Blood-Derived Metabolic Signatures as Biomarkers of Injury Severity in Traumatic Brain Injury: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Characteristics and Study Design
2.2. Clinical Assessment
2.3. NMR Sample Preparation, Data Acquisition, and Processing
2.4. Statistical Analysis
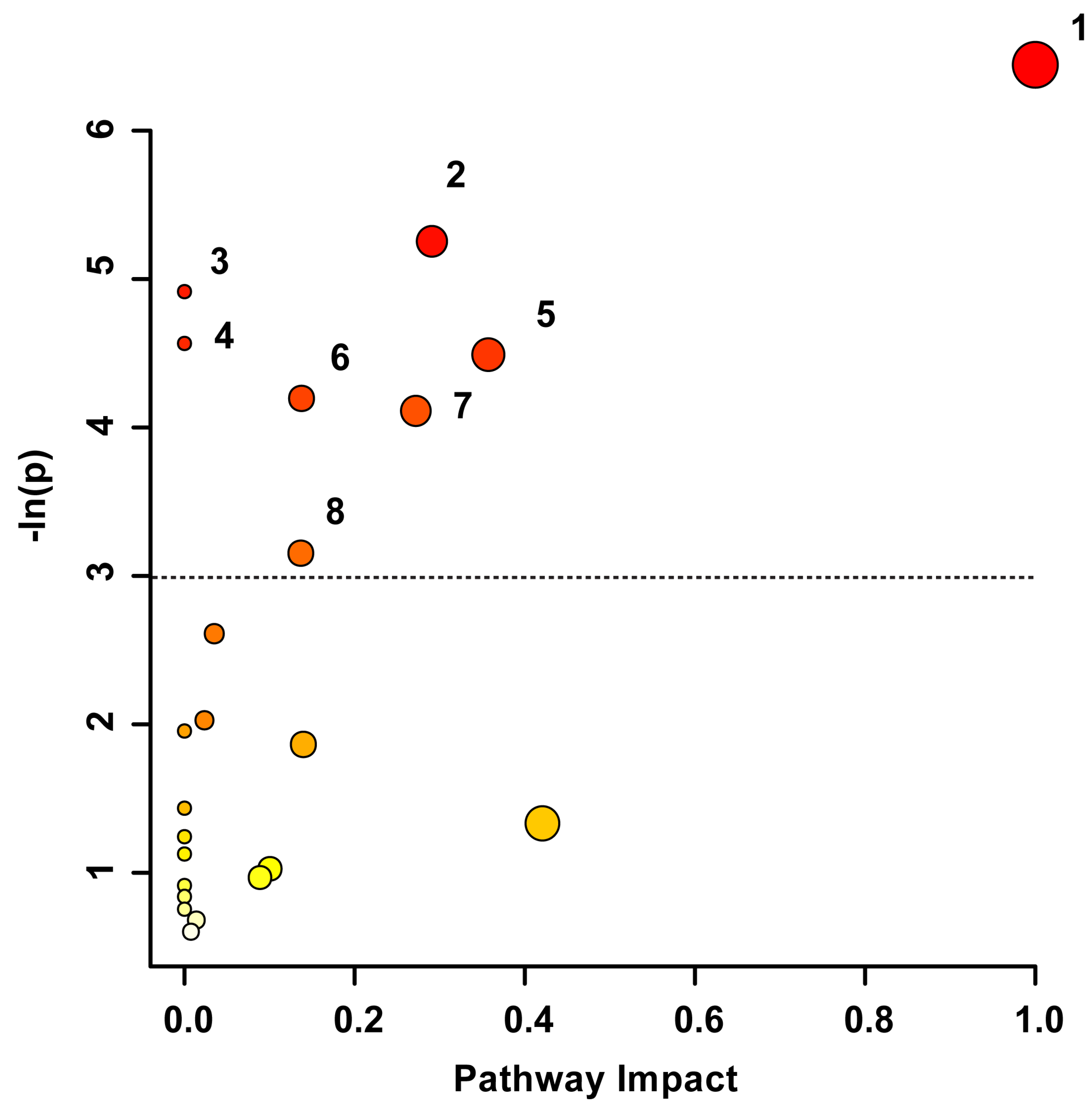
3. Results
3.1. Clinical Demographics
3.2. Metabolomic Profiles Significantly Change over Time following TBI
3.3. Metabolomic Signatures Correlate with Injury Severity
4. Discussion
4.1. General Discussion
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dewan, M.; Rattani, A.; Gupta, S.; Baticulon, R.E.; Hung, Y.; Punchak, M.; Agrawal, A.; Adeleye, A.O.; Shrime, M.G.; Rubiano, A.M.; et al. Estimating the global incidence of traumatic brain injury. J. Neurol. 2019, 130, 1080–1097. [Google Scholar] [CrossRef]
- Meaney, D.F.; Smith, D.H. Biomechanics of concussion. Clin. Sports Med. 2011, 30, 19. [Google Scholar] [CrossRef]
- Siesjo, B.K.; Siesjo, P. Mechanisms of secondary brain injury. Eur. J. Anaesthesiol. 1996, 13, 247–268. [Google Scholar] [CrossRef]
- Abdelhak, A.; Foschi, M.; Abu-Rumeileh, S.; Yue, J.K.; D’Anna, L.; Huss, A.; Oeckl, P.; Ludolph, A.C.; Kuhle, J.; Petzold, A.; et al. Blood GFAP as an emerging biomarker in brain and spinal cord disorders. Nat. Rev. Neurol. 2022, 18, 158–172. [Google Scholar] [CrossRef]
- Oris, C.; Kahouadji, S.; Durif, J.; Bouvier, D.; Sapin, V. S100B, actor and biomarker of mild traumatic brain injury. Int. J. Mol. Sci. 2023, 24, 6602. [Google Scholar] [CrossRef]
- Wanner, Z.R.; Southam, C.G.; Sanghavi, P.; Boora, N.S.; Paxman, E.J.; Dukelow, S.P.; Benson, B.W.; Montina, T.; Metz, G.A.S.; Debert, C.T. Alterations in urine metabolomics following sport-related concussion: A 1H NMR-based analysis. Front. Neurol. 2021, 12, 645829. [Google Scholar] [CrossRef] [PubMed]
- Bykowski, E.A.; Petersson, J.N.; Dukelow, S.; Ho, C.; Debert, C.T.; Montina, T.; Metz, G.A.S. Urinary biomarkers indicative of recovery from spinal cord injury: A pilot study. IBRO Neurosci. Rep. 2021, 10, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Bykowski, E.A.; Petersson, J.N.; Dukelow, S.; Ho, C.; Debert, C.T.; Montina, T.; Metz, G.A.S. Urinary metabolomic signatures as indicators of injury severity following traumatic brain injury: A pilot study. IBRO Neurosci. Rep. 2021, 11, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Bykowski, E.A.; Petersson, J.N.; Dukelow, S.; Ho, C.; Debert, C.T.; Montina, T.; Metz, G.A.S. Identification of serum metabolites as prognostic biomarkers following spinal cord injury: A pilot study. Metabolites 2023, 13, 605. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Ambeskovic, M.; Hopkins, G.; Hoover, T.; Joseph, J.T.; Montina, T.; Metz, G.A.S. Metabolomic signatures of Alzheimer’s disease indicate brain region-specific neurodegenerative progression. Int. J. Mol. Sci. 2023, 24, 14769. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Calabresi, P.A. Metabolomics in multiple sclerosis. Mult. Scler. J. 2016, 22, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Powers, R. NMR metabolomics analysis of parkinson’s disease. Curr. Metabolomics 2013, 1, 191. [Google Scholar] [CrossRef]
