Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Slaughter and Genotyping
2.2. Quantitative Genetic Analysis
2.3. Genomic Association Analysis
- Let D = I in the first step;
- Calculate G = ZDZ′q;
- Calculate GEBVs for the entire data set using ssGBLUP;
- Convert GEBVs to SNP effects.
- 5.
- Calculate the weight for each SNP as follows: , where i represents the i-th SNP;
- 6.
- Normalize SNP weight to preserve constant variance in total genetics.
- 7.
- Loop to step 2.
2.4. Enrichment Analysis
3. Results
3.1. Description of Phenotypic Data and Heritability
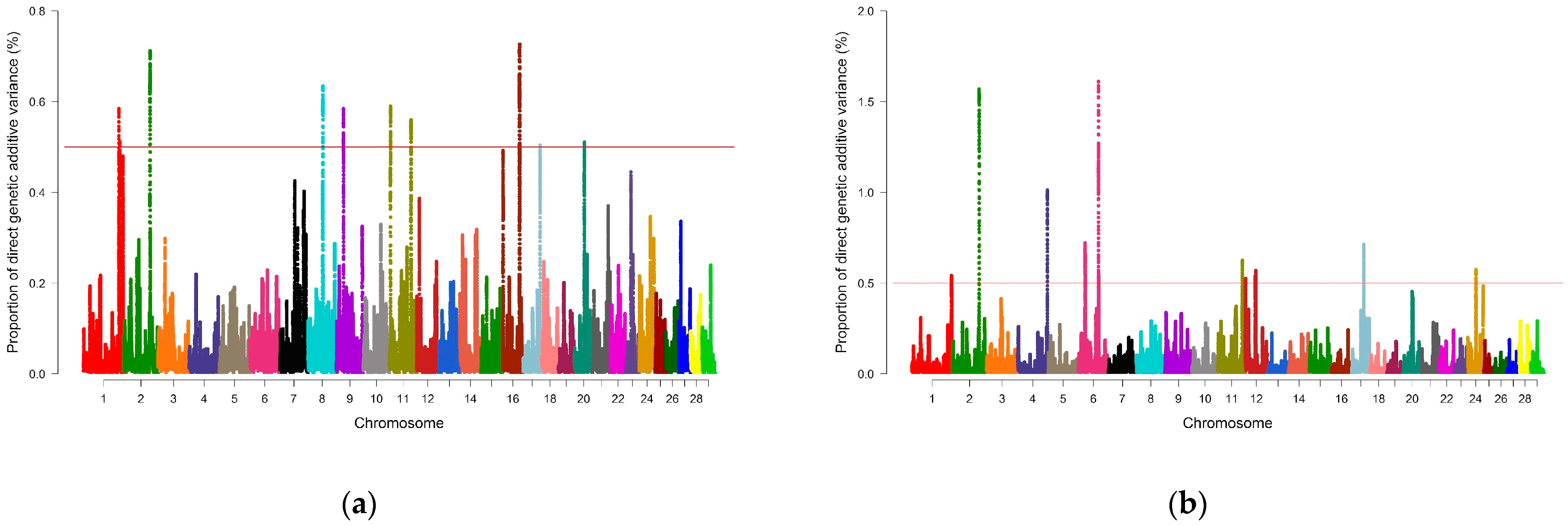
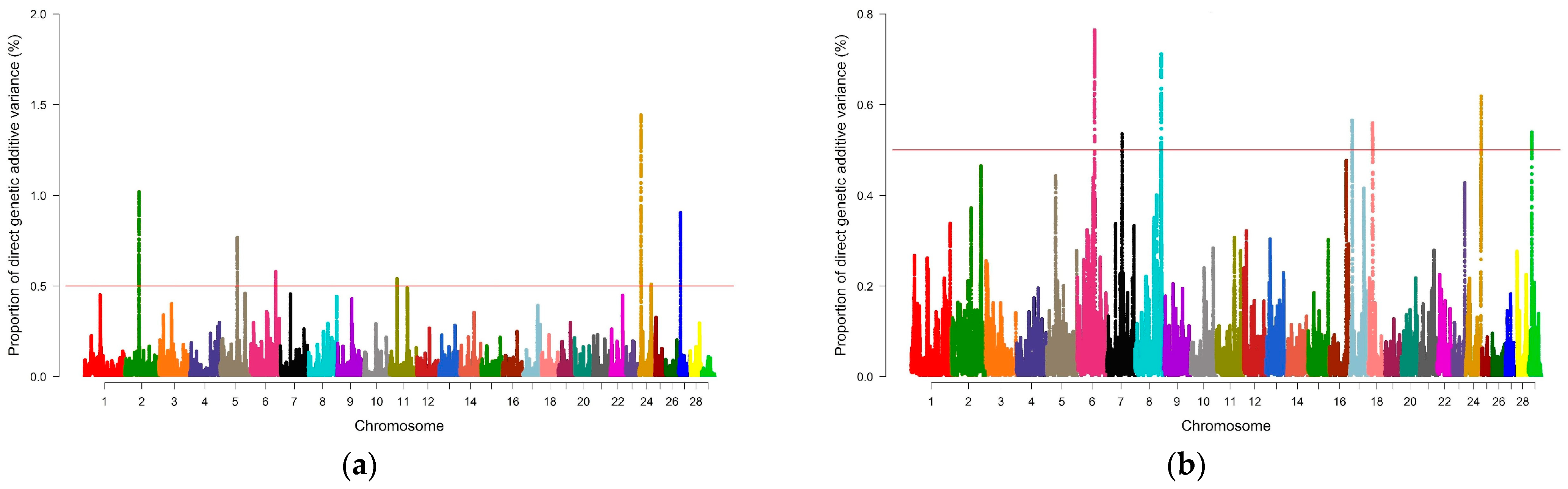
3.2. Genome-Wide Association Study and Identification of Candidate Genes
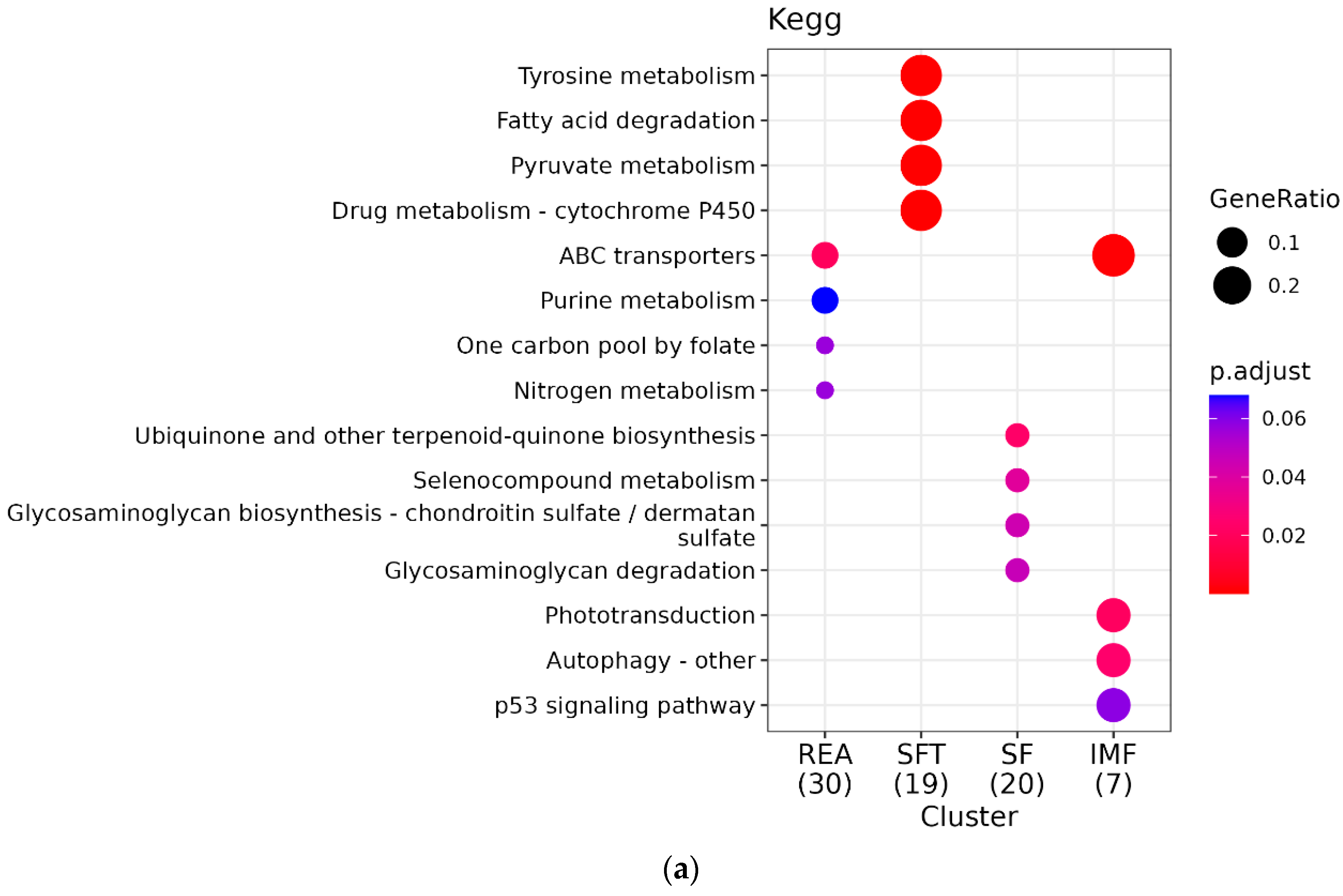
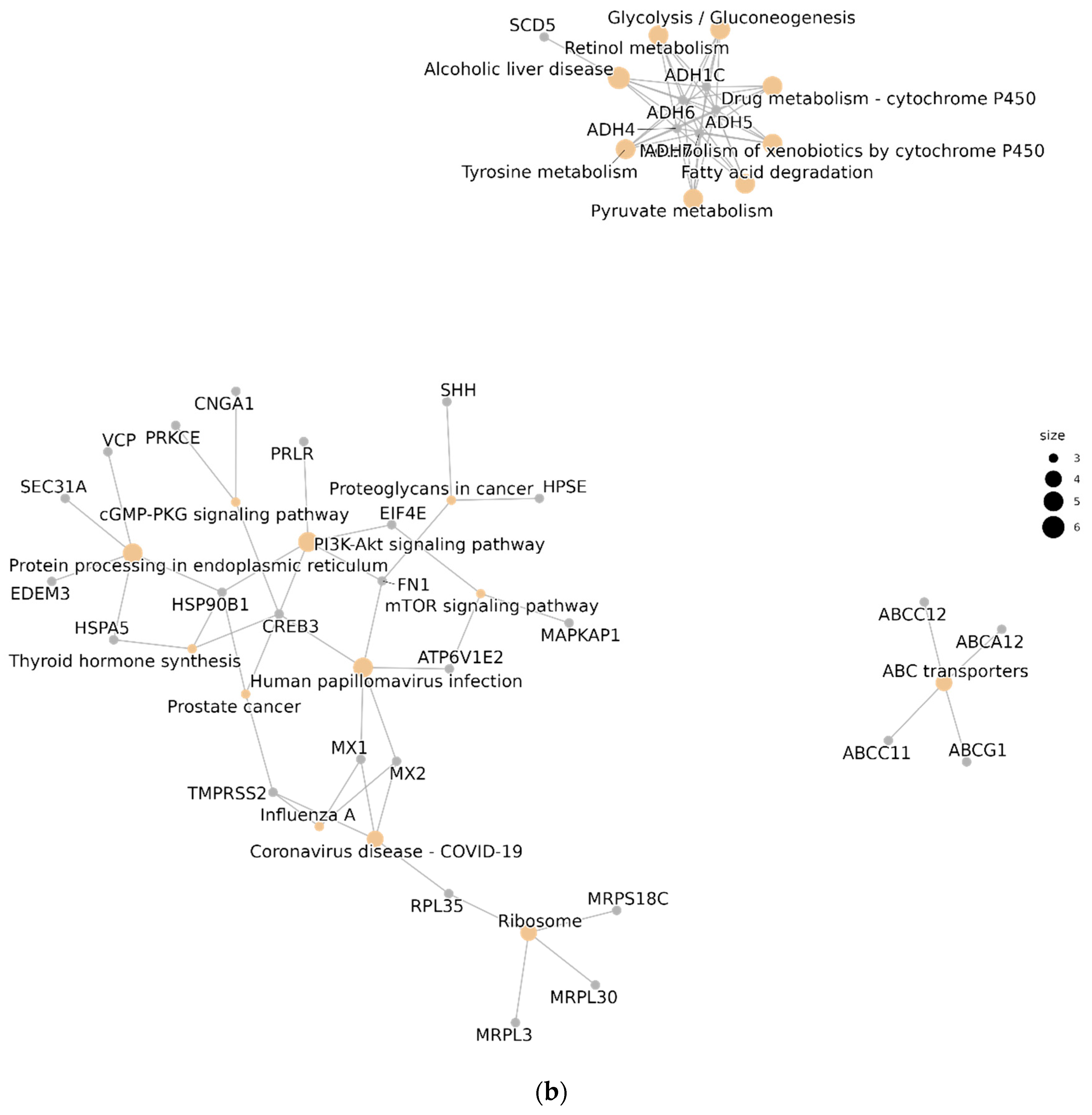
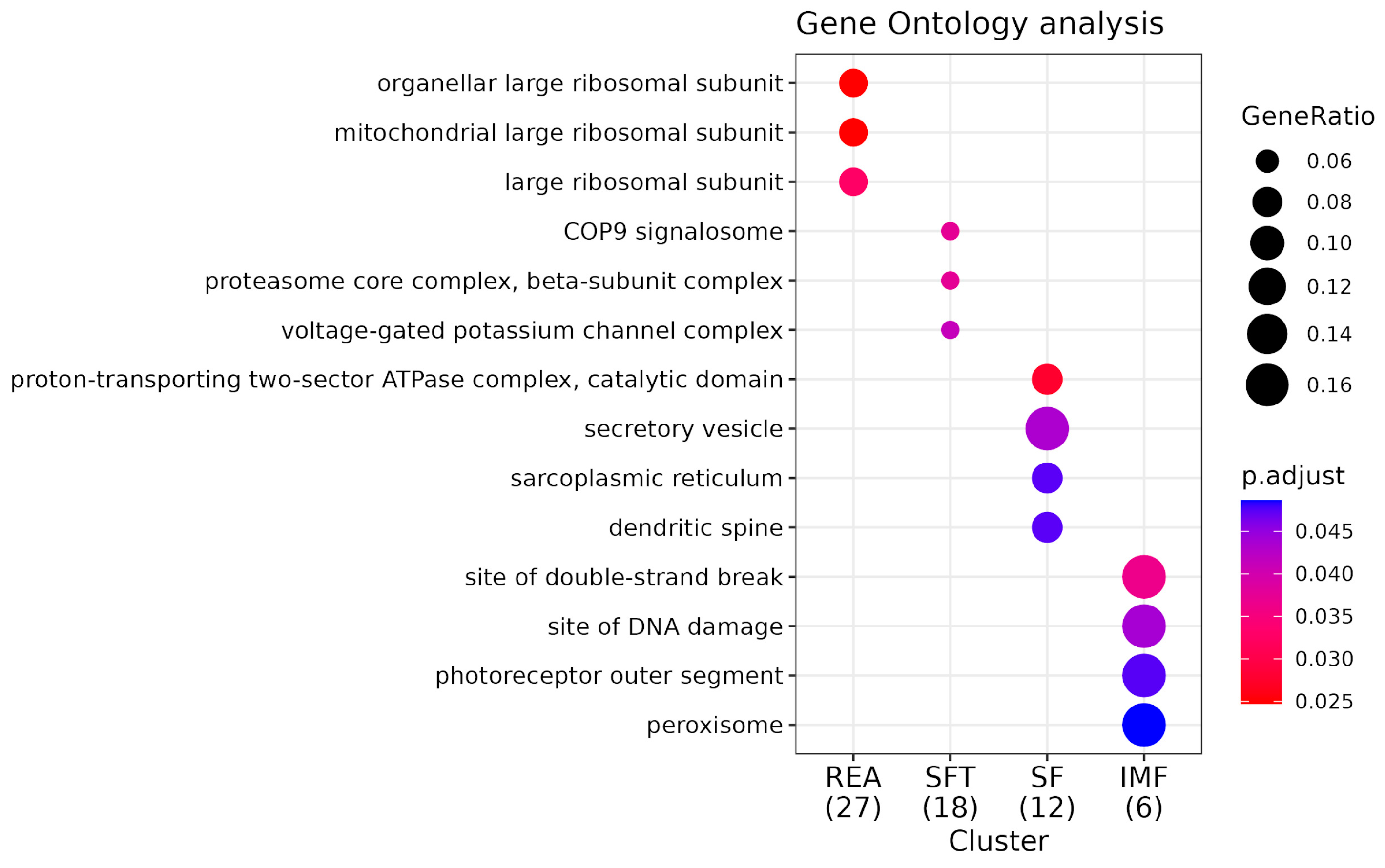
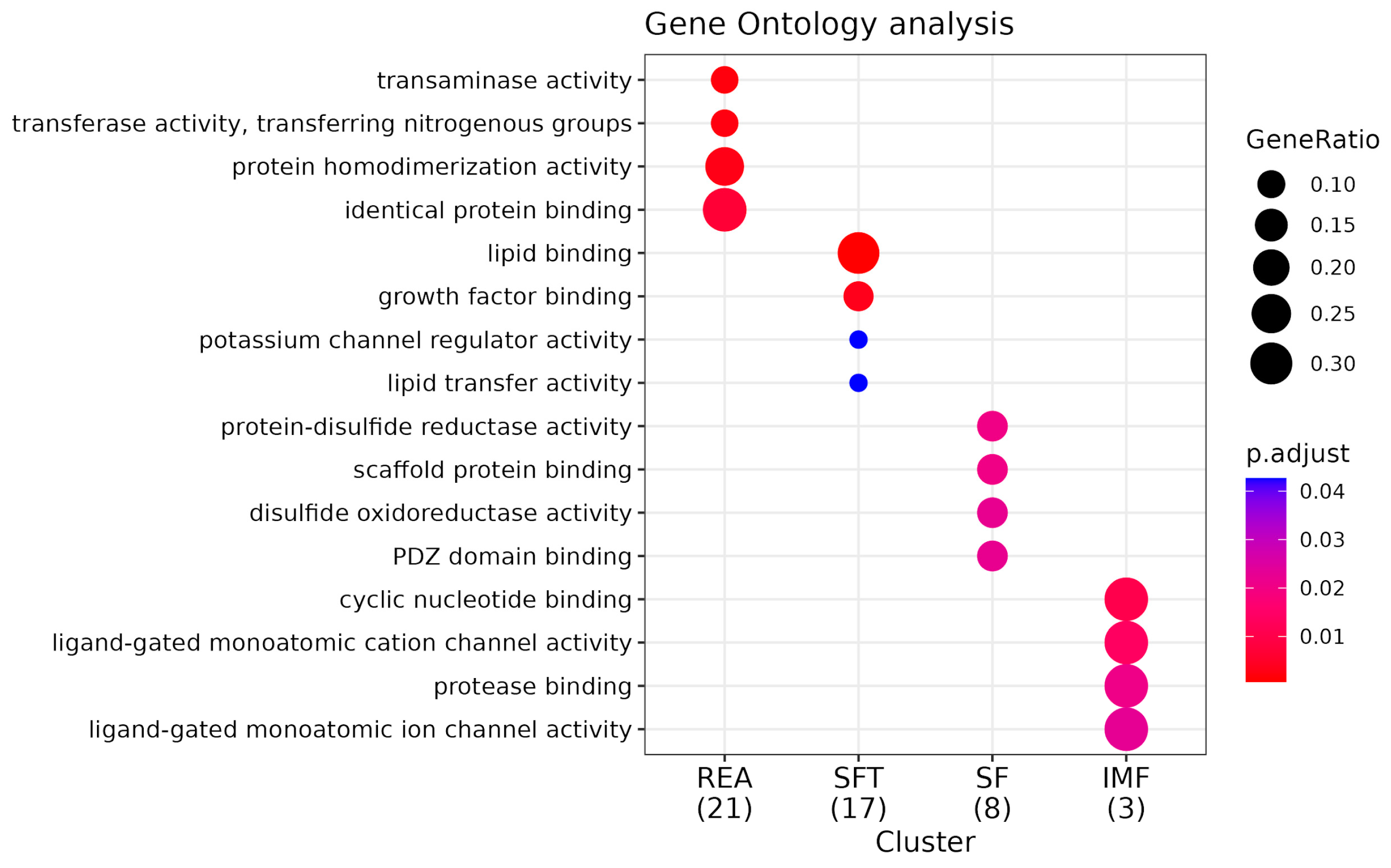
3.3. Functional Analysis and Pathway Enrichment
4. Discussion
4.1. Description of Phenotypic Data and Heritability
4.2. Genome-Wide Association Study and Candidate Genes
4.3. Functional Analysis and Pathway Enrichment
4.3.1. Functional Analysis
4.3.2. Network Enrichment
4.3.3. Pathway Enrichment KEGG
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brazilian Beff. Abiec BEEF REPORT Perfil Da Pecuária No Brasil 2022. In Paper Knowledge: Toward a Media History of Documents; Duke University Press: Durham, NC, USA, 2022; Volume 7, pp. 1–70. [Google Scholar]
- Henchion, M.M.; Mccarthy, M.; Resconi, V.C. Beef Quality Attributes: A Systematic Review of Consumer Perspectives. MESC 2017, 128, 1–7. [Google Scholar] [CrossRef]
