Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves
Abstract
:1. Introduction
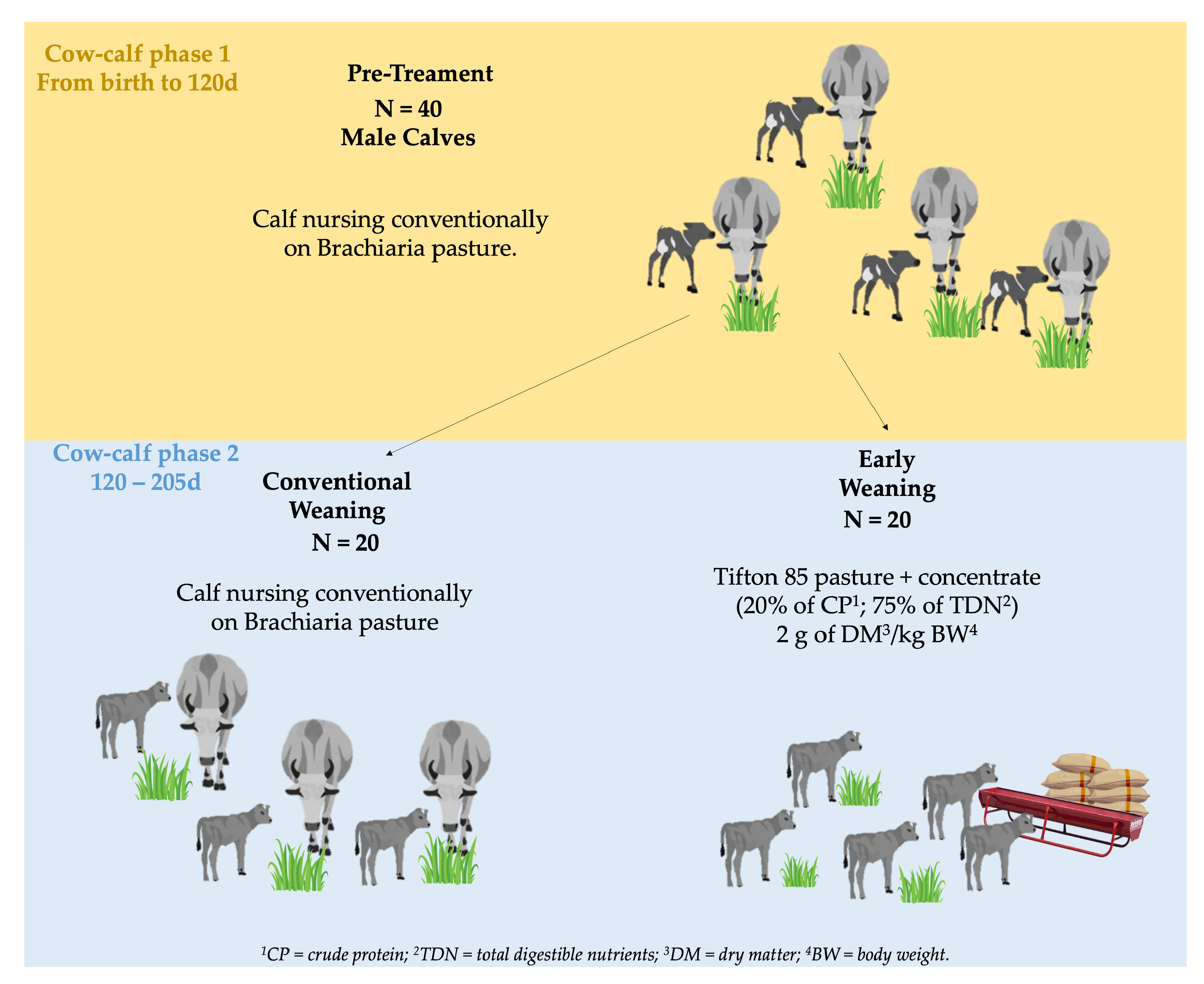
2. Materials and Methods
