Abstract
Background: Carotenoids play essential nutritional and physiological roles in aquatic animals. Since aquatic species cannot synthesize carotenoids de novo, they must obtain these compounds from their diet to meet the physiological and adaptive requirements needed in specific aquaculture stages and conditions. Carotenoid supplementation in Artemia represents a promising strategy to enhance pigmentation, health, and growth in aquaculture species, particularly in larvae and other early developmental stages. Methods: In this study, a β-carotene enrichment process was applied to Artemia metanauplii to investigate the biological fate and potential effects of β-carotene. Results: The results indicated significant β-carotene uptake by Artemia, with peak levels observed at 12 h. Alongside β-carotene, two xanthophylls—canthaxanthin and echinenone—were detected in Artemia, each exhibiting distinct patterns during the enrichment and subsequent depletion phases. The transcriptome analysis identified 2705 differentially expressed genes (DEGs), offering valuable insights into gene functions associated with carotenoid absorption, metabolism, and antioxidant mechanisms. The findings suggest that β-carotene enrichment enhances metabolic activity and energy pathways, supporting the physiological functions of Artemia. Notably, unlike other crustaceans, Artemia lack certain enzymes necessary for converting β-carotene into astaxanthin, restricting them to producing keto-carotenoids like canthaxanthin. Furthermore, the study highlights the upregulation of genes encoding lipid transport proteins, such as CD36 and ABC transporters, which may contribute to carotenoid absorption in Artemia. Additional functional insights are provided by the gene BCO2, which regulates pigmentation by preventing excessive carotenoid accumulation, along with ketolase and hydroxylase enzymes in carotenoid metabolic pathways. Conclusions: This research advances our understanding of carotenoid metabolism in crustaceans, with potential implications for aquaculture nutrition and feed formulation.
1. Introduction
Carotenoids represent a group of conjugated, double-bonded, long-chain terpenoid compounds that are widely distributed in the flesh, skin, and exoskeleton of aquatic animals [1]. The pigmentation and immune function of aquatic animals are closely associated with the composition and concentration of carotenoids within their bodies [2]. In natural ecosystems, carotenoids are synthesized by microorganisms and subsequently accumulate through the food chain, involving zooplankton, crustaceans, and fish, which themselves lack the capability for the de novo synthesis of carotenoids [3]. However, intensive aquaculture practices have disrupted these natural communities, resulting in a scarcity of effective pigment sources. This disruption is a key factor leading to carotenoid deficiencies in cultured species [4]. To address this issue, carotenoid supplementation in feed has become an important strategy in aquaculture to improve coloration, enhance taste, boost disease resistance, and support reproductive performance [2].
Despite advancements in formulated diets, rearing early-stage aquatic animals remains heavily dependent on live feed [5]. Among live feeds, the newly hatched nauplii, metanauplii, and adults of the micro-crustacean Artemia spp. are widely utilized for marine larval rearing, owing to their availability and acceptance across diverse species [6,7]. Traditionally, Artemia has been considered an excellent larval live feed for larvae, primarily due to its provision of digestible protein [6]. Furthermore, Artemia is a non-selective filter feeder capable of filtering particles between 1 and 50 μm [8], which allows it to serve as a live carrier for encapsulating and transporting specific nutrients within its digestive tract [9]. Although previous research has primarily focused on enriching Artemia with essential polyunsaturated fatty acids (PUFAs), phospholipids, amino acids, and fat-soluble vitamins to meet larval nutritional requirements [10], the use of Artemia as carotenoid carriers in the aquaculture food chain is gaining increasing attention [11].
Similar to most crustaceans, Artemia cannot synthesize carotenoids and must acquire them from their diets. While endogenous carotenoids are present in Artemia eggs and newly hatched nauplii, their levels decline significantly during as development progresses in the absence of exogenous carotenoid supplementation [12,13]. Carotenoids from microalgae, serving as a primary source of natural feed, can be efficiently transferred to Artemia, thereby enhancing the growth and health of metanauplii and adults through carotenoid accumulation [9,14]. Although there has been substantial research on the biochemical pathways of carotenoid metabolism in Artemia, studies focused on the underlying genes and transcriptomic data remain limited. RNA sequencing (RNA-seq) has emerged as a cost-effective strategy for exploring carotenoid metabolism-related genes on a genome-wide scale [15]. This study provides fundamental data on qualitative and quantitative changes in carotenoids within Artemia, with and without carotenoid enrichment, and offers insights into the molecular mechanisms underlying carotenoid metabolism in crustaceans.
2. Materials and Methods
2.1. Artemia Cyst Incubation
Artemia metanauplii were obtained by hatching Artemia sinica cysts. The cysts were initially decapsulated with sodium hypochlorite (2 g of Artemia cysts/1 g of active chlorine) and subsequently neutralized with sodium thiosulfate (Na2S2O3) dissolved in filtered seawater (2%, w/v). Following this treatment, the cysts were incubated in a 10 L cylindro-conical PVC container at a density of 2 g/L filtered rearing seawater (salinity: 30 ‰; temperature: 28 °C; pH: 8–8.5) with continuous aeration and 2000 lux illumination over 24 h, in accordance with the protocol of Reis et al. [7]. After hatching, nauplii were separated from hatching debris and transferred to a 20 L cylindrical PVC container with fresh rearing seawater, where they were held until they reached the metanauplius stage (Instars II through V, mouth and anus opening). At this stage, the desired number of metanauplii were randomly selected for subsequent enrichment and starvation procedures.
2.2. Artemia Enrichment Experiment Design
During the enrichment process, Artemia metanauplii were maintained under the previously described experimental conditions, with the diet supplemented with a pure carotenoid emulsion. This emulsion was prepared by dissolving 0 mg (control), 100 mg, 300 mg, or 500 mg of synthesized β-carotene (98% purity, Adamas) in a minimal amount of chloroform, which was subsequently mixed with Tween 80. Following thorough mixing, the chloroform was removed from the carotenoid emulsions using a nitrogen blower (HUXI, Shanghai, China).
The enrichment procedure involved incubating Artemia metanauplii (30 individuals/mL) with β-carotene substrates for 24 h in sixteen 30 L cylindrical PVC containers, each containing 20 L of filter seawater. Four experimental enrichment treatments (0 mg/L, 5 mg/L, 15 mg/L, and 25 mg/L β-carotene) were established using the prepared carotenoid emulsions, with each treatment replicated four times. To examine carotenoid dynamics, metanauplius samples were collected after 6, 12, 18, or 24 h of exposure, then stored at −80 °C for subsequent analyses.