- Webb, E.C.; Neill, H.A.O. The Animal Fat Paradox and Meat Quality. Meat Sci. 2008, 80, 28–36. [Google Scholar] [CrossRef]
- Frank, D.; Ball, A.; Hughes, J.; Krishnamurthy, R.; Piyasiri, U.; Stark, J.; Watkins, P.; Warner, R. Sensory and Flavor Chemistry Characteristics of Australian Beef: In Fl Uence of Intramuscular Fat, Feed, and Breed. J. Agric. Food Chem. 2016, 64, 4299–4311. [Google Scholar] [CrossRef]
- Sainz, R.D.; Araujo, F.R.C. Tipificação de Carcaças de Bovinos e Suínos. In Proceedings of the Congresso Brasileiro de Ciencia e Tecnologia de Carne, São Pedro, SP, Brazil; 2001; Volume 1. [Google Scholar]
- Du, L.; Li, K.; Chang, T.; An, B.; Liang, M.; Deng, T. Genomics Integrating Genomics and Transcriptomics to Identify Candidate Genes for Subcutaneous Fat Deposition in Beef Cattle. Genomics 2022, 114, 110406. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Christensen, O.F.; Aguilar, I.; Misztal, I. Single Step, a General Approach for Genomic Selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Melo, A.F.; Moreira, J.M.; Ataídes, D.S.; Guimarães, R.A.M.; Loiola, J.L.; Oliveira, R.Q. Fatores Que Influenciam Na Qualidade Da Carne Bovina: Revisão. Publicações Em Med. Veterinária E Zootec. 2016, 10, 785–794. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 91l–917. [Google Scholar]
- AMSA. Research Guidelines for Cookery, Sensory Evaluation, and Instrumental Tenderness Measurements of Meat; American Meat Science Association: Savoy, IL, USA, 2015; ISBN 8005172672. [Google Scholar]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. Blupf90 and Related Programs (Bgf90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; pp. 2001–2002. [Google Scholar]
