 200.00 | RHEA:21735 | 200.00 | RHEA:21735 | 6.33 | |
| PD0296 | 2.00 | 200.00 | RHEA:14879 | 200.00 | RHEA:14879 | 6.33 | |
| PD0541 | 2.00 | 200.00 | RHEA:15890 | 200.00 | RHEA:15890 | 6.33 | |
| PD0110 | 2.00 | 200.00 | RHEA:13239 | 200.00 | RHEA:13239 | 6.33 | |
| PD0655 | 2.00 | 200.00 | RHEA:13285 | 200.00 | RHEA:13285 | 6.33 | |
| Glutamine | PD1447 | 2.00 | 200.00 | GMP-SYN-GLUT-RXN | 200.00 | GMP-SYN-GLUT-RXN | 6.33 |
| PD0584 | 2.00 | 200.00 | GLUTAMINE--TRNA-LIGASE-RXN | 200.00 | GLUTAMINE--TRNA-LIGASE-RXN | 6.33 | |
| PD2063 | 2.00 | 200.00 | GLUTAMATE-SYNTHASE-FERREDOXIN-RXN | 200.00 | GLUTAMATE-SYNTHASE-FERREDOXIN-RXN | 6.33 | |
| Glycine | PD1750 | 2.00 | 200.00 | RHEA:15482 | 200.00 | RHEA:15482 | 6.33 |
| PD0620 | 2.00 | 200.00 | GCVMULTI-RXN | 200.00 | GCVMULTI-RXN | 6.33 | |
| 2.00 | 200.00 | GCVP-RXN | 200.00 | GCVP-RXN | 6.33 | ||
| PD1810 | 2.00 | 200.00 | GCVMULTI-RXN | 200.00 | GCVMULTI-RXN | 6.33 | |
| 2.00 | 200.00 | GCVP-RXN | 200.00 | GCVP-RXN | 6.33 | ||
| PD0827 | 2.00 | 200.00 | RHEA:17453 | 200.00 | RHEA:17453 | 6.33 | |
| PD0704 | 2.00 | 200.00 | RHEA:19938 | 200.00 | RHEA:19938 | 6.33 | |
| PD0844 | 2.00 | 200.00 | RHEA:13557 | 200.00 | RHEA:13557 | 6.33 | |
| PD0773 | 2.00 | 200.00 | RXN0-7068 | 200.00 | RXN0-7068 | 6.33 | |
| 2.00 | 200.00 | RXN0-7082 | 200.00 | RXN0-7082 | 6.33 | ||
| PD0841 | 2.00 | 170.04 | GLYCINE--TRNA-LIGASE-RXN | 168.01 | GLYCINE--TRNA-LIGASE-RXN | 6.06 | |
| PD0840 | 2.00 | 164.75 | GLYCINE--TRNA-LIGASE-RXN | 162.87 | GLYCINE--TRNA-LIGASE-RXN | 6.02 | |
| Histidine | PD1267 | 2.00 | 200.00 | RHEA:20641 | 200.00 | RHEA:20641 | 6.33 |
| PD1772 | 2.00 | 60.65 | RHEA:20641 | 58.79 | RHEA:20641 | 4.66 | |
| PD1270 | 2.00 | 26.76 | HISTIDINE--TRNA-LIGASE-RXN | 24.84 | HISTIDINE--TRNA-LIGASE-RXN | 3.76 | |
| Isoleucine | PD1437 | 2.00 | 200.00 | ISOLEUCINE--TRNA-LIGASE-RXN | 200.00 | ISOLEUCINE--TRNA-LIGASE-RXN | 6.33 |
| Leucine | PD1230 | 2.00 | 200.00 | LEUCINE--TRNA-LIGASE-RXN | 200.00 | LEUCINE--TRNA-LIGASE-RXN | 6.33 |
| Lysine | PD0404 | 2.00 | 200.00 | LYSINE--TRNA-LIGASE-RXN | 200.00 | LYSINE--TRNA-LIGASE-RXN | 6.33 |
| PD1514 | 2.00 | 69.38 | RXN-1961 | 78.45 | RXN-1961 | 4.86 | |
| PD2000 | 2.00 | 55.90 | RHEA:15944 | 53.31 | RHEA:15944 | 1.14 | |