2.3. Artemia Starvation Experimental Sampling After β-Carotene Enrichment
An independent experiment was conducted to investigate the carotenoid metabolic conversion in Artemia metanauplii following β-carotene enrichment. After the 12 h enrichment procedure described above, enriched Artemia were transferred to sixteen fresh 30 L cylindrical PVC containers with non-pigmented rearing seawater and were fed appropriate quantities of pigment-free yeast for an additional 24 h. Samples of Artemia were collected every 6 h and stored for further analysis.
2.4. Analysis of the Carotenoid Content and Composition
Carotenoids were extracted from freeze-dried Artemia metanauplii using an acetone and methanol solution (2:1, v/v). The extraction process was repeated at least three times until no further color was observed in the solvent. The resulting extracts were pooled into new tubes and subsequently dried using a nitrogen blower. The carotenoid residues in each tube were redissolved in a mobile phase solution (acetonitrile/methanol, 70:30, v/v) and filtered through 0.22 μm hydrophilic polypropylene discs (Pall Corp) for the carotenoid composition analysis. All extraction procedures were conducted under low light conditions to minimize the photo-oxidation of the carotenoids. For the compositional analysis, samples were analyzed via ultraperformance convergence chromatography (UPC2, Waters ACQUITY, Milford, CT, USA) using a Waters ACQUITY HSS C18 SB column (1.8 μm, 2.1 mm × 150 mm). The UPC2 conditions included a mobile phase consisting of A (100% liquid CO2) and B (100% methanol) with a flow rate of 0.6 mL/min and an injection volume of 10 μL. The gradient program began with an initial ratio of 90% A to 10% B for 7 min, following by a shift to 80% B to 20% C for 5 min, and a return to the initial ratio over 4 min. Carotenoid concentrations in each sample were detected at 475 nm using a photodiode array (PDA) detector and quantified by calculating peak areas against known standards, including astaxanthin, zeaxanthin, β-cryptoxanthin, canthaxanthin, echinenone, and β-carotene (Figure 1). Standards for astaxanthin, canthaxanthin, zeaxanthin, echinenone, and β-carotene were obtained from Sigma-Aldrich (St. Louis, MO, USA), while the β-cryptoxanthin standard was sourced from CaroteNature (Zurich, Switzerland).
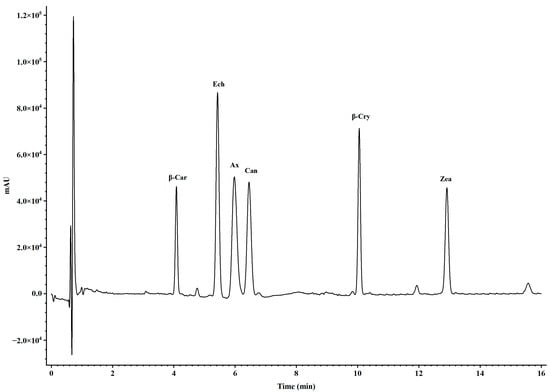
Figure 1.
Peak curves of carotenoid standards detected by UPC2-PDA with gradient elution conditions. The notations above the peaks represent β-car, β-carotene; Ech, echinenone; Ax, astaxanthin; β-Cry, β-cryptoxanthin; and Zea, zeaxanthin.
2.5. Extraction and Sequencing of RNA from Artemia
Total RNA was extracted from Artemia metanauplii after the 12 h enrichment procedure using RNAiso Plus reagent (Takara, Osaka, Japan) following the manufacturer’s protocol. Genomic DNA was subsequently removed using the RapidOut DNA Removal kit (Thermo, Waltham, MA, USA). The RNA quality and concentration were assessed using 1.5% agarose gel electrophoresis (Thermo, Waltham, MA, USA) and quantified with a NanoDrop 2000 microvolume spectrophotometer (Thermo, Waltham, MA, USA). The RNA quality number (RQN) was further evaluated using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), with only high-quality RNA (RQN > 6.5) proceeding to cDNA library construction. The cDNA library was prepared using the Illumina TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA), following the manufacturer’s instructions, by enriching mRNA with immobilized oligo(dT) primers. The mRNA was fragmented using fragmentation buffer and subsequently reverse transcribed into cDNA with random hexamers. The cDNA libraries were sequenced on the Illumina NovaSeq 6000 platform by Majorbio Co., Ltd. (Shanghai, China).
2.6. De Novo Assembly
To obtain high-quality clean reads for the de novo assembly, raw RNA-seq reads were filtered by removing those containing: (1) adapter sequences, (2) more than 10% ambiguous “N” bases, and (3) over 40% of low-quality bases (Q < 20). Clean reads were de novo assembled using Trinity version 2.4.0 [16] To increase the assembly quality, all the assembled sequences were filtered by CD-HIT 4.8.1 [17] to obtain the unigenes. Annotations for the assembled unigenes were conducted via BLAST against multiple databases, including GO, KEGG, eggNog, NR, SwissProt, and Pfam. Differentially expressed genes (DEGs) were identified using DeSeq2 with thresholds of a log2 fold change ≥1 and p < 0.05. An enrichment analysis of DEGs (control vs. EG) was conducted using GO terms and KEGG pathways, with the results visualized in bubble plots and histogram generated by GraphPad Prism 9.0. Principal component analysis (PCA) was also conducted to assess similarities and differences in the expression profiles of all samples.
2.7. Validation of the Transcription Data with qRT-PCR
Thirteen DEGs were randomly selected for qRT-PCR validation to assess the reliability of the transcriptome data. All primers (Table 1) were designed using Primer-BLAST online software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome, accessed on 1 August 2023) The reactions were conducted using an Agilent AriaMX system, with β-actin serving as the internal reference gene. The reaction mixture consisted of 0.8 μL of the forward primer (10 μM), 0.8 μL of the reverse primer (10 μM), 3 μL of the cDNA template, and 10 μL of TB Green Premix Ex Taq II (Takara, Osaka, Japan), for a total reaction volume of 20 μL. The thermal cycling conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 58 °C for 30 s. The relative expression levels were calculated using the 2−ΔΔCt method.

Table 1.
Target gene primers used for qRT-PCR. The gene profiles were normalized that of the β-actin gene.