- Aguilar, I.; Misztal, I.; Legarra, A.; Tsuruta, S. Efficient Computation of the Genomic Relationship Matrix and Other Matrices Used in Single-Step Evaluation. J. Anim. Breed. Genet. 2011, 128, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot Topic: A Unified Approach to Utilize Phenotypic, Full Pedigree, and Genomic Information for Genetic Evaluation of Holstein Final Score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Vitezica, Z.G.; Aguilar, I.; Misztal, I.; Legarra, A. Bias in Genomic Predictions for Populations under Selection. Genet. Res. 2011, 93, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing 2021; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Fonseca, P.A.S.; Suarez-Vega, A.; Marras, G.; Cánovas, Á. GALLO: An R Package for Genomic Annotation and Integration of Multiple Data Sources in Livestock for Positional Candidate Loci. Gigascience 2020, 9, giaa149. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. R Package Version 1.2. 2021. Available online: https://github.com/GuangchuangYu/enrichplot (accessed on 23 August 2023).
- Wickham, H.; Henry, L. tidyr: Tidy Messy Data, R Package Version 1.0.0. 2019. Available online: https://cran.r-project.org/package=tidyr (accessed on 23 August 2023).
- Mailund, T. Manipulating Data Frames: Dplyr. In R Data Science Quick Reference: A Pocket Guide to APIs, Libraries, and Packages; Apress: Berkeley, CA, USA, 2019; pp. 109–160. ISBN 978-1-4842-4894-2. [Google Scholar]
- Wickham, H.; Hester, J.; Francois, R.; Bryan, J.; Bearrows, S.; Jylänki, J.; Jørgensen, M. Package ‘readr.’ Read Rectangular Text Data. Available online: https://cran.r-project.org/web/packages/readr/readr.pdf (accessed on 23 August 2023).
