Untargeted Plasma Metabolomic Profiling in Patients with Depressive Disorders: A Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Study Population and Sample Collection
2.3. Sample Preparation
2.4. LC/MS Parameters
2.5. Analysis Optimization
2.6. Compound Identification and Statistical Analysis
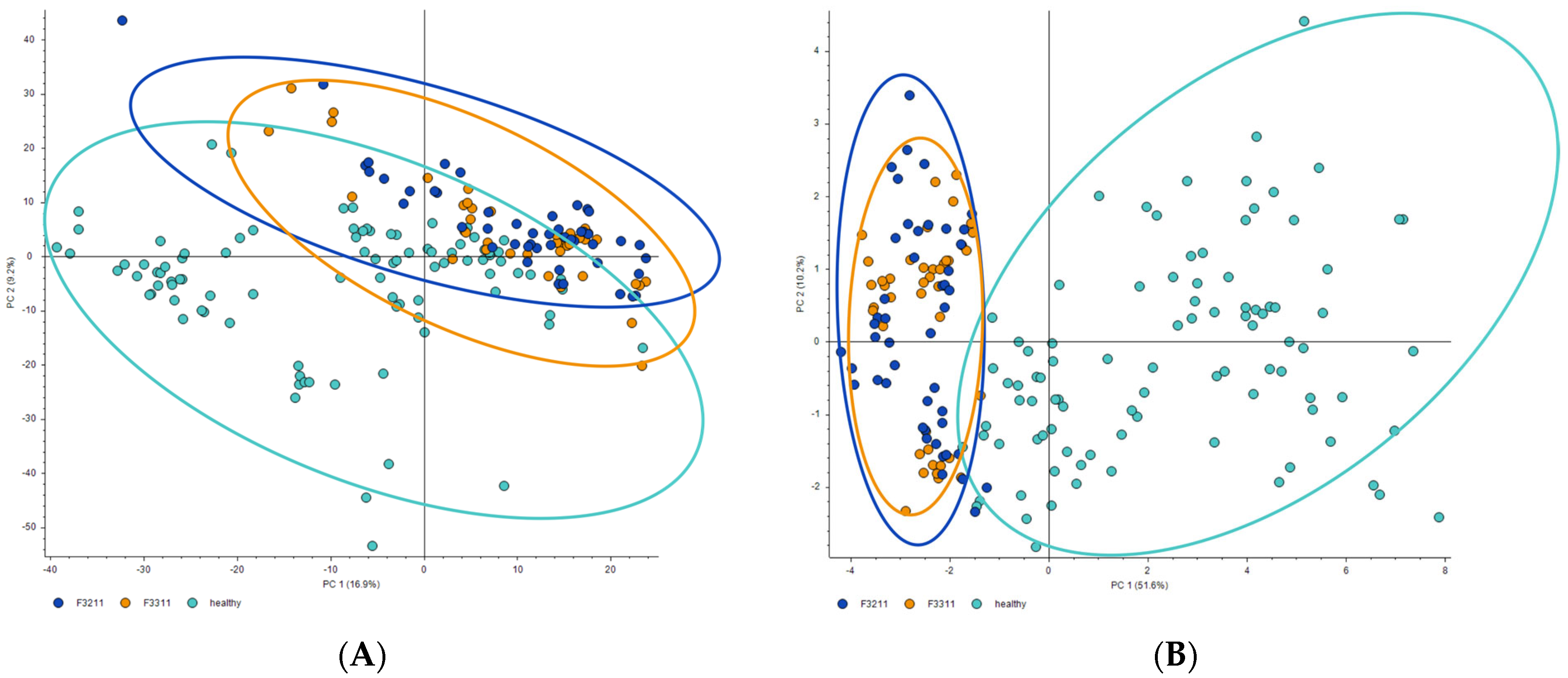
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moffitt, T.E.; Caspi, A.; Taylor, A.; Kokaua, J.; Milne, B.J.; Polanczyk, G.; Poulton, R. How common are common mental disorders? Evidence that lifetime prevalence rates are doubled by prospective versus retrospective ascertainment. Psychol. Med. 2010, 40, 899–909. [Google Scholar] [CrossRef]
- Lim, G.Y.; Tam, W.W.; Lu, Y.; Ho, C.S.; Zhang, M.W.; Ho, R.C. Prevalence of Depression in the Community from 30 Countries between 1994 and 2014. Sci. Rep. 2018, 8, 2861. [Google Scholar] [CrossRef]
- Bueno-Notivol, J.; Gracia-García, P.; Olaya, B.; Lasheras, I.; López-Antón, R.; Santabárbara, J. Prevalence of depression during the COVID-19 outbreak: A meta-analysis of community-based studies. Int. J. Clin. Health Psychol. 2021, 21, 100196. [Google Scholar] [CrossRef]
- van de Velde, S.; Bracke, P.; Levecque, K. Gender differences in depression in 23 European countries. Cross-national variation in the gender gap in depression. Soc. Sci. Med. 2010, 71, 305–313. [Google Scholar] [CrossRef]
- Pu, J.; Liu, Y.; Gui, S.; Tian, L.; Yu, Y.; Song, X.; Zhong, X.; Chen, X.; Chen, W.; Zheng, P.; et al. Metabolomic changes in animal models of depression: A systematic analysis. Mol. Psychiatry 2021, 26, 7328–7336. [Google Scholar] [CrossRef]
- Cipriani, A.; Furukawa, T.A.; Salanti, G.; Chaimani, A.; Atkinson, L.Z.; Ogawa, Y.; Leucht, S.; Ruhe, H.G.; Turner, E.H.; Higgins, J.P.T.; et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: A systematic review and network meta-analysis. Lancet 2018, 391, 1357–1366. [Google Scholar] [CrossRef]
- Schiepers, O.J.G.; Wichers, M.C.; Maes, M. Cytokines and major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2005, 29, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Pruessner, M.; Hellhammer, D.H.; Pruessner, J.C.; Lupien, S.J. Self-reported depressive symptoms and stress levels in healthy young men: Associations with the cortisol response to awakening. Psychosom. Med. 2003, 65, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.; Keshavan, M. The neurobiology of depression: An integrated view. Asian J. Psychiatr. 2017, 27, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Kennis, M.; Gerritsen, L.; van Dalen, M.; Williams, A.; Cuijpers, P.; Bockting, C. Prospective biomarkers of major depressive disorder: A systematic review and meta-analysis. Mol. Psychiatry 2020, 25, 321–338. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.L.; Honer, W.G.; Bergmann, K.; Falkai, P.; Lütjohann, D.; Bayer, T.A. Reductions in cholesterol and synaptic markers in association cortex in mood disorders. Bipolar Disord. 2005, 7, 449–455. [Google Scholar] [CrossRef]