2.8. Statistical Analysis
The results from the current study are reported as means ± standard errors of the means (S.E.Ms., n = 3). The data were subjected to one-way ANOVA using SPSS 20.0 software. Duncan’s multiple range tests were used as post hoc tests. Differences among the treatments’ means were considered significant at p < 0.05.
2.9. Ethics Statement
The present experimental procedures were carried out in strict accordance with the recommendations in the ethical guidelines of EU Directive 2010/63/EU for animal experiments.
3. Results
3.1. Change in the β-Carotene Contents of Artemia During the Enrichment and Starvation Processes
Changes in the β-carotene contents of Artemia metanauplii during the enrichment process are presented in Figure 2a. No β-carotene was detected in metanauplii without/prior to β-carotene enrichment. Over a 24 h period, the β-carotene content in enriched metanauplii was significantly increased (p < 0.05) across the three levels of β-carotene suspensions. The highest β-carotene content (p < 0.05) was observed at the 12th h of enrichment, which remained elevated until the 18th h compared to other time points. Based on these results, β-carotene depletion in metanauplii began after the 12 h enrichment period (Figure 2b). In the absence of further β-carotene intake, β-carotene levels declined significantly (p < 0.05) over time in the enriched treatment groups.
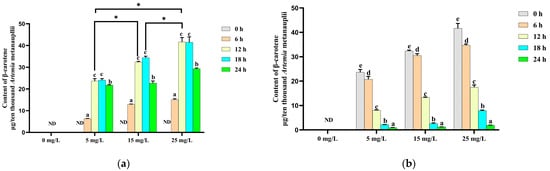
Figure 2.
Changes in the β-carotene content during the β-carotene enrichment and starvation processes. (a) For the Artemia metanauplius enrichment process; (b) for the Artemia metanauplius starvation process. Data are expressed as the means ± S.E.Ms. from triplicate samples from each group. Bars with different letters represent significant differences between various treatments (p < 0.05). The asterisk above the black lines denotes significant differences (p < 0.05) between different enrichment groups at 12 h. ND means not detected.
3.2. Changes in the Endogenous Carotenoid Composition in Artemia During the β-Carotene Enrichment and Starvation Processes
In addition to β-carotene, only canthaxanthin and echinenone were detected in Artemia metanauplii before and after the enrichment process in this study. In the control group, the canthaxanthin content decreased significantly (p < 0.05) over time in the absence of a carotenoid supplement. In contrast, in the β-carotene-enriched treatment groups, canthaxanthin levels initially declined and then significantly increased with time. The highest canthaxanthin levels in each enrichment treatment group were observed at 24 h and were significantly higher (p < 0.05) than in the control group (Figure 3a). During the depletion phase, the canthaxanthin content continued to increase until to 18 h, showing a significantly difference (p < 0.05) from the control group at this time point (Figure 3b).
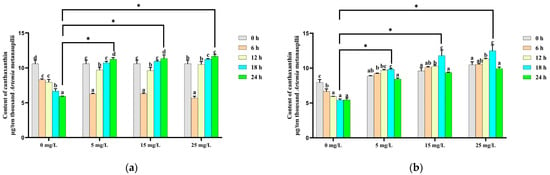
Figure 3.
Change in the canthaxanthin content during the β-carotene enrichment and starvation processes. (a) For the Artemia metanauplius enrichment process; (b) for the Artemia metanauplius starvation process. Data are expressed as the means ± S.E.Ms. from triplicate samples from each group. Bars with different letters represent significant differences between various treatments (p < 0.05). The asterisk above the black lines denotes significant differences (p < 0.05) between different enrichment groups.
The echinenone content in the control group exhibited a similar trend to that of canthaxanthin, decreasing steadily over time. In the enriched treatment groups, the echinenone content was significantly increased during the initial 18 h β-carotene enrichment, followed by a significant decline thereafter (Figure 4a). During the depletion phase, echinenone levels in both the control and enriched treatment groups decreased significantly (p < 0.05) over time (Figure 4b).
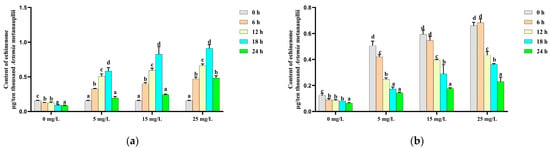
Figure 4.
Change in the echinenone content during the β-carotene enrichment and starvation processes. (a) For the Artemia metanauplius enrichment process; (b) for the Artemia metanauplius starvation process. Data are expressed as the means ± S.E.Ms. from triplicate samples from each group. Bars with different letters represent significant differences between various treatments (p < 0.05).
3.3. De Novo Sequencing and Assembly
Six cDNA libraries were constructed using whole-body Artemia metanauplii from two groups: control and EG. Sequencing on the Illumina Novaseq 6000 sequencing platform yielded 51,061,208, 45,635,052, and 46,527,368 raw reads for the control group, and 54,199,590, 45,287,112, and 47,207,506 raw reads were generated for the EG group. After filtering, 50,706,124, 45,299,184, and 46,208,168 clean reads were obtained for the control group, and 53,788,384, 44,898,224, and 46,891,296 clean reads for the EG group. The Q30 quality score for each sample exceeded 90%, indicating high sequencing quality. Following the merging of reads from all samples for assembly, a total of 58,202 unigenes were obtained, with minimum, maximum, and average lengths of 201 bp, 18,835 bp, and 815 bp, respectively, and an N50 values of 1428 bp. The distribution of unigene lengths is presented in Figure 5a.
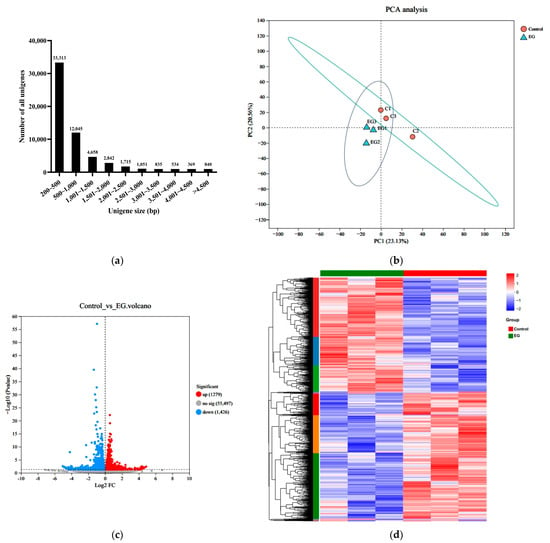
Figure 5.