- Provost, E.; Weier, C.A.; Leach, S.D. Multiple Ribosomal Proteins Are Expressed at High Levels in Developing Zebrafish Endoderm and Are Required for Normal Exocrine Pancreas Development. Zebrafish 2013, 10, 161–169. [Google Scholar] [CrossRef]
- Zeng, L.L.; Qu, K.; Zhang, J.; Huang, B.; Lei, C. Genes Related to Heat Tolerance in Cattle—A Review. Anim. Biotechnol. 2023, 34, 1840–1848. [Google Scholar] [CrossRef]
- Costa, R.G.; Lima, A.G.V.O.; Oliveira, C.F.S.; Azevedo, P.S.; Medeiros, A.N. Using Different Methodologies to Determination the Loin Eye Area in Sheep. Arch. Zootec. 2012, 61, 615–618. [Google Scholar] [CrossRef][Green Version]
- Gordo, D.G.M.; Espigolan, R.; Tonussi, R.L.; Júnior, G.A.F.; Bresolin, T.; Magalhães, A.F.B.; Feitosa, F.L.; Baldi, F.; Carvalheiro, R.; Tonhati, H.; et al. Genetic Parameter Estimates for Carcass Traits and Visual Scores Including or Not Genomic Information1. J. Anim. Sci. 2016, 94, 1821–1826. [Google Scholar] [CrossRef]
- Kluska, S.; Olivieri, B.F.; Bonamy, M.; Chiaia, H.L.J.; Feitosa, F.L.B.; Berton, M.P.; Peripolli, E.; Lemos, M.V.A.; Tonussi, R.L.; Lôbo, R.B.; et al. Estimates of Genetic Parameters for Growth, Reproductive, and Carcass Traits in Nelore Cattle Using the Single Step Genomic BLUP Procedure. Livest. Sci. 2018, 216, 203–209. [Google Scholar] [CrossRef]
- Gordo, D.G.M.; Espigolan, R.; Bresolin, T.; Júnior, G.A.F.; Magalhães, A.F.B.; Braz, C.U.; Fernandes, W.B.; Baldi, F.; Albuquerque, L.G. Genetic Analysis of Carcass and Meat Quality Traits in Nelore Cattle. J. Anim. Sci. 2018, 96, 3558–3564. [Google Scholar] [CrossRef]
- Cesar, A.S.M.; Regitano, L.C.A.; Mourão, G.B.; Tullio, R.R.; Lanna, D.P.D.; Nassu, R.T.; Mudado, M.A.; Oliveira, P.S.N.; do Nascimento, M.L.; Chaves, A.S.; et al. Genome-Wide Association Study for Intramuscular Fat Deposition and Composition in Nellore Cattle. BMC Genet. 2014, 15, 39. [Google Scholar] [CrossRef]
- Kojic, S.; Radojkovic, D.; Faulkner, G. Muscle Ankyrin Repeat Proteins: Their Role in Striated Muscle Function in Health and Disease. Crit. Rev. Clin. Lab. Sci. 2011, 48, 269–294. [Google Scholar] [CrossRef]
- Doyle, J.L.; Berry, D.P.; Veerkamp, R.F.; Carthy, T.R.; Evans, R.D.; Walsh, S.W.; Purfield, D.C. Genomic Regions Associated with Muscularity in Beef Cattle Differ in Five Contrasting Cattle Breeds. Genet. Sel. Evol. 2020, 52, 2. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Puig-Oliveras, A.; Revilla, M.; Castelló, A.; Fernández, A.I.; Folch, J.M.; Ballester, M. Expression-Based GWAS Identifies Variants, Gene Interactions and Key Regulators Affecting Intramuscular Fatty Acid Content and Composition in Porcine Meat. Sci. Rep. 2016, 6, 31803. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Podstawski, P.; Piórkowska, K.; Tyra, M. Association of Missense MTTP Gene Polymorphism with Carcass Characteristics and Meat Quality Traits in Pigs. Czech J. Anim. Sci. 2017, 62, 9–14. [Google Scholar] [CrossRef]
- Cristancho, A.G.; Lazar, M.A. Forming Functional Fat: A Growing Understanding of Adipocyte Differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Ortega Torres, J.; Ariza Botero, M.F. El Mecanismo de Muerte Celular Programada y Su Importancia En El Proceso de Maduración de La Carne Bovina. Rev. Med. Vet. 2012, 83, 83–96. [Google Scholar] [CrossRef]
- Rodrigues, R.T.d.S.; Chizzotti, M.L.; Vital, C.E.; Baracat-Pereira, M.C.; Barros, E.; Busato, K.C.; Gomes, R.A.; Ladeira, M.M.; Martins, T.d.S. Differences in Beef Quality between Angus (Bos Taurus Taurus) and Nellore (Bos Taurus Indicus) Cattle through a Proteomic and Phosphoproteomic Approach. PLoS ONE 2017, 12, e0170294. [Google Scholar] [CrossRef] [PubMed]
- Leal-Gutiérrez, J.D.; Elzo, M.; Johnson, D.; Mateescu, R. Genome-Wide Association and Gene Enrichment Analyses of Meat Tenderness in an Angus-Brahman Cattle Population. In Proceedings of the 11th World Congress on Genefics Applied to Livestock Producfion, Auckland, New Zealand, 11–16 February 2018; pp. 1–5. [Google Scholar]
- Neupane, M.; Kiser, J.N.; Neibergs, H.L. Gene Set Enrichment Analysis of SNP Data in Dairy and Beef Cattle with Bovine Respiratory Disease. Anim. Genet. 2018, 49, 527–538. [Google Scholar] [CrossRef]
- Gaughan, J.B.; Mader, T.L.; Holt, S.M.; Lisle, A. A New Heat Load Index for Feedlot Cattle. J. Anim. Sci. 2008, 86, 226–234. [Google Scholar] [CrossRef]
- Brown-Brandl, T.M. Understanding Heat Stress in Beef Cattle. Rev. Bras. Zootec. 2018, 47, e20160414. [Google Scholar] [CrossRef]
- Hansen, P.J. Prospects for Gene Introgression or Gene Editing as a Strategy for Reduction of the Impact of Heat Stress on Production and Reproduction in Cattle. Theriogenology 2020, 154, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.B.S.; Fonseca, L.F.S.; Pinheiro, D.G.; Magalhães, A.F.B.; Muniz, M.M.M.; Ferro, J.A.; Baldi, F.; Chardulo, L.A.L.; Schnabel, R.D.; Taylor, J.F.; et al. Spliced Genes in Muscle from Nelore Cattle and Their Association with Carcass and Meat Quality. Sci. Rep. 2020, 10, 1470. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Takahashi, H.; Nishimura, T.; Kuchida, K.; Mannen, H. Meat Produced by Japanese Black Cattle and Wagyu. Anim. Front. 2014, 4, 46–54. [Google Scholar] [CrossRef]
- Tor, M.; Vilaró, F.; Ros-Freixedes, R.; Álvarez-Rodríguez, J.; Bosch, L.; Gol, S.; Pena, R.N.; Reixach, J.; Estany, J. Circulating Non-Esterified Fatty Acids as Biomarkers for Fat Content and Composition in Pigs. Animals 2021, 11, 386. [Google Scholar] [CrossRef]
- Morzel, M.; Terlouw, C.; Chambon, C.; Micol, D.; Picard, B. Muscle Proteome and Meat Eating Qualities of Longissimus Thoracis of “Blonde d’Aquitaine” Young Bulls: A Central Role of HSP27 Isoforms. Meat Sci. 2008, 78, 297–304. [Google Scholar] [CrossRef]
- Lopez-Navarro, E.R.; Gutierrez, J. Metalloproteinases and Their Inhibitors in Neurological Disease. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 395, 27–38. [Google Scholar] [CrossRef]
- Bond, N.D.; Nelliot, A.; Bernardo, M.K.; Ayerh, M.A.; Gorski, K.A.; Hoshizaki, D.K.; Woodard, C.T. SSFTZ-F1 and Matrix Metalloproteinase 2 Are Required for Fat-Body Remodeling in Drosophila. Dev. Biol. 2011, 360, 286–296. [Google Scholar] [CrossRef]