- Feyissa, A.M.; Chandran, A.; Stockmeier, C.A.; Karolewicz, B. Reduced levels of NR2A and NR2B subunits of NMDA receptor and PSD-95 in the prefrontal cortex in major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2009, 33, 70–75. [Google Scholar] [CrossRef]
- Kennedy, S.E.; Koeppe, R.A.; Young, E.A.; Zubieta, J.-K. Dysregulation of endogenous opioid emotion regulation circuitry in major depression in women. Arch. Gen. Psychiatry 2006, 63, 1199–1208. [Google Scholar] [CrossRef]
- Cai, S.; Huang, S.; Hao, W. New hypothesis and treatment targets of depression: An integrated view of key findings. Neurosci. Bull. 2015, 31, 61–74. [Google Scholar] [CrossRef]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef]
- Couttas, T.A.; Jieu, B.; Rohleder, C.; Leweke, F.M. Current State of Fluid Lipid Biomarkers for Personalized Diagnostics and Therapeutics in Schizophrenia Spectrum Disorders and Related Psychoses: A Narrative Review. Front. Psychiatry 2022, 13, 885904. [Google Scholar] [CrossRef]
- Al-Sulaiti, H.; Almaliti, J.; Naman, C.B.; Al Thani, A.A.; Yassine, H.M. Metabolomics Approaches for the Diagnosis, Treatment, and Better Disease Management of Viral Infections. Metabolites 2023, 13, 948. [Google Scholar] [CrossRef]
- Lazofsky, A.; Brinker, A.; Rivera-Núñez, Z.; Buckley, B. A comparison of four liquid chromatography-mass spectrometry platforms for the analysis of zeranols in urine. Anal. Bioanal. Chem. 2023, 415, 4885–4899. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chu, S.; Tan, S.; Yin, X.; Jiang, Y.; Dai, X.; Gong, X.; Fang, X.; Tian, D. Towards Higher Sensitivity of Mass Spectrometry: A Perspective from the Mass Analyzers. Front. Chem. 2021, 9, 813359. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Ebbels, T.M.D.; van der Hooft, J.J.J.; Chatelaine, H.; Broeckling, C.; Zamboni, N.; Hassoun, S.; Mathé, E.A. Recent advances in mass spectrometry-based computational metabolomics. Curr. Opin. Chem. Biol. 2023, 74, 102288. [Google Scholar] [CrossRef]
- Misra, B.B. New software tools, databases, and resources in metabolomics: Updates from 2020. Metabolomics 2021, 17, 49. [Google Scholar] [CrossRef]
- Kontou, E.E.; Walter, A.; Alka, O.; Pfeuffer, J.; Sachsenberg, T.; Mohite, O.S.; Nuhamunada, M.; Kohlbacher, O.; Weber, T. UmetaFlow: An untargeted metabolomics workflow for high-throughput data processing and analysis. J. Cheminform. 2023, 15, 52. [Google Scholar] [CrossRef]
- Souza, A.L.; Patti, G.J. A Protocol for Untargeted Metabolomic Analysis: From Sample Preparation to Data Processing. Methods Mol. Biol. 2021, 2276, 357–382. [Google Scholar] [CrossRef]