Transcriptome analysis of the Artemia metanauplius samples from the treated and control groups. (a) Distribution of unigenes with different sizes; (b) principal component analysis (PCA) of the Artemia metanauplius samples; and (c) volcano plot of DEGs. The blue dots indicate that the genes are downregulated. The red dots indicate that the genes are upregulated. The gray dots indicate that there are no significant differences in expression. Each dot represents a gene. (d) Heatmap of DEGs among these groups.
The assembled unigenes were annotated using BLAST searches of six public databases: Nr (10,203), eggNOG (15,338), GO (9449), KEGG (11,144), Pfam (15,256), and Swiss-Prot (13,401, 23.46%).
3.4. Gene Expression and Functional Enrichment Analysis of DEGs
The principal component analysis (PCA) results indicated a clear distinction between the EG group and the control group (Figure 5b). DEGs were identified by comparing the treated and control libraries, focusing on genes with significant changes in expression during the enrichment process. A total of 2705 DEGs, with a false discovery rate (FDR) < 0.05 and |log2 fold change| > 1, exhibited differential transcription. As shown in Figure 5c,d, β-carotene enrichment resulted in 1279 upregulated and 1426 downregulated genes.
To explore the functional implications of these DEGs, GO and KEGG pathway analyses were conducted. The DEGs were categorized into GO terms, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs). The top 30 enriched GO terms in each category are illustrated in Figure 6. In the BP category, “transmembrane transport” and “electron transport chain” were the most significantly enriched terms. In the CC category, the terms “respiratory chain”, “oxidoreductase”, and “mitochondrial protein” were the most enriched. For the MF category, “oxidoreductase activity” and “chitin binding” were the most significant terms. In the KEGG enrichment analysis, the top three pathways were “oxidative phosphorylation”, “ribosome”, and “phagosome”, which are crucial for cellular metabolic processes and homeostasis. Furthermore, the DEGs were predominantly enriched in metabolic pathways associated with the regulation of energy production (Figure 7).
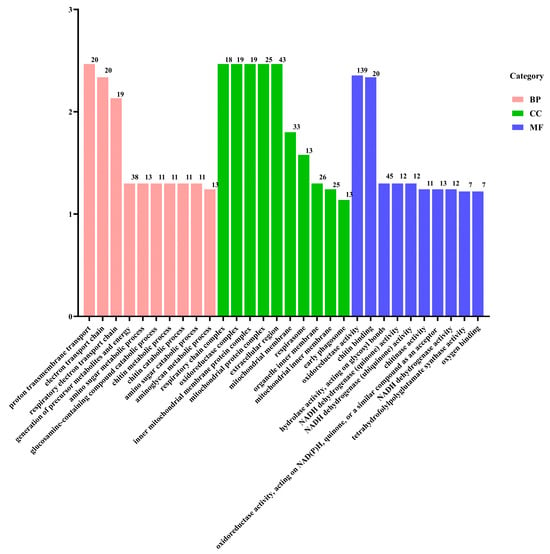
Figure 6.
GO classification of the DEGs.
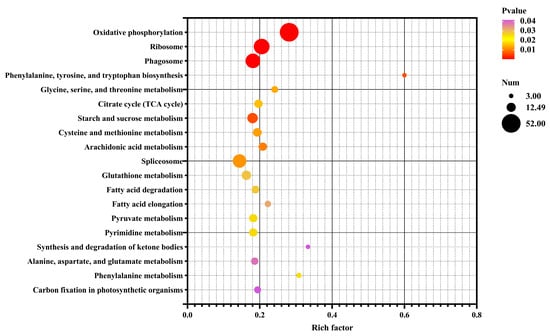
Figure 7.
Bubble chart of enriched KEGG pathways for DEGs in the control vs. EG groups. Rich factor is the ratio of the number of DEGs for a certain KEGG pathway to the total number of genes in that pathway. The significance of identified KEGG pathways was determined as p < 0.05.
3.5. Validation by qRT-PCR
To validate the RNA-seq analysis results, thirteen DEGs were randomly selected for validation using qRT-PCR. As shown in Figure 8, the expression patterns obtained from qRT-PCR closely matched those derived from RNA-seq data, indicating the reliability of the sequencing results and confirming the accuracy of the transcriptome analysis employed in this study.
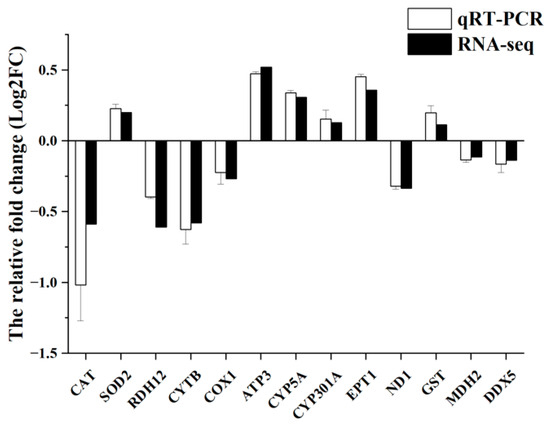
Figure 8.
Validation of RNA-seq data by qRT-PCR. qRT-PCR data are shown as the means ± S.E.Ms. (n = 3) from three biological replicates utilized in RNA-seq.
4. Discussion
4.1. Changes in the Carotenoid Content and Composition in Artemia in Response to β-Carotene Enrichment
Carotenoids in crustaceans are broadly categorized into two types: carotenes and xanthophylls [2]. Among carotenes, β-carotene is the most prominent, while xanthophylls are oxygenated derivatives of carotenes, primarily including β-cryptoxanthin, zeaxanthin, echinenone, canthaxanthin, and astaxanthin [18,19]. The composition and content of carotenoids in crustaceans vary significantly across species, strains, ages, body weights, and physiological stages [20,21], and are closely linked to environmental conditions and dietary factors [22]. Carotenoids in Artemia salina have been isolated and analyzed extensively. Echinenone and canthaxanthin, in a ratio of 19:1, are the only carotenoids present in California Artemia cysts and newly hatched nauplii [23]. From the metanauplius to adult stages, significant carotenoid accumulation occurs in Artemia, primarily derived from the assimilation of carotenoids synthesized by microorganisms within the food chain [13,24]. Consistent with previous findings, the current research demonstrates that, in the absence of exogenous carotenoid intake, environmental factors do not induce the production of xanthophylls beyond canthaxanthin and echinenone throughout the Artemia life cycle [25]. In this study, a method for feeding pure β-carotenoid to carotenoid-depleted Artemia metanauplii is presented. Additionally, feeding β-carotenoid-rich microalgae (Dunaliella salina) to Artemia metanauplii effectively enhanced the carotenoid content [9]. These findings confirm that Artemia metanauplii and adults can act as bio-encapsulated carotenoid carriers, offering a natural carotenoid source for seed breeding and broodstock [6,14]. Furthermore, Artemia exhibit efficient hydrolysis of fatty acid-esterified forms into free compounds, particularly from algal and plant-derived dietary carotenoids, thereby improving the bioavailability and absorption of these compounds by predators [14,25].