- PAGEMCCAW, A. Remodeling the Model Organism: Matrix Metalloproteinase Functions in Invertebrates. Semin. Cell Dev. Biol. 2008, 19, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; Purslow, P.P. The Role of Matrix Metalloproteinases in Muscle and Adipose Tissue Development and Meat Quality: A Review. Meat Sci. 2016, 119, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Bai, Y.; Liu, J.; Cai, W.; Liu, L.; He, Y.; Song, J. Metabolic Regulations by LncRNA, MiRNA, and CeRNA Under Grass-Fed and Grain-Fed Regimens in Angus Beef Cattle. Front. Genet. 2021, 12, 579393. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, Z.; Pan, C.; Wang, S. Characterization of Feed Efficiency-Related Key Signatures Molecular in Different Cattle Breeds. PLoS ONE 2023, 18, e0289939. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.S.; Zhang, F.K.; Ma, Q.N.; Ehsan, M.; Zhao, Q.; Zhu, X. Transcriptomic Landscape of Hepatic Lymph Nodes, Peripheral Blood Lymphocytes and Spleen of Swamp Buffaloes Infected with the Tropical Liver Fluke Fasciola Gigantica. PLoS Neglected Trop. Dis. 2022, 16, e0010286. [Google Scholar] [CrossRef] [PubMed]
- Illa, S.K.; Mukherjee, S.; Nath, S.; Mukherjee, A. Genome-Wide Scanning for Signatures of Selection Revealed the Putative Genomic Regions and Candidate Genes Controlling Milk Composition and Coat Color Traits in Sahiwal Cattle. Front. Genet. 2021, 12, 699422. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, J.; Ma, Q.; Liu, A.; Li, Y.; Guan, G.; Luo, J.; Yin, H. Screening and Identification of Theileria Annulata Subtelomere-Encoded Variable Secreted Protein-950454 (SVSP454) Interacting Proteins from Bovine B Cells. Parasites Vectors 2021, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.S.; Kristensen, T.N.; Loeschcke, V. Effects of Inbreeding and Rate of Inbreeding in Drosophila Melanogaster–Hsp70 Expression and Fitness. J. Evol. Biol. 2005, 18, 756–762. [Google Scholar] [CrossRef]
- Arshad, U.; Sagheer, M.; González-Silvestry, F.B.; Hassan, M.; Sosa, F. Vitrification Improves In-Vitro Embryonic Survival in Bos Taurus Embryos without Increasing Pregnancy Rate Post Embryo Transfer When Compared to Slow-Freezing: A Systematic Meta-Analysis. Cryobiology 2021, 101, 1–11. [Google Scholar] [CrossRef]
- Hammond, K. Breeding Strategies for the Development of the Australian Beef Industry: An Overview. Aust. J. Exp. Agric. 2006, 46, 183–198. [Google Scholar] [CrossRef]
- Gonzalez-Rivas, P.A.; Chauhan, S.S.; Ha, M.; Fegan, N.; Dunshea, F.R.; Warner, R.D. Effects of Heat Stress on Animal Physiology, Metabolism, and Meat Quality: A Review. Meat Sci. 2020, 162, 108025. [Google Scholar] [CrossRef]
- Gomes, R.; Silva, S.; Carvalho, M.; Rezende, F.; Pinto, L.; Santana, M.; Stella, T.; Meirelles, F.; Júnior, P.R.; Leme, P.; et al. Protein synthesis and degradation gene SNPs related to feed intake, feed efficiency, growth, and ultrasound carcass traits in Nellore cattle. Genet. Mol. Res. 2013, 12, 2923–2936. [Google Scholar] [CrossRef]
- Oh, E.; Lee, B.; Choi, Y.M. Associations of Heat-Shock Protein Expression with Meat Quality and Sensory Quality Characteristics in Highly Marbled Longissimus Thoracis Muscle from Hanwoo Steers Categorized by Warner–Bratzler Shear Force Value. Foods 2019, 8, 638. [Google Scholar] [CrossRef]
- Silva-Vignato, B.; Coutinho, L.L.; Cesar, A.S.M.; Poleti, M.D.; Regitano, L.C.A.; Balieiro, J.C.C. Comparative Muscle Transcriptome Associated with Carcass Traits of Nellore Cattle. BMC Genom. 2017, 18, 506. [Google Scholar] [CrossRef]
- Hocquette, J.F. Endocrine and Metabolic Regulation of Muscle Growth and Body Composition in Cattle. Animal 2010, 4, 1797–1809. [Google Scholar] [CrossRef]
- Sedykh, T.A.; Dolmatova, I.Y.; Valitov, F.R.; Gizatullin, R.S.; Kalashnikova, L.A. The Influence of Growth Hormone Gene Polymorphism on Growth Rate of Young Cattle. Iran. J. Appl. Anim. Sci. 2020, 10, 445–451. [Google Scholar]
- Ariyasu, D.; Kubo, E.; Higa, D.; Shibata, S.; Takaoka, Y.; Sugimoto, M.; Imaizumi, K.; Hasegawa, T.; Araki, K. Decreased Activity of the Ghrhr and Gh Promoters Causes Dominantly Inherited GH Deficiency in Humanized GH1 Mouse Models. Endocrinology 2019, 160, 2673–2691. [Google Scholar] [CrossRef]
- Zia, M.; Hansen, J.; Hjort, K.; Valdes, C. Brazil Once Again Becomes the World’s Largest Beef Exporter. Amber Waves:Econ. Food Farming Nat. Resour. Rural Am. 2019, 2019, 2019–2021. [Google Scholar] [CrossRef]
- Voeltz, G.K.; Rolls, M.M.; Rapoport, T.A. Structural Organization of the Endoplasmic Reticulum. EMBO Rep. 2002, 3, 944–950. [Google Scholar] [CrossRef]
- Nakanishi, K.; Dohmae, N.; Morishima, N. O Estresse Do Retículo Endoplasmático Aumenta a Formação de Miofibras In Vitro. FASEB J. 2007, 21, 2994–3003. [Google Scholar] [CrossRef]
- Liljas, A. Ribosomes. Brenner’s Encycl. Genet. Second Ed. 2013, 6, 248–253. [Google Scholar] [CrossRef]
- Chaillou, T.; Kirby, T.J.; McCarthy, J.J. Ribosome Biogenesis: Emerging Evidence for a Central Role in the Regulation of Skeletal Muscle Mass. J. Cell. Physiol. 2014, 229, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Alwan, R.; Bruel, A.L.; Da Silva, A.; Blanquet, V.; Bouhouche, K. An SiRNA-Based Screen in C2C12 Myoblasts Identifies Novel Genes Involved in Myogenic Differentiation. Exp. Cell Res. 2017, 359, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the Human 80S Ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Chaillou, T. Ribosome Specialization and Its Potential Role in the Control of Protein Translation and Skeletal Muscle Size. J. Appl. Physiol. 2019, 127, 599–607. [Google Scholar] [CrossRef]
- Figueiredo, V.C.; McCarthy, J.J. Regulation of Ribosome Biogenesis in Skeletal Muscle Hypertrophy. Physiology 2019, 34, 30–42. [Google Scholar] [CrossRef]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase Superfamily: Conserved Structure and Molecular Mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef]
- Bryan, B.A.; Li, D.; Wu, X.; Liu, M. The Rho Family of Small GTPases: Crucial Regulators of Skeletal Myogenesis. Cell. Mol. Life Sci. CMLS 2005, 62, 1547–1555. [Google Scholar] [CrossRef]
- Boureux, A.; Vignal, E.; Faure, S.; Fort, P. Evolution of the Rho Family of Ras-like GTPases in Eukaryotes. Mol. Biol. Evol. 2007, 24, 203–216. [Google Scholar] [CrossRef]
- Hindi, S.M.; Tajrishi, M.M.; Kumar, A. Signaling Mechanisms in Mammalian Myoblast Fusion. Sci. Signal. 2013, 6, re2. [Google Scholar] [CrossRef]
- Rodríguez-Fdez, S.; Bustelo, X.R. Rho GTPases in Skeletal Muscle Development and Homeostasis. Cells 2021, 10, 2984. [Google Scholar] [CrossRef] [PubMed]
- Passetti, R.A.C.; Macedo, F.d.A.F.d.; Santos, G.R.d.A.; Bonin, E.; Vital, A.C.P.; Ramos, T.R.; Passetti, L.C.G.; Ornaghi, M.G.; Costa, I.C.A.; Prado, I.N.d. Sensorial, Color, Lipid Oxidation, and Visual Acceptability of Dry-Aged Beef from Young Bulls with Different Fat Thickness. Anim. Sci. J. 2020, 91, e13498. [Google Scholar] [CrossRef]