- Kostikova, V.A.; Chernonosov, A.A.; Kuznetsov, A.A.; Petrova, N.V.; Krivenko, D.A.; Chernysheva, O.A.; Wang, W.; Erst, A.S. Identification of Flavonoids in the Leaves of Eranthis longistipitata (Ranunculaceae) by Liquid Chromatography with High-Resolution Mass Spectrometry (LC-HRMS). Plants 2021, 10, 2146. [Google Scholar] [CrossRef] [PubMed]
- Erst, A.S.; Chernonosov, A.A.; Petrova, N.V.; Kulikovskiy, M.S.; Maltseva, S.Y.; Wang, W.; Kostikova, V.A. Investigation of Chemical Constituents of Eranthis longistipitata (Ranunculaceae): Coumarins and Furochromones. Int. J. Mol. Sci. 2021, 23, 406. [Google Scholar] [CrossRef]
- Kostikova, V.A.; Petrova, N.V.; Shaldaeva, T.M.; Koval, V.V.; Chernonosov, A.A. Non-Targeted Screening of Metabolites in Aqueous-Ethanol Extract from Spiraea hypericifolia (Rosaceae) Using LC-HRMS. Int. J. Mol. Sci. 2023, 24, 13872. [Google Scholar] [CrossRef] [PubMed]
- Petrova, N.V.; Chernonosov, A.A.; Koval, V.V.; Andreeva, V.Y.; Erst, A.S.; Kuznetsov, A.A.; Kulikovskiy, M.S.; Wang, W.; Yu, S.-X.; Kostikova, V.A. LC-HRMS for the Identification of Quercetin and Its Derivatives in Spiraea hypericifolia (Rosaceae) and Anatomical Features of Its Leaves. Plants 2023, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, M.-I.; Termentzi, A.; Kasiotis, K.M.; Cheilari, A.; Stathopoulou, K.; Machera, K.; Aligiannis, N. Untargeted Ultrahigh-Performance Liquid Chromatography-Hybrid Quadrupole-Orbitrap Mass Spectrometry (UHPLC-HRMS) Metabolomics Reveals Propolis Markers of Greek and Chinese Origin. Molecules 2021, 26, 456. [Google Scholar] [CrossRef] [PubMed]
- Elessawy, F.M.; Wright, D.; Vandenberg, A.; El-Aneed, A.; Purves, R.W. Mass Spectrometry-Based Untargeted Metabolomics Reveals the Importance of Glycosylated Flavones in Patterned Lentil Seed Coats. J. Agric. Food Chem. 2023, 71, 3541–3549. [Google Scholar] [CrossRef]
- Li, B.; Fu, Y.; Xi, H.; Liu, S.; Zhao, W.; Li, P.; Fan, W.; Wang, D.; Sun, S. Untargeted Metabolomics Using UHPLC-HRMS Reveals Metabolic Changes of Fresh-Cut Potato during Browning Process. Molecules 2023, 28, 3375. [Google Scholar] [CrossRef]
- Züllig, T.; Zandl-Lang, M.; Trötzmüller, M.; Hartler, J.; Plecko, B.; Köfeler, H.C. A Metabolomics Workflow for Analyzing Complex Biological Samples Using a Combined Method of Untargeted and Target-List Based Approaches. Metabolites 2020, 10, 342. [Google Scholar] [CrossRef]