In crustaceans, a natural metabolic pathway exists whereby structurally similar carotenoids can be interconverted through a series of ketonization or hydroxylation reactions on their ionone rings [2,4]. Astaxanthin is generally regarded as the terminal product of this metabolic conversion pathway, with the transformation from β-carotene to astaxanthin represents the key metabolic axis in Penaeus monodon [2]. However, the carotenoid composition kinetics observed in the current study indicated that β-carotene in Artemia can only be converted into echinenone (4-keto-β-carotene), with canthaxanthin (4,4′-diketo-β-carotene) as the final product. This provides a theoretical foundation for understanding why Artemia cannot metabolize β-carotene into astaxanthin, likely due to the absence of hydroxylase enzymes required for this pathway. Additionally, dietary precursors such as zeaxanthin and lutein do not yield astaxanthin in Artemia, further underscoring the substrate specificity of the β-carotene ketolase in this species [25]. Given the ease of preservation and hatching of Artemia cysts, establishing a metabolic model for the conversion of β-carotene to canthaxanthin in Artemia could provide valuable insights into carotenoid metabolic mechanisms in crustaceans. Previous studies have also documented geometric and structural isomers of canthaxanthin, with their relative proportions varying significantly at different developmental stages [13]. The selective accumulation of carotenoids, including distinct isomers with sub-molecular structural variations, in various crustacean tissues suggests that these structural differences may have specific physiological effects on the growth, development, and health of aquatic organisms. This finding also underscores the complexity of interactions between carotenoids and other biomolecules.
4.2. Genes Associated with Carotenoid Physiological Function
Carotenoids play essential nutritional and physiological roles in crustaceans, impacting not only pigmentation but also promoting growth, enhancing immunity, and improving reproductive performance and stress resistance [4,26]. Supplementation with carotenoids in feed has been shown to significantly enhance growth performance and survival rates in crustaceans [27]. Additionally, carotenoids have been reported to positively influence survival and metamorphosis during the larval stages [2,28]. Studies further indicates that carotenoid intake is often positively correlated with pigment deposition and growth performance in crustaceans [29]. Moreover, compared to fatty acids, carotenoids contribute more significantly to the growth and health of Artemia, leading to improvements in the survival rate, average body length, total effective body length, and total antioxidant capacity [9]. Consistent with these findings, metabolic pathways related to growth, such as amino acid metabolism and ketone body metabolism, were significantly upregulated, suggesting that dietary carotenoids may positively impact Artemia growth. Research has shown that lipid transport proteins, rather than specific carotenoid-binding proteins, are primarily involved in carotenoid absorption, secretion, and intracellular transport [30]. Consequently, the KEGG enrichment analysis revealed upregulated pathways related to lipid metabolism, including fatty acids, sphingolipids, glycerides, and fatty acid biosynthesis, which aligns with the requirement of lipid transport proteins for carotenoid transfer. Additionally, the significant upregulation of the “ribosome” and “oxidative phosphorylation” pathways suggests that carotenoid metabolism demands substantial amounts of energy and protein, supporting the hypothesis that the carotenoid concentration reflects mitochondrial function [15]. Carotenoids are widely used as supplements in aquaculture due to their antioxidant and anti-inflammatory properties [31]. Glutathione, a crucial antioxidant, plays an fundamental role in protecting cells from oxidative damage [32]. The gsto1 gene, which is essential for glutathione biosynthesis, was significantly upregulated among the DEGs. Other antioxidant-related genes, such as gpx, also showed significant upregulation, consistent with previous studies [33]. Moreover, several genes involved in ferroptosis and the MAPK signaling pathway, such as fgh1 and hsp90, exhibited significant upregulation. Ferroptosis, a form of regulated cell death caused by the accumulation of lipid peroxides within cellular membrane, is closely associated with growth, oxidative stress, and immunity [30,34].
4.3. Genes Associated with Carotenoid Absorption and Metabolism
Carotenoids, as large organic molecules, are absorbed in aquatic animals through mechanisms similar to those used for lipid absorption [2]. Digestive enzymes facilitate the release of free carotenoids from protein-bound and esterified forms, enabling them to form chylomicrons with other lipids and bile salts, which enhances their bioavailability [35]. Carotenoids are then absorbed by enterocytes at the brush border of the intestinal mucosa, either through passive diffusion or active transport across the membrane bilayer, where they perform various physiological functions [36]. The remaining carotenoids are temporarily stored in various tissues and organs, such as the liver, in the form of free carotenoids, monoesters, diesters, or protein complexes, and are transported via the lymphatic and circulatory systems [37]. Depending on the intake of exogenous carotenoids, free carotenoids can be hydrolyzed and released to maintain a dynamic balance of carotenoid levels across different tissues, thereby supporting normal cellular functions [2].
The efficiency of carotenoid absorption is regulated by various lipid transport proteins rather than by any single specific protein [38]. Three main receptors are believed to facilitate carotenoid transmembrane transport [39]. Scavenger receptor class B1 (Scarb1) is a key mediator of carotenoid-based coloration in both vertebrates and invertebrates [40]. However, in the present study, no significant differences in scarb1 expression levels were observed between the two groups. Cluster determinant 36 (CD36), also known as fatty acid transporter, is implicated in the cellular uptake of β-carotene [41]. A significant up-regulation of cd36 was observed in the treated libraries compared to the control library in this study. Additionally, several ATP-binding cassette transporters (ABC), recognized for their roles in lipid transport, have been linked to carotenoid transport in previous studies [42]. The significant upregulation of Abca1 and Abcb1 in the β-carotene-enriched group suggests that ABC transporters may also contribute to carotenoid transport in Artemia, similar to observations in Crassostrea gigas [43].