- Sternbach, G.L. The glasgow coma scale. J. Emerg. Med. 2000, 19, 67–71. [Google Scholar] [CrossRef]
- Nasreddine, Z.S.; Phillips, N.A.; Bédirian, V.; Charbonneau, S.; Whitehead, V.; Collin, I.; Cummings, J.L.; Chertkow, H. The montreal cognitive assessment, MoCA: A brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc. 2005, 53, 695–699. [Google Scholar] [CrossRef]
- Kidd, D.; Stewart, G.; Baldry, J.; Johnson, J.; Rossiter, D.; Petruckevitch, A.; Thompson, A. The functional independence measure—A comparative validity and reliability study. Disabil. Rehabil. 1995, 17, 10–14. [Google Scholar] [CrossRef]
- Anderson, P.E.; Mahle, D.A.; Doom, T.E.; Reo, N.V.; DelRaso, N.J.; Raymer, M.L. Dynamic adaptive binning: An improved quantification technique for NMR spectroscopic data. Metabolomics 2011, 7, 179–190. [Google Scholar] [CrossRef]
- Box, G.E.P.; Cox, D.R. An analysis of transformations. J. R. Stat. Soc. 1964, 26, 211–252. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Craig, A.; Cloarec, O.; Holmes, E.; Nicholson, J.K.; Lindon, J.C. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal. Chem. 2006, 78, 2262–2267. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Yun, Y.; Liang, F.; Deng, B.; Lai, G.; Vicente Gonçalves, C.M.; Lu, H.; Yan, J.; Huang, X.; Yi, L.; Liang, Y. Informative metabolites identification by variable importance analysis based on random variable combination. Metabolomics 2015, 11, 1539–1551. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Szymanska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Goodpaster, A.M.; Romick-Rosendale, L.E.; Kennedy, M.A. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal. Biochem. 2010, 401, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Boison, D.; Schwarzschild, M.A.; Kochanek, P.M. Purines: Forgotten mediators in traumatic brain injury. J. Neurochem. 2016, 137, 142–153. [Google Scholar] [CrossRef]
- Daines, S.A. The therapeutic potential and limitations of ketones in traumatic brain injury. Front. Neurol. 2021, 12, 723148. [Google Scholar] [CrossRef] [PubMed]
- Fernstrom, J.D. Effects of precursors on brain neurotransmitter synthesis and brain functions. Diabetologia 1981, 20, 281–289. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Price, L.; Wilson, C.; Grant, G. Blood-Brain Barrier Pathophysiology following Traumatic Brain Injury; Laskowitz, D., Grant, G., Eds.; Taylor & Francis: Abingdon, UK, 2015; pp. 85–96. [Google Scholar] [CrossRef]