- Feng, Y.-L.; Baesu, A. Influence of data acquisition modes and data analysis approaches on non-targeted analysis of phthalate metabolites in human urine. Anal. Bioanal. Chem. 2023, 415, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Courraud, J.; Ernst, M.; Svane Laursen, S.; Hougaard, D.M.; Cohen, A.S. Studying Autism Using Untargeted Metabolomics in Newborn Screening Samples. J. Mol. Neurosci. 2021, 71, 1378–1393. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Xie, P. The potential for metabolomics in the study and treatment of major depressive disorder and related conditions. Expert Rev. Proteom. 2020, 17, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro-Costa, L.N.F.; Carneiro, B.A.; Alves, G.S.; Silva, D.H.L.; Guimaraes, D.F.; Souza, L.S.; Bandeira, I.D.; Beanes, G.; Scippa, A.M.; Quarantini, L.C. Metabolomics of Major Depressive Disorder: A Systematic Review of Clinical Studies. Cureus 2022, 14, e23009. [Google Scholar] [CrossRef]
- Bot, M.; Milaneschi, Y.; Al-Shehri, T.; Amin, N.; Garmaeva, S.; Onderwater, G.L.J.; Pool, R.; Thesing, C.S.; Vijfhuizen, L.S.; Vogelzangs, N.; et al. Metabolomics Profile in Depression: A Pooled Analysis of 230 Metabolic Markers in 5283 Cases with Depression and 10,145 Controls. Biol. Psychiatry 2020, 87, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Folberth, J.; Begemann, K.; Jöhren, O.; Schwaninger, M.; Othman, A. MS2 and LC libraries for untargeted metabolomics: Enhancing method development and identification confidence. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1145, 122105. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.-H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Zaric, B.L.; Obradovic, M.; Bajic, V.; Haidara, M.A.; Jovanovic, M.; Isenovic, E.R. Homocysteine and Hyperhomocysteinaemia. Curr. Med. Chem. 2019, 26, 2948–2961. [Google Scholar] [CrossRef]
- Setién-Suero, E.; Suárez-Pinilla, M.; Suárez-Pinilla, P.; Crespo-Facorro, B.; Ayesa-Arriola, R. Homocysteine and cognition: A systematic review of 111 studies. Neurosci. Biobehav. Rev. 2016, 69, 280–298. [Google Scholar] [CrossRef] [PubMed]
- Momin, M.; Jia, J.; Fan, F.; Li, J.; Dou, J.; Chen, D.; Huo, Y.; Zhang, Y. Relationship between plasma homocysteine level and lipid profiles in a community-based Chinese population. Lipids Health Dis. 2017, 16, 54. [Google Scholar] [CrossRef]
- Ganguly, P.; Alam, S.F. Role of homocysteine in the development of cardiovascular disease. Nutr. J. 2015, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Moradi, F.; Lotfi, K.; Armin, M.; Clark, C.C.T.; Askari, G.; Rouhani, M.H. The association between serum homocysteine and depression: A systematic review and meta-analysis of observational studies. Eur. J. Clin. Investig. 2021, 51, e13486. [Google Scholar] [CrossRef] [PubMed]
- Ueland, P.M. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2011, 34, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gui, X.; Wu, L.; Tian, S.; Wang, H.; Xie, L.; Wu, W. Amino acid metabolism, lipid metabolism, and oxidative stress are associated with post-stroke depression: A metabonomics study. BMC Neurol. 2020, 20, 250. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.; Singh, N. Homocysteine excess: Delineating the possible mechanism of neurotoxicity and depression. Fundam. Clin. Pharmacol. 2015, 29, 522–528. [Google Scholar] [CrossRef]