Carotenoids can be classified into two categories based on function: provitamin A carotenoids (such as β-carotene and cryptoxanthin) and non-provitamin A carotenoids (such as lutein, canthaxanthin, and zeaxanthin). Among these, β-carotene exhibits 100% provitamin A activity and serves as the primary source of vitamin A [2]. In animals, carotenoids undergo symmetric and asymmetric cleavage by enzymes to produce vitamin A and its derivatives. β-Carotene-15, 15′-dioxygenase (BCO1) is an intracellular enzyme with high substrate specificity, exclusively catalyzing carotenoids that act as vitamin A precursors [44]. However, BCO1 was not detected in our transcriptome data. In contrast, β-carotene-9, 10-oxygenase (BCO2) is a mitochondrial enzyme with broader substrate specificity and is capable of metabolizing non-vitamin A precursor carotenoids and participating in biological processes beyond vitamin A synthesis [45]. BCO2 has been significantly associated with fish flesh pigmentation and carotenoid accumulation [46]. Nonetheless, we observed no differences in BCO2 expression levels between the control and β-carotene-enriched groups, aligning with the enzyme’s established role in promoting white pigmentation in various animals.
The functional roles of ketolase and hydroxylase are critical in regulating carotenoid metabolic conversion [47]. Research on the key enzyme genes and mechanisms involved in the conversion pathway from β-carotene to astaxanthin has been relatively systematic and comprehensive, though primarily limited to microalgae, bacteria, and fungi capable of synthesizing astaxanthin autonomously [48,49]. In Haematococcus pluvialis, which shares an astaxanthin biosynthesis pathway similar to that of crustaceans, β-carotene is converted into astaxanthin through the hydroxylation of the 3,3′ positions of the β-ionone rings catalyzed by β-carotene hydroxylase encoded by the Crt-b gene, and the addition of two keto groups at the 4,4′ positions by β-carotene ketolase encoded by the bkt gene [50]. Further studies have identified multiple types of bkt genes (bkt1, bkt2, and bkt3), where gene duplications add specificity, complexity, and flexibility to the carotenoid metabolic pathway [51]. Recent research has implicated cytochrome P450 family members, CYP2J19 and CYP3A80, as ketolases responsible for keto-carotenoid formation in animals [52]. Although these two genes were not present among the DEGs in this study, several P450 family genes exhibited varied expression profiles. Research on carotenoid metabolism in crustaceans remains limited; however, a bioinformatic analysis of high-throughput transcriptome data in copepods (Acartia fossae) has identified differentially expressed β-carotene hydroxylase genes involved in metabolic conversion pathways [53]. Nevertheless, the functional mechanisms of these genes remain unexplored. The absorption, metabolism, transport, and deposition of carotenoids in crustaceans are highly complex processes, with many aspects yet to be fully elucidated. Further investigations are necessary to clarify the detailed mechanisms of carotenoid esterification and hydrolysis, the specific metabolic pathways of carotenoid transformation, the cellular mechanisms of carotenoid uptake, and the genetic regulation underlying carotenoid transport and deposition. A more comprehensive understanding of these processes could facilitate the optimization of feed formulations and feeding strategies, ultimately enhancing the growth, health, and nutritional quality of carotenoid-rich crustaceans in aquaculture.
In conclusion, the observed increases in the β-carotene, canthaxanthin, and echinenone contents during enrichment, along with the depletion dynamics during starvation, underscore Artemia’s potential as a bio-encapsulated carotenoid source in aquaculture. Furthermore, the gene expression analysis illuminates the intricate metabolic pathways and gene regulations associated with carotenoid transport, absorption, and conversion, indicating potential avenues for further research on carotenoid metabolism in crustaceans.
Author Contributions
W.W. prepared the initial draft of the manuscript. Z.M. conducted the data analysis and prepared the figures. W.L. oversaw the Artemia feeding trials, while Y.X. performed the carotenoid detection analysis. A.S.M. and M.W. provided experimental guidance. All authors have read and agreed to the published version of the manuscript.
Funding
The current research was financially supported by supported by the National Key Research and Development Program of China, 2022YFE0203900; And APC was funded by National Natural Science Foundation of China, 31902385.
Institutional Review Board Statement
The animal study protocol adhered to the ethical guidelines of Shanghai Ocean University (approval number SHOU-DW-2022-044) for the care and use of experimental animals throughout the experimental procedures.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We are deeply grateful for the collaborative research project between the University of Stirling and Shanghai Ocean University, titled “Study on metabolic pathway of carotenoid enhancement in Artemia and discovery of key enzyme genes”.
The current research was financially supported by supported by the National Key Research and Development Program of China, 2022YFE0203900; National Natural Science Foundation of China, 31902385; collaborative research project funding between the University of Stirling and Shanghai Ocean University titled “Study on metabolic pathway of carotenoid enhancement in Artemia and discovery of key enzyme genes”.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fawzy, S.; Wang, W.L.; Zhou, Y.; Xue, Y.C.; Yi, G.F.; Wu, M.Q.; Huang, X.X. Can dietary β-carotene supplementation provide an alternative to astaxanthin on the performance of growth, pigmentation, biochemical, and immune-physiological parameters of Litopenaeus vannamei? Aquacult. Rep. 2022, 23, 101054. [Google Scholar]