2.1. Samples and Weight Recording
2.2. Growing Performance Statistics
2.3. Collection of Muscle Tissue and Total RNA Extraction
2.4. RNA Sequencing
2.5. Mapping of Sequences to the Reference Genome
2.6. Identification of Differentially Expressed Genes
2.7. Enrichment of Functional Terms
3. Results
3.1. Pre- and Post-Early Weaning Performance
3.2. RNA Extraction and Sequencing
3.3. Count Matrix and Quality
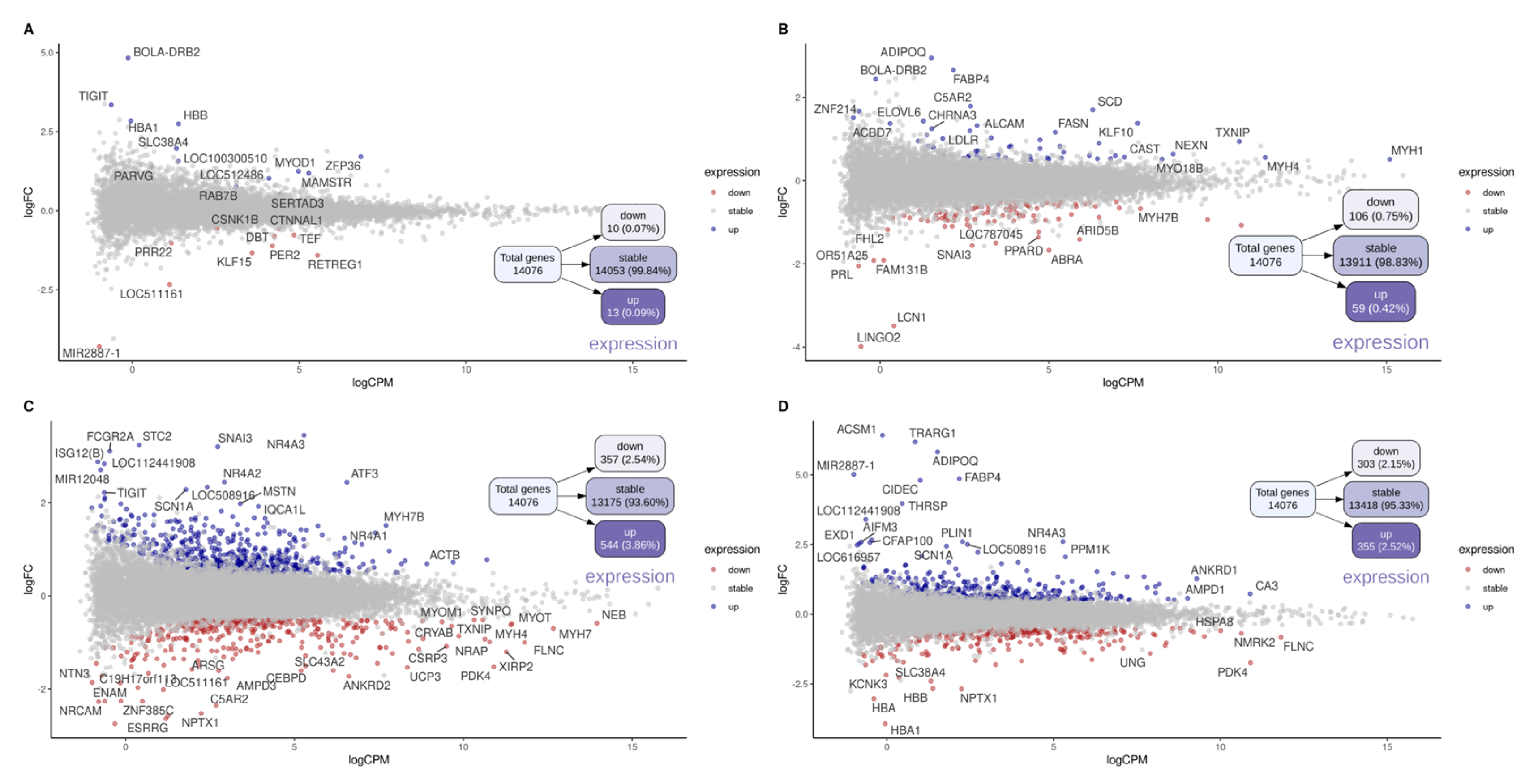