- Setoyama, D.; Kato, T.A.; Hashimoto, R.; Kunugi, H.; Hattori, K.; Hayakawa, K.; Sato-Kasai, M.; Shimokawa, N.; Kaneko, S.; Yoshida, S.; et al. Plasma Metabolites Predict Severity of Depression and Suicidal Ideation in Psychiatric Patients-A Multicenter Pilot Analysis. PLoS ONE 2016, 11, e0165267. [Google Scholar] [CrossRef]
- Di Pierro, F.; Orsi, R.; Settembre, R. Role of betaine in improving the antidepressant effect of S-adenosyl-methionine in patients with mild-to-moderate depression. J. Multidiscip. Healthc. 2015, 8, 39–45. [Google Scholar] [CrossRef]
- Di Pierro, F.; Settembre, R. Preliminary results of a randomized controlled trial carried out with a fixed combination of S-adenosyl-L-methionine and betaine versus amitriptyline in patients with mild depression. Int. J. Gen. Med. 2015, 8, 73–78. [Google Scholar] [CrossRef]
- Liu, R.T.; Rowan-Nash, A.D.; Sheehan, A.E.; Walsh, R.F.L.; Sanzari, C.M.; Korry, B.J.; Belenky, P. Reductions in anti-inflammatory gut bacteria are associated with depression in a sample of young adults. Brain Behav. Immun. 2020, 88, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Jin, K.; Jiang, C.; Pan, F.; Wu, J.; Luan, H.; Zhao, Z.; Chen, J.; Mou, T.; Wang, Z.; et al. A pilot exploration of multi-omics research of gut microbiome in major depressive disorders. Transl. Psychiatry 2022, 12, 8. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, P.; Li, Y.; Wu, J.; Tan, X.; Zhou, J.; Sun, Z.; Chen, X.; Zhang, G.; Zhang, H.; et al. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Sci. Adv. 2020, 6, eaba8555. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.I.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef] [PubMed]
- Lirong, W.; Mingliang, Z.; Mengci, L.; Qihao, G.; Zhenxing, R.; Xiaojiao, Z.; Tianlu, C. The clinical and mechanistic roles of bile acids in depression, Alzheimer’s disease, and stroke. Proteomics 2022, 22, e2100324. [Google Scholar] [CrossRef]
- Caspani, G.; Kennedy, S.; Foster, J.A.; Swann, J. Gut microbial metabolites in depression: Understanding the biochemical mechanisms. Microb. Cell 2019, 6, 454–481. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, J.; Wang, J.; Liu, Z.; Wang, X.; Kang, P.; Yang, C.; Liu, P.; Zhang, K. Abnormal gut microbiota and bile acids in patients with first-episode major depressive disorder and correlation analysis. Psychiatry Clin. Neurosci. 2022, 76, 321–328. [Google Scholar] [CrossRef]
- MahmoudianDehkordi, S.; Bhattacharyya, S.; Brydges, C.R.; Jia, W.; Fiehn, O.; Rush, A.J.; Dunlop, B.W.; Kaddurah-Daouk, R. Gut Microbiome-Linked Metabolites in the Pathobiology of Major Depression with or without Anxiety-A Role for Bile Acids. Front. Neurosci. 2022, 16, 937906. [Google Scholar] [CrossRef]