- Werner, C.; Engelhard, K. Pathophysiology of traumatic brain injury. Br. J. Anaesth. 2007, 99, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Timofeev, I.; Carpenter, K.L.H.; Nortje, J.; Al-Rawi, P.G.; O’Connell, M.T.; Czosnyka, M.; Smielewski, P.; Pickard, J.D.; Menon, D.K.; Kirkpatrick, P.J.; et al. Cerebral extracellular chemistry and outcome following traumatic brain injury: A microdialysis study of 223 patients. Brain 2011, 134, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Babu, M.M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef]
- Di Pietro, V.; O’Halloran, P.; Watson, C.N.; Begum, G.; Acharjee, A.; Yakoub, K.M.; Bentley, C.; Davies, D.J.; Iliceto, P.; Candilera, G.; et al. Unique diagnostic signatures of concussion in the saliva of male athletes: The study of concussion in rugby union through MicroRNAs (SCRUM). Br. J. Sports Med. 2021, 55, 1395–1404. [Google Scholar] [CrossRef]
- Xu, X.; Yang, M.; Zhang, B.; Ge, Q.; Niu, F.; Dong, J.; Zhuang, Y.; Liu, B. Genome-wide interrogation of transfer RNA-derived small RNAs in a mouse model of traumatic brain injury. Neural Regen. Res. 2022, 17, 386–394. [Google Scholar] [CrossRef]
- Puhakka, N.; Das Gupta, S.; Vuokila, N.; Pitkanen, A. Transfer RNA-derived fragments and isomiRs are novel components of chronic TBI-induced neuropathology. Biomedicines 2022, 10, 136. [Google Scholar] [CrossRef]
- Faden, A.I.; Demediuk, P.; Panter, S.S.; Vink, R. The role of excitatory amino acids and NMDA receptors in traumatic brain injury. Science 1989, 244, 798–800. [Google Scholar] [CrossRef]
- Panter, S.S.; Yum, S.W.; Faden, A.I. Alteration in extracellular amino acids after traumatic spinal cord injury. Ann. Neurol. 1990, 27, 96–99. [Google Scholar] [CrossRef]
- Saransaari, P.; Oja, S.S. Beta-alanine release from the adult and developing hippocampus is enhanced by ionotropic glutamate receptors agonists and cell-damaging conditions. Neurochem. Res. 1999, 24, 407–414. [Google Scholar] [CrossRef]
- Flydal, M.I.; Martinez, A. Phenylalanine hydroxylase: Function, structure, and regulation. IUBMB Life 2013, 65, 341–349. [Google Scholar] [CrossRef] [PubMed]
- McGuire, J.L.; Ngwenya, L.B.; McCullumsmith, R.E. Neurotransmitter changes after traumatic brain injury: An update for new treatment strategies. Mol. Psychiatry 2019, 24, 995–1012. [Google Scholar] [CrossRef]
- Ahn, S.; Jung, J.; Jang, I.; Madsen, E.L.; Park, W. Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 2016, 291, 11928–11938. [Google Scholar] [CrossRef]
- Das, D.K.; Engelman, R.M.; Rousou, J.A.; Breyer, R.H. Aerobic vs anaerobic metabolism during ischemia in heart muscle. Ann. Chir. Gynaecol. 1987, 76, 68–75. [Google Scholar]
- Elder, C.; Apple, D.; Bickel, C.; Meyer, R.; Dudley, G. Intramuscular fat and glucose tolerance after spinal cord injury—A cross-sectional study. Spinal Cord 2004, 42, 711–716. [Google Scholar] [CrossRef]