| Methionine | PD1590 | 2.00 | 200.00 | METHIONINE--TRNA-LIGASE-RXN | 200.00 | METHIONINE--TRNA-LIGASE-RXN | 6.33 |
| 2.00 | 200.00 | RXN-16165 | 200.00 | RXN-16165 | 6.33 | ||
| Phenylalanine | PD1911 | 2.00 | 200.00 | PHENYLALANINE--TRNA-LIGASE-RXN | 200.00 | PHENYLALANINE--TRNA-LIGASE-RXN | 6.33 |
| PD0665 | 2.00 | 200.00 | RXN-15898 | 200.00 | RXN-15898 | 1.58 | |
| Proline | PD1635 | 2.00 | 200.00 | PROLINE--TRNA-LIGASE-RXN | 200.00 | PROLINE--TRNA-LIGASE-RXN | 6.33 |
| Serine | PD0612 | 2.00 | 200.00 | RHEA:26437 | 200.00 | RHEA:26437 | 6.33 |
| 2.00 | 200.00 | RHEA:10532 | 200.00 | RHEA:10532 | 6.33 | ||
| PD1750 | 2.00 | 200.00 | RHEA:15482 | 200.00 | RHEA:15482 | 6.33 | |
| PD1318 | 2.00 | 200.00 | RXN0-2161 | 200.00 | RXN0-2161 | 6.33 | |
| 2.00 | 200.00 | SERINE--TRNA-LIGASE-RXN | 200.00 | SERINE--TRNA-LIGASE-RXN | 6.33 | ||
| PD0613 | 2.00 | 200.00 | RHEA:26437 | 200.00 | RHEA:26437 | 6.33 | |
| 2.00 | 200.00 | RHEA:10532 | 200.00 | RHEA:10532 | 6.33 | ||
| Threonine | PD1916 | 2.00 | 200.00 | THREONINE--TRNA-LIGASE-RXN | 200.00 | THREONINE--TRNA-LIGASE-RXN | 6.33 |
| Tryptophan | PD0612 | 2.00 | 200.00 | RHEA:10532 | 200.00 | RHEA:10532 | 6.33 |
| 2.00 | 200.00 | RHEA:26437 | 200.00 | RHEA:26437 | 6.33 | ||
| PD0613 | 2.00 | 200.00 | RHEA:10532 | 200.00 | RHEA:10532 | 6.33 | |
| 2.00 | 200.00 | RHEA:26437 | 200.00 | RHEA:26437 | 6.33 | ||
| PD1650 | 2.00 | 47.82 | TRYPTOPHAN--TRNA-LIGASE-RXN | 45.82 | TRYPTOPHAN--TRNA-LIGASE-RXN | 4.38 | |
| Tyrosine | PD0665 | 2.00 | 200.00 | RXN-15898 | 200.00 | RXN-15898 | 6.33 |
| PD0132 | 2.00 | 33.23 | TYROSINE--TRNA-LIGASE-RXN | 31.35 | TYROSINE--TRNA-LIGASE-RXN | 3.98 | |
| Valine | PD0102 | 2.00 | 200.00 | VALINE--TRNA-LIGASE-RXN | 200.00 | VALINE--TRNA-LIGASE-RXN | 6.33 |
| Compound Name | Gene ID | Reciprocal Score | e-Score Reaction-to-Gene | Database ID Reaction-to-Gene | e-Score Gene-to-Reaction | Database ID Gene-to-Reaction | MAGI Score |
|---|---|---|---|---|---|---|---|
| Cysteine | Bphyt_3930 | 2.00 | 200.00 | RHEA:13285 | 200.00 | RHEA:13285 | 6.33 |
| Bphyt_4072 | 2.00 | 200.00 | RXN-17172 | 200.00 | RXN-17172 | 6.33 | |
| Bphyt_2579 | 2.00 | 200.00 | RXN-15881 | 200.00 | RXN-15881 | 6.33 | |
| 2.00 | 200.00 | RXN0-308 | 200.00 | RXN0-308 | 6.33 | ||
| Bphyt_3068 | 2.00 | 200.00 | RHEA:28784 | 200.00 | RHEA:28784 | 6.33 | |