- Hosein, E.A.; Smart, M.; Hawkinsk, K.; Rochon, S.; Strasberg, Z. The enzymic synthesis of gamma-butyrobetaine and its CoA ester derivative. Arch. Biochem. Biophys. 1962, 96, 246–251. [Google Scholar] [CrossRef]
- Möhler, H. The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology 2012, 62, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Lydiard, R.B. The role of GABA in anxiety disorders. J. Clin. Psychiatry 2003, 64 (Suppl. 3), 21–27. [Google Scholar] [PubMed]
- Liu, T.; Deng, K.; Xue, Y.; Yang, R.; Yang, R.; Gong, Z.; Tang, M. Carnitine and Depression. Front. Nutr. 2022, 9, 853058. [Google Scholar] [CrossRef] [PubMed]
- Vaz, F.M.; Wanders, R.J.A. Carnitine biosynthesis in mammals. Biochem. J. 2002, 361, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, P.; Zhao, X.; Zhang, Y.; Hu, C.; Li, J.; Zhao, J.; Zhou, J.; Xie, P.; Xu, G. Discovery and validation of plasma biomarkers for major depressive disorder classification based on liquid chromatography-mass spectrometry. J. Proteome Res. 2015, 14, 2322–2330. [Google Scholar] [CrossRef]
- Nie, L.-J.; Liang, J.; Shan, F.; Wang, B.-S.; Mu, Y.-Y.; Zhou, X.-H.; Xia, Q.-R. L-Carnitine and Acetyl-L-Carnitine: Potential Novel Biomarkers for Major Depressive Disorder. Front. Psychiatry 2021, 12, 671151. [Google Scholar] [CrossRef]
- Gall, W.E.; Beebe, K.; Lawton, K.A.; Adam, K.-P.; Mitchell, M.W.; Nakhle, P.J.; Ryals, J.A.; Milburn, M.V.; Nannipieri, M.; Camastra, S.; et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS ONE 2010, 5, e10883. [Google Scholar] [CrossRef]
- Moradi, Y.; Albatineh, A.N.; Mahmoodi, H.; Gheshlagh, R.G. The relationship between depression and risk of metabolic syndrome: A meta-analysis of observational studies. Clin. Diabetes Endocrinol. 2021, 7, 4. [Google Scholar] [CrossRef]
- Fernandes, B.S.; Salagre, E.; Enduru, N.; Grande, I.; Vieta, E.; Zhao, Z. Insulin resistance in depression: A large meta-analysis of metabolic parameters and variation. Neurosci. Biobehav. Rev. 2022, 139, 104758. [Google Scholar] [CrossRef]
- Kim, H.G.; Han, E.H.; Jang, W.-S.; Choi, J.H.; Khanal, T.; Park, B.H.; Tran, T.P.; Chung, Y.C.; Jeong, H.G. Piperine inhibits PMA-induced cyclooxygenase-2 expression through downregulating NF-κB, C/EBP and AP-1 signaling pathways in murine macrophages. Food Chem. Toxicol. 2012, 50, 2342–2348. [Google Scholar] [CrossRef]
- Mittal, R.; Gupta, R.L. In vitro antioxidant activity of piperine. Methods Find. Exp. Clin. Pharmacol. 2000, 22, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Koul, I.B.; Kapil, A. Evaluation of the liver protective potential of piperine, an active principle of black and long peppers. Planta Med. 1993, 59, 413–417. [Google Scholar] [CrossRef] [PubMed]