- Pophiwa, P.; Webb, E.C.; Frylinck, L. A Review of Factors Affecting Goat Meat Quality and Mitigating Strategies. Small Rumin. Res. 2020, 183, 106035. [Google Scholar] [CrossRef]
- Pacheco, R.F.; Machado, D.S.; Restle, J.; Sartori, D.B.S.; Costa, P.T.; Vaz, R.Z. Meta-Analysis of Meat Quality of Cattle Slaughtered with Different Subcutaneous Fat Thicknesses. Pesqui. Agropecu. Bras. 2023, 58, e03110. [Google Scholar] [CrossRef]
- Owens, F.N.; Dubeski, P.; Hanson, C.F. Factors That Alter the Growth and Development of Ruminants. J. Anim. Sci. 1993, 71, 3138–3150. [Google Scholar] [CrossRef] [PubMed]
- Naserkheil, M.; Mehrban, H.; Lee, D.; Park, M.N. Genome-Wide Association Study for Carcass Primal Cut Yields Using Single-Step Bayesian Approach in Hanwoo Cattle. Front Genet 2021, 12, 752424. [Google Scholar] [CrossRef]
- Iqbal, J.; Jahangir, Z.; Al-Qarni, A.A. Microsomal Triglyceride Transfer Protein: From Lipid Metabolism to Metabolic Diseases. In Lipid Transfer in Lipoprotein Metabolism and Cardiovascular Disease; Jiang, X.-C., Ed.; Springer: Singapore, 2020; pp. 37–52. ISBN 978-981-15-6082-8. [Google Scholar]
- Wilson, M.H.; Rajan, S.; Danoff, A.; White, R.J.; Hensley, M.R.; Quinlivan, V.H.; Recacha, R.; Thierer, J.H.; Tan, F.J.; Busch-Nentwich, E.M.; et al. A Point Mutation Decouples the Lipid Transfer Activities of Microsomal Triglyceride Transfer Protein. PLoS Genet 2020, 16, e1008941. [Google Scholar] [CrossRef]
- Wilcox, G. Insulin and Insulin Resistance. Clin. Biochem. Rev. 2005, 26, 19–39. [Google Scholar]
- De Sousa, O.A.; Cappellozza, B.I.; Fonseca, V.G.L.; Cooke, R.F. Insulin Resistance Increases as Days on Feed Advance in Feedlot Bos Indicus Beef Cattle Offered a High-Concentrate Finishing Diet. J Anim Sci 2022, 100, skac182. [Google Scholar] [CrossRef]
- Donicht, P.A.M.M. Efeitos Da Espessura De Gordura, Conformação, Peso De Carcaça E Idade Sobre a Qualidade Da Carcaça E Da Carne De Vacas De Descarte. Ph.D. Dissertation, Universidade Federal de Santa Maria, Santa Maria, Brazil, 2011; p. 175. [Google Scholar]
- Beld, J.; Lee, D.J.; Burkart, M.D. Fatty Acid Biosynthesis Revisited: Structure Elucidation and Metabolic Engineering. Mol. BioSystems 2015, 11, 38–59. [Google Scholar] [CrossRef]
- Kullmann, S.; Valenta, V.; Wagner, R.; Tschritter, O.; Machann, J.; Häring, H.U.; Preissl, H.; Fritsche, A.; Heni, M. Brain Insulin Sensitivity Is Linked to Adiposity and Body Fat Distribution. Nat. Commun. 2020, 11, 1841. [Google Scholar] [CrossRef]
- Schwarzmann, G.; Breiden, B.; Sandhoff, K. Membrane-Spanning Lipids for an Uncompromised Monitoring of Membrane Fusion and Intermembrane Lipid Transfer. J. Lipid Res. 2015, 56, 1861–1879. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.; Anderson, A.K.; Cowart, L.A. Sphingolipids in Adipose Tissue: What’s Tipping the Scale? Adv. Biol. Regul. 2018, 70, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Machado, P.C.; Pinto, L.F.B.; Silva, M.R.; Schenkel, F.S.; Brito, L.F.; Pedrosa, V.B. Genome-Wide Association Study and Pathway Analysis for Fat Deposition Traits in Nellore Cattle Raised in Pasture–Based Systems. J. Anim. Breed. Genet. 2021, 138, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Na, S.W.; Guan, L.L. Understanding the Role of Rumen Epithelial Host-Microbe Interactions in Cattle Feed Efficiency. Anim. Nutr. 2022, 10, 41–53. [Google Scholar] [CrossRef]
- Urbano, S.A.; Ferreira, M.A.; Oliveira, J.P.F.; de Lima Júnior, D.M.; Andrade, R.P.X. Lipid Sources on Modulation of Fatty Acid Profile of Small Ruminants Meat. Arch. Zootec. 2014, 63, 147–171. [Google Scholar] [CrossRef][Green Version]
- Yu, H.; Zhao, Z.; Yu, X.; Li, J.; Lu, C.; Yang, R. Bovine Lipid Metabolism Related Gene GPAM: Molecular Characterization, Function Identification, and Association Analysis with Fat Deposition Traits. Gene 2017, 609, 9–18. [Google Scholar] [CrossRef]
- Deprince, A.; Haas, J.T.; Staels, B. Dysregulated Lipid Metabolism Links NAFLD to Cardiovascular Disease. Mol. Metab. 2020, 42, 101092. [Google Scholar] [CrossRef]
- Magri, M.C.; Prata, T.V.G.; Manchiero, C.; Dantas, B.P.; Mazza, C.C.; Tengan, F.M. Genetic Variation in the Microsomal Triglyceride Transfer Protein (−493G/T) Is Associated with Hepatic Steatosis in Patients Infected with Hepatitis C Virus. BMC Infect. Dis. 2017, 17, 235. [Google Scholar] [CrossRef]
- Higuchi, N.; Kato, M.; Tanaka, M.; Miyazaki, M.; Takao, S.; Kohjima, M.; Kotoh, K.; Enjoji, M.; Nakamuta, M.; Takayanagi, R. Effects of Insulin Resistance and Hepatic Lipid Accumulation on Hepatic MRNA Expression Levels of ApoB, MTP and L-FABP in Non-Alcoholic Fatty Liver Disease. Exp. Ther. Med. 2011, 2, 1077–1081. [Google Scholar] [CrossRef]
- Ackman, R.G. Fatty Acids. Mar. Biog. Lipids Fats Oils 1989, 1, 103–137. [Google Scholar]
- Scollan, N.; Hocquette, J.-F.; Nuernberg, K.; Dannenberger, D.; Richardson, I.; Moloney, A. Innovations in Beef Production Systems That Enhance the Nutritional and Health Value of Beef Lipids and Their Relationship with Meat Quality. Meat Sci. 2006, 74, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.; Magistrali, A.; Butler, G.; Stergiadis, S. Nutritional Benefits from Fatty Acids in Organic and Grass-Fed Beef. Foods 2022, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.D.; Richardson, R.I.; Nute, G.R.; Fisher, A.V.; Campo, M.M.; Kasapidou, E.; Sheard, P.R.; Enser, M. Effects of Fatty Acids on Meat Quality: A Review. Meat Sci. 2004, 66, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Das, U.N. Essential Fatty Acids: Biochemistry, Physiology and Pathology. Biotechnol. J. Healthc. Nutr. Technol. 2006, 1, 420–439. [Google Scholar] [CrossRef] [PubMed]
- Christie, W.W.; Harwood, J.L. Oxidation of Polyunsaturated Fatty Acids to Produce Lipid Mediators. Essays Biochem. 2020, 64, 401–421. [Google Scholar] [PubMed]
- Lengi, A.J.; Corl, B.A. Comparison of Pig, Sheep and Chicken SCD5 Homologs: Evidence for an Early Gene Duplication Event. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 150, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, L.M. School of Agricultural and Veterinarian Sciences Campus of Jaboticabal Genomic Study for Female Sexual Precocity, Carcass, and Meat Quality Traits in Nellore Cattle; UNESP: São Paulo, Brazil, 2022. [Google Scholar]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M.; Balasubramanian, S. Meat Quality Assessment by Electronic Nose (Machine Olfaction Technology). Sensors 2009, 9, 6058–6083. [Google Scholar] [CrossRef]