- Shahidi, B.; Shah, S.B.; Esparza, M.; Head, B.P.; Ward, S. Skeletal muscle atrophy and degeneration in a mouse model of traumatic brain injury. J. Neurotrauma 2018, 35, 398–401. [Google Scholar] [CrossRef]
- Felig, P. The glucose-alanine cycle. Metabolism 1973, 22, 179–207. [Google Scholar] [CrossRef]
- Conger, K.A.; Garcia, J.H.; Kauffman, F.C.; Lust, W.D.; Murakami, N.; Passonneau, J.V. Alanine:Glutamate ratios as an index of reversibility of cerebral ischemia in gerbils. Exp. Neurol. 1981, 71, 370–382. [Google Scholar] [CrossRef]


| Participant Code | TBI Type | Glasgow Coma Scale Score | Age | Co-Morbidities | Medications | Blood Collection (Days Post-Injury) | Clinical Assessments (Days Post-Injury) | MoCA | FIM | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial | 6 Month | Initial | 6 Month | Initial | 6 Month | Initial | 6 Month | ||||||
| TBI_02 | Frontal | 3 | 18 | Injury to right ear, right fracture petrous temporal bone | Tylenol | 3 | 218 | 4 | 218 | 25 | 30 | 126 | 126 |
| TBI_03 | Frontal | 10 | 49 | Depression, asthma, EtOH abuse | Docusate sodium, fentanyl, lorazepam, phenytoin, senokot, thiomine, tobradex, multi-vits | 1 | 312 | 20 | 312 | 23 | 26 | 113 | 122 |
| TBI_07 | SDH | 6 | 18 | None | None | 4 | 226 | 59 | 226 | 26 | 27 | 124 | 125 |
| TBI_13 | DAI-Left | 13 | 64 | Multiple face lacerations, nasal fracture, liver laceration, dental injuries | Acetaminophen, docusate sodium, heparin, quetiapine | 4 | 200 | 33 | 200 | 20 | 27 | 112 | 121 |
| TBI_19 | SDH/SAH Bifrontal | 8 | 46 | None | Trazadone, testosterone, seroquel | 2 | 197 | 31 | 197 | 25 | 26 | 113 | 124 |
| TBI_24 | SDH/SAH | 15 | 68 | Chronic lower back pain, liver laceration, bilateral shoulder injuries, torn right rotator cuff | None | 3 | 198 | 70 | 198 | 21 | 23 | 126 | 123 |
| TBI_26 | SDH/SAH | 14 | 48 | None | Tylenol | 2 | 184 | 29 | 184 | 27 | 27 | 124 | 126 |
| TBI_29 | SAH-Right Frontal | 12 | 48 | L2, L4, L5 fracture, sciatic nerve damage, eczema, history of smoking | Tylenol, baclofen, panoloc | 2 | NaN | 16 | NaN | 23 | 23 | 115 | 122 |
| Metabolite | Chemical Shift (ppm) | Paired t/Wilcoxon p-Value | Regulation (% Difference) |
|---|---|---|---|
| L-Phenylalanine.1 | 7.362 | 0.0002 | Down (−60.140%) |
| 1,9-Dimethyluric Acid | 3.290 | 0.0005 | Up (25.651%) |
| Phosphonoacetate.1 | 2.657 | 0.0012 | Up (42.262%) |
| p-Cresol.1 | 7.159 | 0.0013 | Down (−84.436%) |
| L-Phenylalanine.2 | 7.379 | 0.0031 | Down (−30.914%) |
| Glycine.1 | 3.582 | 0.0031 | Up (18.260%) |
| Citric Acid | 2.536 | 0.0040 | Up (35.800%) |
| Phosphonoacetate.2 | 2.672 | 0.0045 | Up (37.428%) |
| 1,3-Dimethyluric Acid.1 | 3.295 | 0.0046 | Up (22.159%) |
| p-Cresol.2 | 7.144 | 0.0050 | Down (−65.625%) |
| 2-Hydroxybutyrate †† | 0.904 | 0.0050 | Down (−79.804%) |
| Glycine.2 | 3.567 | 0.0061 | Up (19.568%) |
| Trimethylamine-N-Oxide | 3.285 | 0.0066 | Up (26.092%) |
| 3-Methyl-2-Oxovaleric Acid | 0.915 | 0.0066 | Down (−75.379%) |