- D’Hooge, R.; Pei, Y.Q.; Raes, A.; Lebrun, P.; van Bogaert, P.P.; de Deyn, P.P. Anticonvulsant activity of piperine on seizures induced by excitatory amino acid receptor agonists. Arzneimittelforschung 1996, 46, 557–560. [Google Scholar] [PubMed]
- Fu, M.; Sun, Z.-H.; Zuo, H.-C. Neuroprotective effect of piperine on primarily cultured hippocampal neurons. Biol. Pharm. Bull. 2010, 33, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, C.; Wang, M.; Li, W.; Matsumoto, K.; Tang, Y. Antidepressant like effects of piperine in chronic mild stress treated mice and its possible mechanisms. Life Sci. 2007, 80, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Wattanathorn, J.; Chonpathompikunlert, P.; Muchimapura, S.; Priprem, A.; Tankamnerdthai, O. Piperine, the potential functional food for mood and cognitive disorders. Food Chem. Toxicol. 2008, 46, 3106–3110. [Google Scholar] [CrossRef]
- Lee, S.A.; Hong, S.S.; Han, X.H.; Hwang, J.S.; Oh, G.J.; Lee, K.S.; Lee, M.K.; Hwang, B.Y.; Ro, J.S. Piperine from the fruits of Piper longum with inhibitory effect on monoamine oxidase and antidepressant-like activity. Chem. Pharm. Bull. 2005, 53, 832–835. [Google Scholar] [CrossRef]
- Mao, Q.-Q.; Huang, Z.; Zhong, X.-M.; Xian, Y.-F.; Ip, S.-P. Brain-derived neurotrophic factor signalling mediates the antidepressant-like effect of piperine in chronically stressed mice. Behav. Brain Res. 2014, 261, 140–145. [Google Scholar] [CrossRef]
| Indicators | Patients with Depressive Disorders | Healthy Controls | p-Value | |||
|---|---|---|---|---|---|---|
| Overall | Depressive Episode (F32.11) | Recurrent Depressive Disorders (F33.11) | p-Value | |||
| Age, years | 40.5 (37; 48) | 42 (39; 49) | 39 (37; 45,5) | 0.759 | 40 (29; 47) | 0.359 |
| Gender (male, n (%)/female, n (%)) | 2(6.7%)/28(93.3%) | 1(7.1%)/13(92.9%) | 1(6.7%)/15(93.3%) | 0.922 | 2(6.7%)/28(93.3%) | 1.0 |
| Duration of disease, years | 0.67 (0.33; 4.5) | 0.42 (0.25; 0.58) | 5 (3.5; 10) | 0.0001 * | - | - |
| Number of depressive episodes experienced (excluding the current one) | 2 (2; 2) | 0 | 2 (1.5; 2.5) | 0.0001 * | - | - |
| Duration of the current affective episode, months | - | 6 (3; 10) | 3 (2; 8) | 0.089 | - | - |
| BMI | 25.1 (22.3; 27.3) | 25.1 (22.9; 27.4) | 25.1 (21.6; 27.1) | 0.786 | 24.7 (22.5; 28.8) | 0.531 |
| No. | Calc. MW, Da | Formula | Name | mzCloud Score | F3211/ Healthy (p-Value) | F3311/ Healthy (p-Value) |
|---|---|---|---|---|---|---|
| 1 | 117,07860 | C5H11NO2 | Betaine | 93.2 | 0.37 (0.0003) | 0.31 (0.0017) |
| 2 | 145,10947 | C7H15NO2 | g-Butyrobetaine | - | 0.56 (0.002) | 0.53 (0.015) |
| 3 | 172,07044 | C6H10N3O3 | 6-Diazonio-5-oxo-L-norleucine | - | 0.39 (0.0006) | 0.39 (0.002) |