| Bphyt_0110 | 2.00 | 200.00 | RHEA:13285 | 200.00 | RHEA:13285 | 6.33 | |
| Bphyt_3953 | 2.00 | 200.00 | RHEA:13285 | 200.00 | RHEA:13285 | 6.33 | |
| Bphyt_2579 | 2.00 | 177.70 | RXN-14385 | 175.79 | RXN-14385 | 6.13 | |
| Bphyt_2510 | 2.00 | 175.50 | CYSTEINE--TRNA-LIGASE-RXN | 171.77 | CYSTEINE--TRNA-LIGASE-RXN | 6.10 | |
| Gibberellic acid | Bphyt_5231 | 0.01 | 18.39 | RXN-14318 | 39.82 | RXN-16827 | 0.23 |
| 0.01 | 18.39 | RXN-14317 | 39.82 | RXN-16827 | 0.23 | ||
| 0.01 | 18.39 | RXN-7617 | 39.82 | RXN-16827 | 0.23 | ||
| Nicotinamide | Bphyt_5413 | 2.00 | 200.00 | RHEA:16150 | 200.00 | RHEA:16150 | 6.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feitosa-Junior, O.R.; Lubbe, A.; Kosina, S.M.; Martins-Junior, J.; Barbosa, D.; Baccari, C.; Zaini, P.A.; Bowen, B.P.; Northen, T.R.; Lindow, S.E.; et al. The Exometabolome of Xylella fastidiosa in Contact with Paraburkholderia phytofirmans Supernatant Reveals Changes in Nicotinamide, Amino Acids, Biotin, and Plant Hormones. Metabolites 2024, 14, 82. https://doi.org/10.3390/metabo14020082
Feitosa-Junior OR, Lubbe A, Kosina SM, Martins-Junior J, Barbosa D, Baccari C, Zaini PA, Bowen BP, Northen TR, Lindow SE, et al. The Exometabolome of Xylella fastidiosa in Contact with Paraburkholderia phytofirmans Supernatant Reveals Changes in Nicotinamide, Amino Acids, Biotin, and Plant Hormones. Metabolites. 2024; 14(2):82. https://doi.org/10.3390/metabo14020082
Chicago/Turabian StyleFeitosa-Junior, Oseias R., Andrea Lubbe, Suzanne M. Kosina, Joaquim Martins-Junior, Deibs Barbosa, Clelia Baccari, Paulo A. Zaini, Benjamin P. Bowen, Trent R. Northen, Steven E. Lindow, and et al. 2024. "The Exometabolome of Xylella fastidiosa in Contact with Paraburkholderia phytofirmans Supernatant Reveals Changes in Nicotinamide, Amino Acids, Biotin, and Plant Hormones" Metabolites 14, no. 2: 82. https://doi.org/10.3390/metabo14020082
APA StyleFeitosa-Junior, O. R., Lubbe, A., Kosina, S. M., Martins-Junior, J., Barbosa, D., Baccari, C., Zaini, P. A., Bowen, B. P., Northen, T. R., Lindow, S. E., & da Silva, A. M. (2024). The Exometabolome of Xylella fastidiosa in Contact with Paraburkholderia phytofirmans Supernatant Reveals Changes in Nicotinamide, Amino Acids, Biotin, and Plant Hormones. Metabolites, 14(2), 82. https://doi.org/10.3390/metabo14020082