- Wade, N.M.; Gabaudan, J.; Glencross, B.D. A review of carotenoid utilisation and function in crustacean aquaculture. Rev. Aquacult. 2017, 9, 141–156. [Google Scholar] [CrossRef]
- Lim, K.C.; Yusoff, F.M.; Shariff, M.; Kamarudin, M.S. Astaxanthin as feed supplement in aquatic animals. Rev. Aquacult. 2017, 10, 738–773. [Google Scholar] [CrossRef]
- Fawzy, S.; Wang, W.L.; Wu, M.Q.; Yi, G.F.; Huang, X.X. Effects of dietary different canthaxanthin levels on growth performance, antioxidant capacity, biochemical and immune-physiological parameters of white (Litopenaeus vannamei). Aquaculture 2022, 556, 738276. [Google Scholar] [CrossRef]
- Hamre, K.; Yúfera, M.; Rønnestad, I.; Boglione, C.; Conceição, L.E.C.; Izquierdo, M. Fish larval nutrition and feed formulation: Knowledge gaps and bottlenecks for advances in larval rearing. Rev. Aquacult. 2013, 5, S26–S58. [Google Scholar] [CrossRef]
- Sorgeloos, P.; Dhert, P.; Candreva, P. Use of brine shrimp, Artemia spp., in marinefish laryiculture. Rev. Fish. Sci. 2001, 6, 55–68. [Google Scholar] [CrossRef]
- Reis, D.B.; Acosta, N.G.; Almansa, E.; Navarro, J.C.; Tocher, D.R.; Andrade, J.P.; Sykes, A.V.; Rodriguez, C. Comparative study on fatty acid metabolism of early stages of two crustacean species: Artemia sp. metanauplii and Grapsus adscensionis zoeae, as live prey for marine animals. Comp. Biochtem. Phys. B. 2017, 204, 53–60. [Google Scholar] [CrossRef]
- Lavens, P.; Sorgeloos, P. The history, present status and prospects of the availability of Artemia cysts for aquaculture. Aquaculture 2000, 181, 397–403. [Google Scholar] [CrossRef]
- Gui, L.; Xu, L.; Liu, Z.Y.; Zhou, Z.G.; Sun, Z. Carotenoid-rich microalgae promote growth and health conditions of Artemia nauplii. Aquaculture 2022, 546, 737289. [Google Scholar] [CrossRef]
- Xie, W.; Ma, Y.C.; Ren, B.; Gao, M.R.; Sui, L.Y. Artemia nauplii enriched with archaea Halorubrum increased survival and challenge tolerance of Litopenaeus vannamei postlarvae. Aquaculture 2021, 533, 736087. [Google Scholar] [CrossRef]
- Domínguez, A.; Ferreira, M.; Coutinho, P.; Fabregas, J.; Otero, A. Delivery of astaxanthin from Haematococcus pluvialis to the aquaculture food chain. Aquaculture 2005, 250, 424–430. [Google Scholar] [CrossRef]
- Guinot, D.; Monroig, Ó.; Navarro, J.C.; Varó, I.; Amat, F.; Hontoria, F. Enrichment of Artemia metanauplii in phospholipids and essential fatty acids as a diet for common octopus (Octopus vulgaris) paralarvae. Aquacult. Nutr. 2013, 19, 837–844. [Google Scholar] [CrossRef]
- Nelis, H.J.C.F.; Lavens, P.; Van Steenberge, M.M.Z.; Sorgeloos, P.; Leenheer, A.P.D. Qualitative and quantitative changes in the carotenoids during development of the brine shrimp Artemia. J. Lipid. Res. 1988, 29, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, Y.; Ahmadifard, N.; Agh, N.; Rahmanifarah, K.; Hejazi, M.A. β-Carotene-enriched Artemia as a natural carotenoid improved skin pigmentation and enhanced the mucus immune responses of platyfish Xiphophorus maculatus. Aquacult. Int. 2019, 27, 1847–1858. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Zhang, L.L.; Wang, G.D.; Huang, S. De novo assembly transcriptome analysis reveals the genes associated with body color formation in the freshwater ornamental shrimps Neocaridina denticulate sinensis. Gene 2022, 806, 145929. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Moghadam, A.; Taghizadeh, M.S.; Haghi, R.; Tahmasebi, A.; Niazi, A.; Ebrahimie, E. Exploring novel insights: Methyl jasmonate treatment reveals novel lncRNA-mediated regulation of secondary metabolite biosynthesis pathways in Echinacea purpurea. Food. Bios. 2024, 57, 103457. [Google Scholar] [CrossRef]
- Su, F.; Huang, B.; Liu, J.G. The carotenoids of shrimps (Decapoda: Caridea and Dendrobranchiata) cultured in China. J. Crustacean. Biol. 2018, 38, 523–530. [Google Scholar] [CrossRef]
- Wade, N.M.; Anderson, M.; Sellars, M.J.; Tume, R.K.; Preston, N.P.; Glencross, B.D. Mechanisms of colour adaptation in the prawn Penaeus monodon. J. Exp. Biol. 2012, 215, 343–350. [Google Scholar] [CrossRef]
- Ribeiro, E.A.; Genofre, G.C.; McNamara, J.C. Identification and quantification of carotenoid pigments during the embryonic development of the freshwater shrimp Macrobrachium olfersii (Crustacea, Decapoda). Mar. Freshw. Behav. Phy. 2001, 34, 105–116. [Google Scholar] [CrossRef]
- Sachindra, N.M.; Bhaskar, N.; Mahendrakar, N.S. Carotenoids in different body components of Indian shrimps. J. Sci. Food. Agric. 2005, 85, 167–172. [Google Scholar] [CrossRef]
- Flores, M.; Diaz, F.; Medina, R.; Licea, A.; Medina, R. Physiological, metabolic and haematological responses in white shrimp Litopenaeus vannamei (Boone) juveniles fed diets supplemented with astaxanthin acclimated to low-salinity water. Aquac. Res. 2007, 38, 740–747. [Google Scholar] [CrossRef]
- Hsu, W.J.; Chichester, C.O.; Davies, B.H. The metabolism of β-carotene and other carotenoids in the brine shrimp, Artemia salina L. (Crustacea: Branchiopoda). Comp. Biochem. Phys. 1970, 32, 69–79. [Google Scholar] [CrossRef]
- Dhankhar, J.; Kadian, S.S.; Sharma, A. Astaxanthin: A potential carotenoid. Int. J. Pharm. 2012, 3, 1246–1259. [Google Scholar]
- Huang, J.; Hui, B. Feed-induced variation in the carotenoid composition of brine shrimp. eFood 2020, 3, 247–253. [Google Scholar] [CrossRef]
- Pereira da Costa, D.; Campos Miranda-Filho, K. The use of carotenoid pigments as food additives for aquatic organisms and their functional roles. Rev. Aquacult. 2020, 12, 1567–1578. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.J.; Tian, L.X.; Yang, H.J.; Liang, G.Y.; Yue, Y.R.; Xu, D.H. Effects of dietary astaxanthin on growth, antioxidant capacity and gene expression in Pacific white shrimp Litopenaeus vannamei. Aquacult. Nutr. 2013, 19, 917–927. [Google Scholar] [CrossRef]