| 4 | 201,17209 | C11H23NO2 | 11-Aminoundecanoic acid | - | 0.07 (0.002) | 0.08 (0.004) |
| 5 | 214,11747 | C9H16N3O3 | Methyl N-acetyl-2-diazonionorleucinate | - | 0.31 (0.004) | 0.30 (0.002) |
| 6 | 216,09639 | C10H16O5 | (4S,5S,8S,10R)-4,5,8-trihydroxy-10-methyl- 3,4,5,8,9,10-hexahydro-2H-oxecin-2-one | 60.2 | 0.36 (0.001) | 0.42 (0.005) |
| 7 | 272,15857 | C10H20N6O3 | Glycyl-glycyl-argininal | - | 0.07 (0.00007) | 0.08 (0.001) |
| 8 | 285,13511 | C17H19NO3 | Piperine | 87.5 | 0.59(0.03) | 0.51 (0.005) |
| 9 | 295,24900 | C18H33NO2 | (2E,4E)-N-(2-Hydroxy-2-methylpropyl)- 2,4-tetradecadienamide | 52.2 * | 7.2 (0.0007) | 8.8 (0.003) |
| 10 | 319,24639 | C20H33NO2 | 17α-Methyl-androstan-3-hydroxyimine-17β-ol | 55.2 | 20.3 (0.0004) | 26.2 (0.0002) |
| 11 | 367,41679 | C25H53N | Dilaurylmethylamine | - | 0.28 (0.003) | 0.37 (0.003) |
| 12 | 390,2752 | C24H38O4 | 12-Ketodeoxycholic acid | - | 0.42 (0.005) | 0.34 (0.007) |
| 13 | 465,52609 | C32H67N | Dicetylamine | - | 0.02 (0.004) | 0.02 (0.006) |
| 14 | 465,52684 | C32H67N | Dicetylamine | - | 0.11(0.1) | 0.13(0.1) |
| 15 | 497,89136 | C8H7N2O17PS2 | - | - | 0.48 (0.002) | 0.54 (0.014) |
| 16 | 519,33092 | C26H50NO7P | 1-Linoleoyl-2-hydroxy-sn-glycero-3-PC | - | 0.17 (0.006) | 0.14 (0.0006) |
| 17 | 701,85267 | C12H5N10O16P3S2 | - | - | 0.49 (0.002) | 0.57 (0.019) |
| 18 | 837,82763 | - | - | - | 0.46 (0.0005) | 0.54 (0.012) |
| 19 | 905,81407 | - | - | - | 0.47 (0.002) | 0.44 (0.013) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernonosov, A.A.; Mednova, I.A.; Levchuk, L.A.; Mazurenko, E.O.; Roschina, O.V.; Simutkin, G.G.; Bokhan, N.A.; Koval, V.V.; Ivanova, S.A. Untargeted Plasma Metabolomic Profiling in Patients with Depressive Disorders: A Preliminary Study. Metabolites 2024, 14, 110. https://doi.org/10.3390/metabo14020110
Chernonosov AA, Mednova IA, Levchuk LA, Mazurenko EO, Roschina OV, Simutkin GG, Bokhan NA, Koval VV, Ivanova SA. Untargeted Plasma Metabolomic Profiling in Patients with Depressive Disorders: A Preliminary Study. Metabolites. 2024; 14(2):110. https://doi.org/10.3390/metabo14020110
Chicago/Turabian StyleChernonosov, Alexander A., Irina A. Mednova, Lyudmila A. Levchuk, Ekaterina O. Mazurenko, Olga V. Roschina, German G. Simutkin, Nikolay A. Bokhan, Vladimir V. Koval, and Svetlana A. Ivanova. 2024. "Untargeted Plasma Metabolomic Profiling in Patients with Depressive Disorders: A Preliminary Study" Metabolites 14, no. 2: 110. https://doi.org/10.3390/metabo14020110
APA StyleChernonosov, A. A., Mednova, I. A., Levchuk, L. A., Mazurenko, E. O., Roschina, O. V., Simutkin, G. G., Bokhan, N. A., Koval, V. V., & Ivanova, S. A. (2024). Untargeted Plasma Metabolomic Profiling in Patients with Depressive Disorders: A Preliminary Study. Metabolites, 14(2), 110. https://doi.org/10.3390/metabo14020110