- Volp, A.C.P.; Renhe, I.R.T.; Barra, K.; Stringueta, P.C. Flavonoids Anthocyanins: Characteristics and Properties in Nutrition and Health. Rev. Bras. Nutr. Clínica 2008, 23, 141–149. [Google Scholar]
- Zhang, W.; Liu, Y.; Zhang, H. Extracellular Matrix: An Important Regulator of Cell Functions and Skeletal Muscle Development. Cell Biosci. 2021, 11, 65. [Google Scholar] [CrossRef]
- Boles, J.A.; Shand, P.J. Effect of Muscle Location, Fiber Direction, and Slice Thickness on the Processing Characteristics and Tenderness of Beef Stir-Fry Strips from the Round and Chuck. Meat Sci. 2008, 78, 369–374. [Google Scholar] [CrossRef]
- Bravo-Sagua, R.; Parra, V.; Muñoz-Cordova, F.; Sanchez-Aguilera, P.; Garrido, V.; Contreras-Ferrat, A.; Chiong, M.; Lavandero, S. Chapter Five—Sarcoplasmic Reticulum and Calcium Signaling in Muscle Cells: Homeostasis and Disease. In Biology of the Endoplasmic Reticulum; Kepp, O., Galluzzi, L., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 350, pp. 197–264. ISBN 1937-6448. [Google Scholar]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L. Integrative Genomics Identifies MCU as an Essential Component of the Mitochondrial Calcium Uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, C.; Agnoletto, C.; Bononi, A.; Bonora, M.; De Marchi, E.; Marchi, S.; Missiroli, S.; Patergnani, S.; Poletti, F.; Rimessi, A. Mitochondrial Calcium Homeostasis as Potential Target for Mitochondrial Medicine. Mitochondrion 2012, 12, 77–85. [Google Scholar] [CrossRef]
- Dang, D.S.; Buhler, J.F.; Davis, H.T.; Thornton, K.J.; Scheffler, T.L.; Matarneh, S.K. Inhibition of Mitochondrial Calcium Uniporter Enhances Postmortem Proteolysis and Tenderness in Beef Cattle. Meat Sci. 2020, 162, 108039. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, J.E., Jr. What Do Scaffold Proteins Really Do? Sci. STKE 2000, 2000, pe1. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Gao, X.; Shen, Q.W.; Du, M.; Zhang, D. Phosphorylation Prevents In Vitro Myofibrillar Proteins Degradation by μ-Calpain. Food Chem. 2017, 218, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Parvatiyar, M.S.; Pinto, J.R. Pathogenesis Associated with a Restrictive Cardiomyopathy Mutant in Cardiac Troponin T Is Due to Reduced Protein Stability and Greatly Increased Myofilament Ca2+ Sensitivity. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC Transporters: The Power to Change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Thomas, C.; Aller, S.G.; Beis, K.; Carpenter, E.P.; Chang, G.; Chen, L.; Dassa, E.; Dean, M.; Duong Van Hoa, F.; Ekiert, D. Structural and Functional Diversity Calls for a New Classification of ABC Transporters. FEBS Lett. 2020, 594, 3767–3775. [Google Scholar] [CrossRef]
- Bunting, K.D. ABC Transporters as Phenotypic Markers and Functional Regulators of Stem Cells. Stem Cells 2002, 20, 11–20. [Google Scholar] [CrossRef]
- Zhao, C.; Ji, G.; Carrillo, J.A.; Li, Y.; Tian, F.; Baldwin, R.L.; Zan, L.; Song, J. The Profiling of DNA Methylation and Its Regulation on Divergent Tenderness in Angus Beef Cattle. Front. Genet. 2020, 11, 939. [Google Scholar] [CrossRef]
- Zhao, C.; Tian, F.; Yu, Y.; Luo, J.; Hu, Q.; Bequette, B.J.; Baldwin VI, R.L.; Liu, G.; Zan, L.; Scott Updike, M. Muscle Transcriptomic Analyses in Angus Cattle with Divergent Tenderness. Mol. Biol. Rep. 2012, 39, 4185–4193. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, R.; Pateiro, M.; Gagaoua, M.; Barba, F.J.; Zhang, W.; Lorenzo, J.M. A Comprehensive Review on Lipid Oxidation in Meat and Meat Products. Antioxidants 2019, 8, 429. [Google Scholar] [CrossRef] [PubMed]
- Daley, C.A.; Abbott, A.; Doyle, P.S.; Nader, G.A.; Larson, S. A Review of Fatty Acid Profiles and Antioxidant Content in Grass-Fed and Grain-Fed Beef. Nutr. J. 2010, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Horcada, A.; Polvillo, O.; González-Redondo, P.; López, A.; Tejerina, D.; Garcia-Torres, S. Stability of Fatty Acid Composition of Intramuscular Fat from Pasture- and Grain-Fed Young Bulls during the First 7d Postmortem. Arch. Anim. Breed. 2020, 63, 45–52. [Google Scholar] [CrossRef]
- Gurr, M.I.; Harwood, J.L.; Frayn, K.N. Lipid Biochemistry; Springer: Berlin/Heidelberg, Germany, 2002; Volume 409. [Google Scholar]
- Omi, S.; Nakata, R.; Okamura-Ikeda, K.; Konishi, H.; Taniguchi, H. Contribution of Peroxisome-Specific Isoform of Lon Protease in Sorting PTS1 Proteins to Peroxisomes. J. Biochem. 2008, 143, 649–660. [Google Scholar] [CrossRef]
- Yokota, S.; Dariush Fahimi, H. Degradation of Excess Peroxisomes in Mammalian Liver Cells by Autophagy and Other Mechanisms. Histochem. Cell Biol. 2009, 131, 455–458. [Google Scholar] [CrossRef]
- Mannaerts, G.P.; Van Veldhoven, P.P.; Casteels, M. Peroxisomal Lipid Degradation via β-and α-Oxidation in Mammals. Cell Biochem. Biophys. 2000, 32, 73–87. [Google Scholar] [CrossRef]
- Kersten, S.; Mandard, S.; Escher, P.; Gonzalez, F.J.; Tafuri, S.; Desvergne, B.; Wahli, W. The Peroxisome Proliferator-Activated Receptor α Regulates Amino Acid Metabolism. FASEB J. 2001, 15, 1971–1978. [Google Scholar] [CrossRef]
- Van Veldhoven, P.P. Biochemistry and Genetics of Inherited Disorders of Peroxisomal Fatty Acid Metabolism. J. Lipid Res. 2010, 51, 2863–2895. [Google Scholar] [CrossRef]
- Schuchardt, J.P.; Hahn, A. Bioavailability of Long-Chain Omega-3 Fatty Acids. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, R.N.; King, I.B. Very Long-Chain Saturated Fatty Acids and Diabetes and Cardiovascular Disease. Curr. Opin. Lipidol. 2022, 33, 76. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, N.; Morales de los Ríos, L.; Lindahl, M.; Quintero, F.J.; Pardo, J.M. Coordinated Transport of Nitrate, Potassium, and Sodium. Front. Plant Sci. 2020, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Zakany, F.; Kovacs, T.; Panyi, G.; Varga, Z. Direct and Indirect Cholesterol Effects on Membrane Proteins with Special Focus on Potassium Channels. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158706. [Google Scholar] [CrossRef]
- Zhai, M.; Yang, D.; Yi, W.; Sun, W. Involvement of Calcium Channels in the Regulation of Adipogenesis. Adipocyte 2020, 9, 132–141. [Google Scholar] [CrossRef]
- Ren, J.; Cheng, Y.; Wen, X.; Liu, P.; Zhao, F.; Xin, F.; Wang, M.; Huang, H.; Wang, W. BKCa Channel Participates in Insulin-induced Lipid Deposition in Adipocytes by Increasing Intracellular Calcium. J. Cell. Physiol. 2021, 236, 5818–5831. [Google Scholar] [CrossRef]