| Creatinine.1 | 3.054 | 0.0078 (W) | Up (21.179%) |
| Levulinate † | 2.456 | 0.0089 | Up (20.412%) |
| Unidentified Multiplet | 0.980 | 0.0107 | Down (−53.026%) |
| Citramalic Acid.1 | 2.478 | 0.0112 | Up (18.255%) |
| 4-Pyridoxate | 2.445 | 0.0153 | Up (16.975%) |
| 1,5-Anhydrosorbitol.1 | 3.360 | 0.0156 (W) | Up (27.891%) |
| 1,3-Dimethyluric Acid.2 † | 3.300 | 0.0156 (W) | Up (20.480%) |
| Citramalic Acid.2 † | 2.489 | 0.0158 | Up (17.716%) |
| Pyruvic Acid | 2.467 | 0.0169 | Up (17.951%) |
| L-Alanine †† | 1.493 | 0.0172 | Up (35.621%) |
| 1,5-Anhydrosorbitol.2 | 3.280 | 0.0178 | Up (20.012%) |
| Guanidoacetate † | 3.804 | 0.0209 | Up (15.256%) |
| 5-Hydroxyindole-3-acetate | 3.572 | 0.0221 | Up (20.959%) |
| Tyrosine | 6.930 | 0.0234 | Down (−33.767%) |
| Glucose.1 | 3.461 | 0.0274 | Down (−14.882%) |
| Glucose.2 | 3.821 | 0.0322 | Down (−9.187%) |
| Unidentified Singlet | 1.212 | 0.0352 | Down (−86.139%) |
| D-Mannose | 5.196 | 0.0391 (W) | Down (−76.053%) |
| Hydroxyphenylacetylglycine | 3.600 | 0.0391 (W) | Up (11.043%) |
| Theophylline | 3.564 | 0.0421 | Up (14.543%) |
| Lactate.1 | 4.109 | 0.0428 | Up (25.843%) |
| Glucose.3 | 3.856 | 0.0438 | Down (−7.688%) |
| π-methylhistidine | 7.970 | 0.0449 | Up (18.003%) |
| Unidentified Multiplet | 2.433 | 0.0476 | Up (15.001%) |
| Creatinine.2 | 4.065 | 0.0487 | Up (24.941%) |
| Acetylphosphate | 2.132 | 0.0490 | Up (14.059%) |
| Methylsuccinic Acid | 2.149 | 0.0497 | Up (13.903%) |
| Lactate.2 † | 4.135 | 0.0571 | Up (29.606%) |
| 1,3,7-Trimethyluric Acid †† | 3.337 | 0.0799 | Up (21.040%) |
| α-Ketoisovaleric Acid † | 1.131 | 0.1389 | Down (−30.615%) |
| Pantothenic Acid † | 0.943 | 0.2278 | Down (−26.969%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bykowski, E.A.; Petersson, J.N.; Dukelow, S.P.; Ho, C.; Debert, C.T.; Montina, T.; Metz, G.A.S. Blood-Derived Metabolic Signatures as Biomarkers of Injury Severity in Traumatic Brain Injury: A Pilot Study. Metabolites 2024, 14, 105. https://doi.org/10.3390/metabo14020105
Bykowski EA, Petersson JN, Dukelow SP, Ho C, Debert CT, Montina T, Metz GAS. Blood-Derived Metabolic Signatures as Biomarkers of Injury Severity in Traumatic Brain Injury: A Pilot Study. Metabolites. 2024; 14(2):105. https://doi.org/10.3390/metabo14020105
Chicago/Turabian StyleBykowski, Elani A., Jamie N. Petersson, Sean P. Dukelow, Chester Ho, Chantel T. Debert, Tony Montina, and Gerlinde A. S. Metz. 2024. "Blood-Derived Metabolic Signatures as Biomarkers of Injury Severity in Traumatic Brain Injury: A Pilot Study" Metabolites 14, no. 2: 105. https://doi.org/10.3390/metabo14020105
APA StyleBykowski, E. A., Petersson, J. N., Dukelow, S. P., Ho, C., Debert, C. T., Montina, T., & Metz, G. A. S. (2024). Blood-Derived Metabolic Signatures as Biomarkers of Injury Severity in Traumatic Brain Injury: A Pilot Study. Metabolites, 14(2), 105. https://doi.org/10.3390/metabo14020105