- Wang, W.L.; Ishikawa, M.; Koshio, S.; Yokoyama, S.; Dawood, M.A.O.; Zhang, Y.K. Effects of dietary astaxanthin supplementation on survival growth and stress resistance in larval and post-larval kuruma shrimp, Marsupenaeus japonicus. Aquac. Res. 2018, 49, 2225–2232. [Google Scholar] [CrossRef]
- Joshi, K.; Kumar, P.; Kataria, R. Microbial carotenoid production and their potential applications as antioxidants: A current update. Process. Biochem. 2023, 128, 190–205. [Google Scholar] [CrossRef]
- Liu, M.; Kong, X.Y.; Yao, Y.; Wang, X.A.; Yang, W.; Wu, H.; Li, S.; Ding, J.W.; Yang, J. The critical role and molecular mechanisms of ferroptosis in antioxidant systems: A narrative review. Ann. Transl. Med. 2022, 10, 368. [Google Scholar] [CrossRef]
- Wang, W.L.; Ishikawa, M.; Koshio, S. Effects of dietary astaxanthin supplementation on juvenile kuruma shrimp, Marsupenaeus japonicus. Aquaculture 2018, 491, 197–204. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.Q.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Aspects. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.Y.; Van Remmen, H.; Frohlich, V.; Lechleiter, J.; Richardson, A.; Ran, Q. Gpx4 protects mitochondrial ATP generation against oxidative damage. Biochem. Bioph. Res. Commun. 2007, 356, 893–898. [Google Scholar] [CrossRef]
- Kang, Y.P.; Mockabee-Macias, A.; Jiang, C. Non-canonical glutamate-cysteine ligase activity protects against ferroptosis. Cell. Metab. 2021, 33, 174–189. [Google Scholar] [CrossRef]
- Von Lintig, J.; Moon, J.; Lee, J.; Ramkumar, S. Carotenoid metabolism at the intestinal barrier. BBA-Mol. Cell. Biol. Lipids 2020, 1865, 158580. [Google Scholar] [CrossRef]
- During, A.; Hussain, M.M.; Morel, D.W. Carotenoid uptake and secretion by CaCo-2 cells: Beta-carotene isomer selectivity and carotenoid interactions. J. Lipid. Res. 2002, 43, 1086–1095. [Google Scholar] [CrossRef]
- Rajasingh, H.; Øyehaug, L.; Våge, D.I.; Omholt, S.W. Carotenoid dynamics in Atlantic salmon. BMC. Biol. 2006, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Borel, P.; Lietz, G.; Goncalves, A.; Szabo, D.; Lecompte, S.; Curtis, P.; Goumidi, L.; Caslake, M.; Miles, E.; Packard, C. CD36 and SR-B1 are involved in cellular uptake of provitamin A carotenoids by caco-2 and HEK cells, and some of their genetic variants are associated with plasma concentrations of these micronutrients in humans. J. Nutr. 2013, 143, 448–456. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araújo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C.; et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef]
- Sundvold, H.; Helgeland, H.; Baranski, M.; Omholt, S.W.; Våge, D.I. Characterisation of a novel paralog of scavenger receptor class B member (SCARB1) in Atlantic salmon (Salmo salar). BMC. Genet. 2011, 12, 52. [Google Scholar] [CrossRef]
- Van Bennekum, A.; Werder, M.; Thuahnai, S.T. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry 2005, 44, 4517–4525. [Google Scholar] [CrossRef]
- Hu, Z.; Song, H.; Zhou, C.; Yu, Z.L.; Yang, M.J.; Zhang, T. De novo assembly transcriptome analysis reveals the preliminary molecular mechanism of pigmentation in juveniles of the hard clam Mercenaria mercenaria. Genomics 2020, 112, 3635–3647. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Li, Q.; Yu, H.; Zhao, X.; Kong, L.; Song, L. Comparative transcriptome analysis of the Pacific Oyster Crassostrea gigas characterized by shell colors: Identification of genetic bases potentially involved in pigmentation. PLoS ONE 2015, 10, e0145257. [Google Scholar] [CrossRef]
- Jin, Y.; Yu, Y.; Zhang, C.S.; Li, S.H.; Zhang, X.J.; Li, F.H. Characterization and function analysis of the beta-carotene oxygenase-like genes in carotenoids metabolism of the ridgetail white prawn Exopalaemon carinicauda. Front. Physiol. 2020, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Vachali, P.P.; Gorusupudi, A. Inactivity of human β, β-carotene-9′,10′-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. Sci. USA 2014, 111, 10173–10178. [Google Scholar] [CrossRef]
- Lehnert, S.J.; Christensen, K.A.; Vandersteen, W.E.; Sakhrani, D.; Pitcher, T.E.; Heath, J.W.; Koop, B.F.; Heath, D.D.; Devlin, R.H. Carotenoid pigmentation in salmon: Variation in expression at BCO2-1 locus controls a key fitness trait affecting red coloration. Proc. R. Soc. B 2019, 286, 1471–2954. [Google Scholar] [CrossRef]
- Martin, J.F.; Gudina, E.; Barredo, J.L. Conversion of β-carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylase-ketolase protein? Microb. Cell. Fact. 2008, 7, 3. [Google Scholar] [CrossRef]
- Hara, K.; Morita, T.; Mochizuki, M.; Yamamoto, K.; Ogino, C.; Araki, M.; Kondo, A. Development of a multi-gene expression system in Xanthophyllomyces dendrorhous. Microb. Cell. Fact. 2014, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Han, D.X.; Li, Y.T.; Hu, Q. Astaxanthin in microalgae: Pathways, functions and biotechnological implications. Algae 2013, 28, 131–147. [Google Scholar] [CrossRef]
- Haque, F.; Dutta, A.; Thimmanagari, M.; Yi, W.C. Intensified green production of astaxanthin from Haematococcus pluvialis. Food. Bioprod. Process. 2016, 99, 1–11. [Google Scholar] [CrossRef]
- Wang, N.; Guan, B.; Kong, Q.; Sun, H.; Geng, Z.Y.; Duan, L.F. Enhancement of astaxanthin production from Haematococcus pluvialis mutants by three-stage mutagenesis breeding. J. Biotechnol. 2016, 236, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Twomey, E.; Melo-Sampaio, P.; Schulte, L.M.; Bossuyt, F.; Brown, J.L. Multiple Routes to Color Convergence in a Radiation of Neotropical Poison Frogs. Syst. Biol. 2023, 72, 1247–1261. [Google Scholar] [CrossRef] [PubMed]
- Mojib, N.; Amad, M.; Thimma, M.; Aldanondo, N.; Kumaran, M.; Irigoien, X. Carotenoid metabolic profiling and transcriptome-genome mining reveal functional equivalence among blue-pigmented copepods and appendicularia. Mol. Ecol. 2014, 23, 2740–2756. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).