| Trait | N | CG | Mean | Min | Max | NAG | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REA | 2417 | 45 | 46.85 | 18.59 | 122.88 | 3.84 | 32.65 | 0.11 ± 0.05 | 2181 | 5.12 | 31.64 | 0.14 ± 0.03 |
| SFT | 2368 | 45 | 3.53 | 0.20 | 10.28 | 0.64 | 2.56 | 0.20 ± 0.07 | 2181 | 0.39 | 2.81 | 0.12 ± 0.03 |
| SF7 | 2431 | 45 | 6.64 | 1.43 | 13.98 | 0.74 | 3.33 | 0.18 ± 0.08 | 2181 | 0.41 | 3.65 | 0.10 ± 0.04 |
| IMF | 1854 | 33 | 1.76 | 0.01 | 4.52 | 0.12 | 0.47 | 0.21 ± 0.09 | 2181 | 0.13 | 0.47 | 0.21 ± 0.05 |
| Trait | BTA_Mb | Genomic Window (First–Last SNP) | #SNPs | %Var |
|---|---|---|---|---|
| REA | 1_139 | rs135422759–rs43160114 | 318 | 0.58 |
| 1_143 | rs108998682–rs110047615 | 225 | 0.51 | |
| 2_103 | rs133661878–rs110353021 | 179 | 0.71 | |
| 8_59 | rs135591081–rs134301958 | 206 | 0.63 | |
| 9_27 | rs133075840–rs135200101 | 319 | 0.58 | |
| 11_4 | rs41632961–rs110740485 | 250 | 0.59 | |
| 11_85 | rs136279193–rs133538333 | 245 | 0.56 | |
| 16_67 | rs135309995–rs41617334 | 257 | 0.73 | |
| 17_67 | rs133453006–rs109252298 | 202 | 0.50 | |
| 20_39 | rs133317890–rs135526251 | 295 | 0.51 | |
| SFT | 1_155 | rs42869710–rs109687330 | 276 | 0.54 |
| 2_105 | rs134465164–rs132698232 | 220 | 1.57 | |
| 4_117 | rs110440609–rs134013499 | 178 | 1.01 | |
| 6_26 | rs109588250–rs43453787 | 275 | 0.72 | |
| 6_78 | rs43057211–rs109068276 | 303 | 1.61 | |
| 11_95 | rs110260566–rs109229702 | 190 | 0.62 | |
| 12_2 | rs135898108–rs137788754 | 362 | 0.52 | |
| 12_39 | rs133691144–rs109792923 | 184 | 0.54 | |
| 12_41 | rs136100155–rs109234911 | 162 | 0.57 | |
| 17_47 | rs133188023–rs110546764 | 323 | 0.71 | |
| 24_31 | rs109644776–rs109246799 | 270 | 0.57 | |
| SF7 | 2_57 | rs43312105–rs135234804 | 253 | 1.02 |
| 5_68 | rs133020210–rs132713848 | 210 | 0.77 | |
| 6_99 | rs135511664–rs134451951 | 228 | 0.58 | |
| 11_28 | rs43677070–rs136376171 | 257 | 0.54 | |
| 24_48 | rs136072970–rs134381469 | 204 | 0.51 | |
| 27_7 | rs134965123–rs42619553 | 266 | 0.90 | |
| IMF | 6_68 | rs135331134–rs41653747 | 303 | 0.76 |
| 7_59 | rs109901778–rs133094202 | 228 | 0.54 | |
| 8_103 | rs134125299–rs136759870 | 258 | 0.71 | |
| 17_9 | rs42655606–rs135933217 | 290 | 0.57 | |
| 18_16 | rs109942482–rs133384244 | 264 | 0.56 | |
| 24_61 | rs133529820–rs135629046 | 268 | 0.62 | |
| 29_12 | rs42670744–rs133896635 | 189 | 0.54 |
| Chr | Genomic Range (Start–End Position) | Genes | %Var |
|---|---|---|---|
| 1 | 139678041–140081419 | CPNE4, MRPL3, NUDT16, NEK11, RF00026 | 0.58 |
| 143048262–144071071 | FAM3B, M X2, MX1, TMPRSS2, RIPK4, PRDM15, C2CD2, ZBTB21, UMODL1, ABCG1, TFF3 | 0.51 | |
| 2 | 103520024–103950562 | ABCA12, RF00156, ATIC, FN1 | 0.71 |
| 8 | 59427078–60332994 | RF00410, FAM205C, PHF24, RF00026, DNAJB5, VCP, FANCG, PIGO, STOML2, FAM214B, UNC13B, RUSC2, FAM166B, TESK1, CD72, SIT1, RF00030, CCDC107, ARHGEF39, CA9, TPM2, TLN1, CREB3, GBA2, RGP1, MSMP | 0.63 |
| 9 | 26690578–27348055 | RF00026, NKAIN2 | 0.58 |
| 11 | 3686196–4599212 | INPP4A, COA5, UNC50, MGAT4A, RF00425, TSGA10, C2orf15, LIPT1, MITD1, MRPL30, LYG2, TXNDC9, EIF5B, REV1 | 0.59 |
| 84872440–85408763 | RF00004, TRIB2, RF00279 | 0.56 | |
| 16 | 66960790–68078846 | C16H1orf21, EDEM3, FAM129A, RNF2, TRMT1L, SWT1, IVNS1ABP | 0.73 |
| 17 | 67357452–68355258 | - | 0.50 |
| 20 | 39073246–39413736 | PRLR, AGXT2, DNAJC21, BRIX1, RAD1, TTC23L, RF00003, RAI14 | 0.51 |
| Chr | Genomic Range (Start–End Position) | Genes | %Var |
|---|---|---|---|
| 1 | 155383458–156185921 | DAZL, PLCL2, RF00026, TBC1D5 | 0.54 |
| 2 | 105338358–106321176 | IGFBP2, IGFBP5, TNP1 | 1.57 |
| 4 | 117153120–118274566 | HTR5A, PAXIP1, INSIG1, CNPY1, RF00026, RBM33, SHH | 1.01 |
| 6 | 26167969–27080766 | DAPP1, C4orf54, MTTP, TRMT10A, C6H4orf17, ADH7, ADH6, ADH4, ADH5, METAP1, EIF4E | 0.72 |
| 78991885–79313522 | ADGRL3 | 1.61 | |
| 11 | 95314593–96511560 | NEK6, ‘PSMB7, ADGRD2, NR5A1, NR6A1, bta-mir-181a-2, bta-mir-181b-2, OLFML2A, RF00026, WDR38, RPL35, ARPC5L, GOLGA1, SCAI, RF00264, PPP6C, RABEPK, HSPA5, GAPVD1, RF00026, RF00020, MAPKAP1 | 0.62 |
| 12 | 1527007–2695362 | TDRD3, DIAPH3 | 0.52 |
| 39640396–39640466 | RF01161 | 0.54 | |
| 41873452–42872104 | - | 0.57 | |
| 17 | 47976840–48975164 | - | 0.71 |
| 24 | 31906526–32905155 | - | 0.57 |
| Chr | Genomic Range (Start–End Position) | Genes | %Var |
|---|---|---|---|
| 2 | 56711720–57011911 | - | 1.02 |
| 5 | 67612447–68813411 | STAB2, NT5DC3, HSP90B1, C5H12orf73, TDG, GLT8D2, HCFC2, NFYB, TXNRD1, CHST11, SLC41A2 | 0.77 |
| 6 | 99233279–100341274 | SCD5, SEC31A, THAP9, LIN54, bta-mir-2447, COPS4, RF00156, PLAC8B, PLAC8, COQ2, HPSE, bta-mir-2446, MRPS18C, ABRAXAS1, GPAT3, RF00265 | 0.58 |
| 11 | 27935104–29210371 | PRKCE, EPAS1, TMEM247, ATP6V1E2, RHOQ, PIGF, CRIPT, SOCS5, MCFD2 | 0.54 |
| 24 | 48278442–49277504 | - | 0.51 |
| 27 | 7955174–8952985 | - | 0.90 |
| Chr | Genomic Range (Start–End Position) | Genes | %Var |
|---|---|---|---|
| 6 | 67925293–69167282 | CORIN, NFXL1, CNGA1, NIPAL1, TXK TEC, SLAIN2, SLC10A4, ZAR1, FRYL, RF00001, OCIAD1 | 0.76 |
| 7 | 59547160–60543762 | - | 0.54 |
| 8 | 103059951–104010320 | RF00001, SUSD1, RF00026, PTBP3, HSDL2, KIAA1958, INIP, SNX30, SLC46A2 | 0.71 |
| 17 | 9882597–10881624 | - | 0.57 |
| 18 | 16921890–17921036 | ABCC12, ABCC11, LONP2, SIAH1 | 0.56 |
| 24 | 61023178–62021049 | - | 0.62 |
| 29 | 12754057–13751988 | PRCP, FAM181B | 0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reis, H.B.D.; Carvalho, M.E.; Espigolan, R.; Poleti, M.D.; Ambrizi, D.R.; Berton, M.P.; Ferraz, J.B.S.; de Mattos Oliveira, E.C.; Eler, J.P. Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle. Metabolites 2024, 14, 6. https://doi.org/10.3390/metabo14010006
Reis HBD, Carvalho ME, Espigolan R, Poleti MD, Ambrizi DR, Berton MP, Ferraz JBS, de Mattos Oliveira EC, Eler JP. Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle. Metabolites. 2024; 14(1):6. https://doi.org/10.3390/metabo14010006
Chicago/Turabian StyleReis, Hugo Borges Dos, Minos Esperândio Carvalho, Rafael Espigolan, Mirele Daiana Poleti, Dewison Ricardo Ambrizi, Mariana Piatto Berton, José Bento Sterman Ferraz, Elisângela Chicaroni de Mattos Oliveira, and Joanir Pereira Eler. 2024. "Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle" Metabolites 14, no. 1: 6. https://doi.org/10.3390/metabo14010006
APA StyleReis, H. B. D., Carvalho, M. E., Espigolan, R., Poleti, M. D., Ambrizi, D. R., Berton, M. P., Ferraz, J. B. S., de Mattos Oliveira, E. C., & Eler, J. P. (2024). Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle. Metabolites, 14(1), 6. https://doi.org/10.3390/metabo14010006