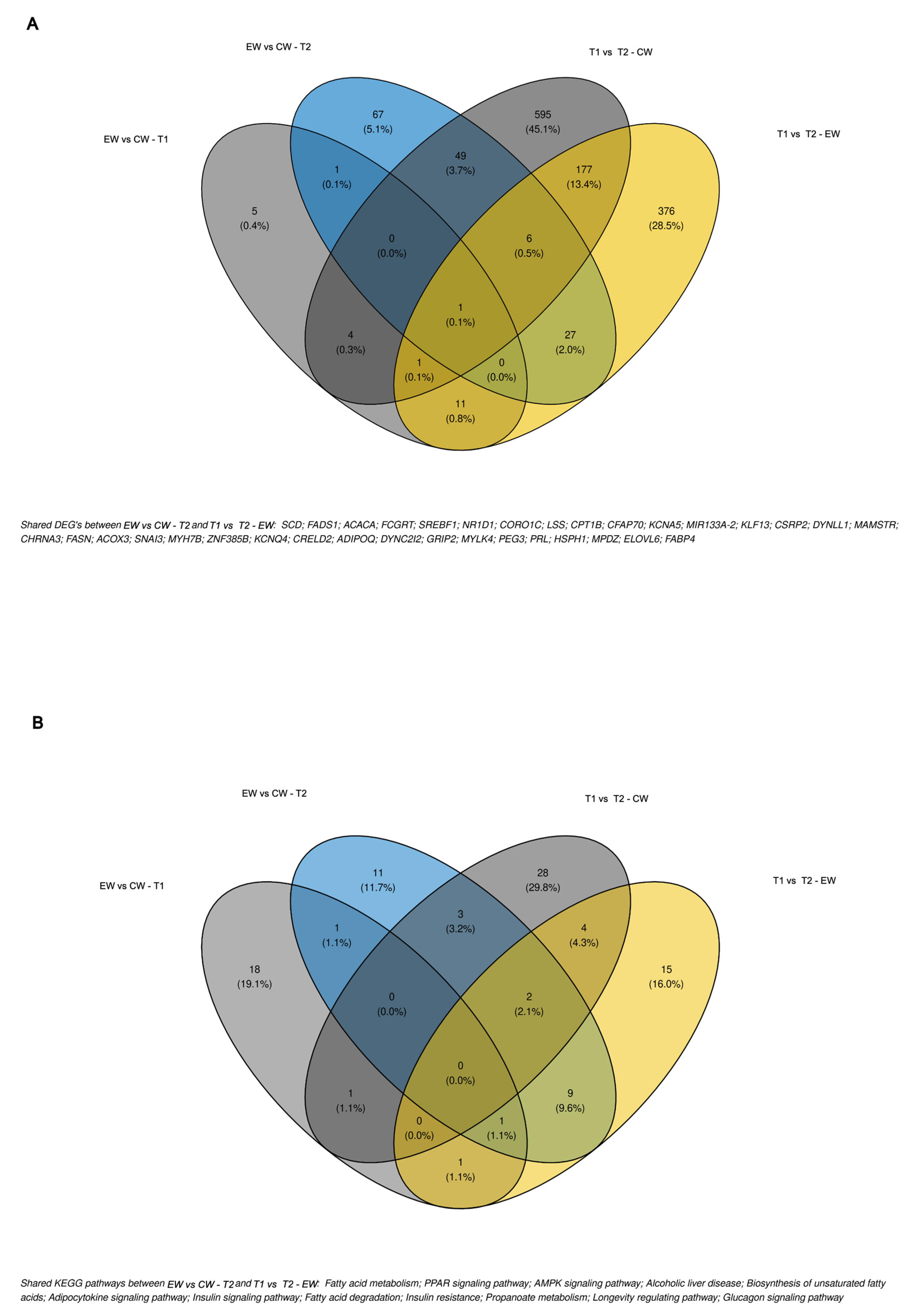
3.4. Differential Gene Expression
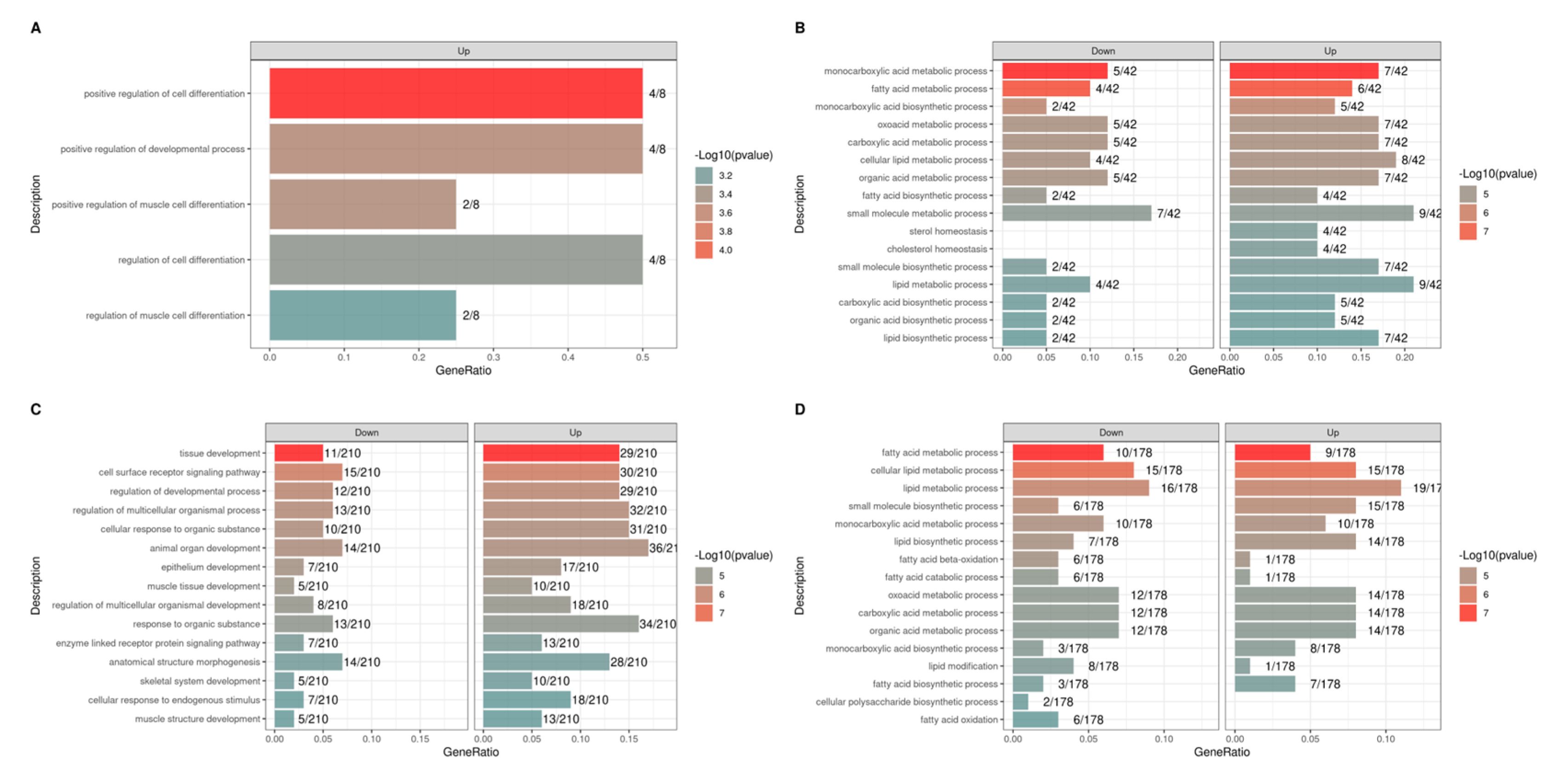
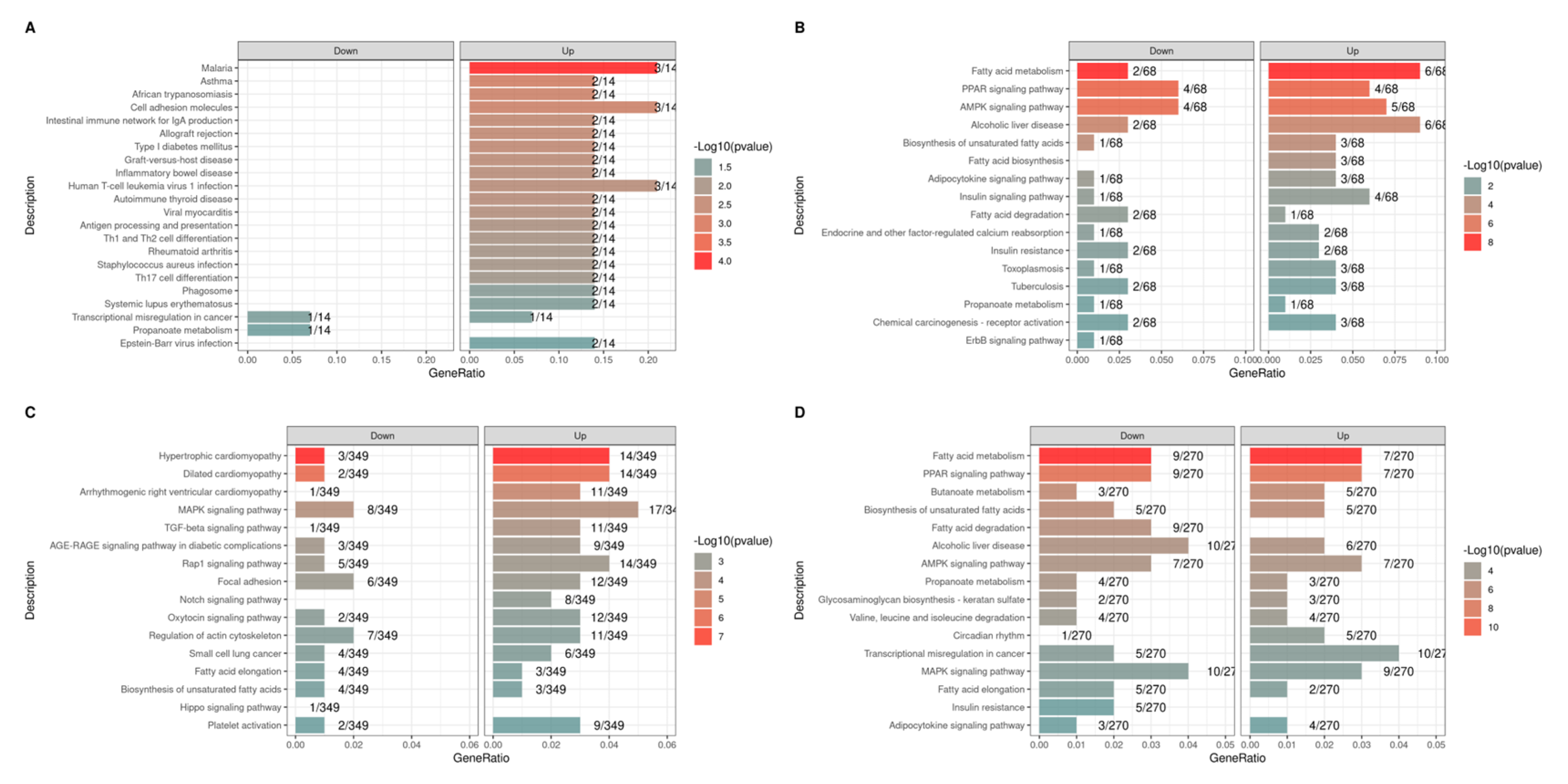
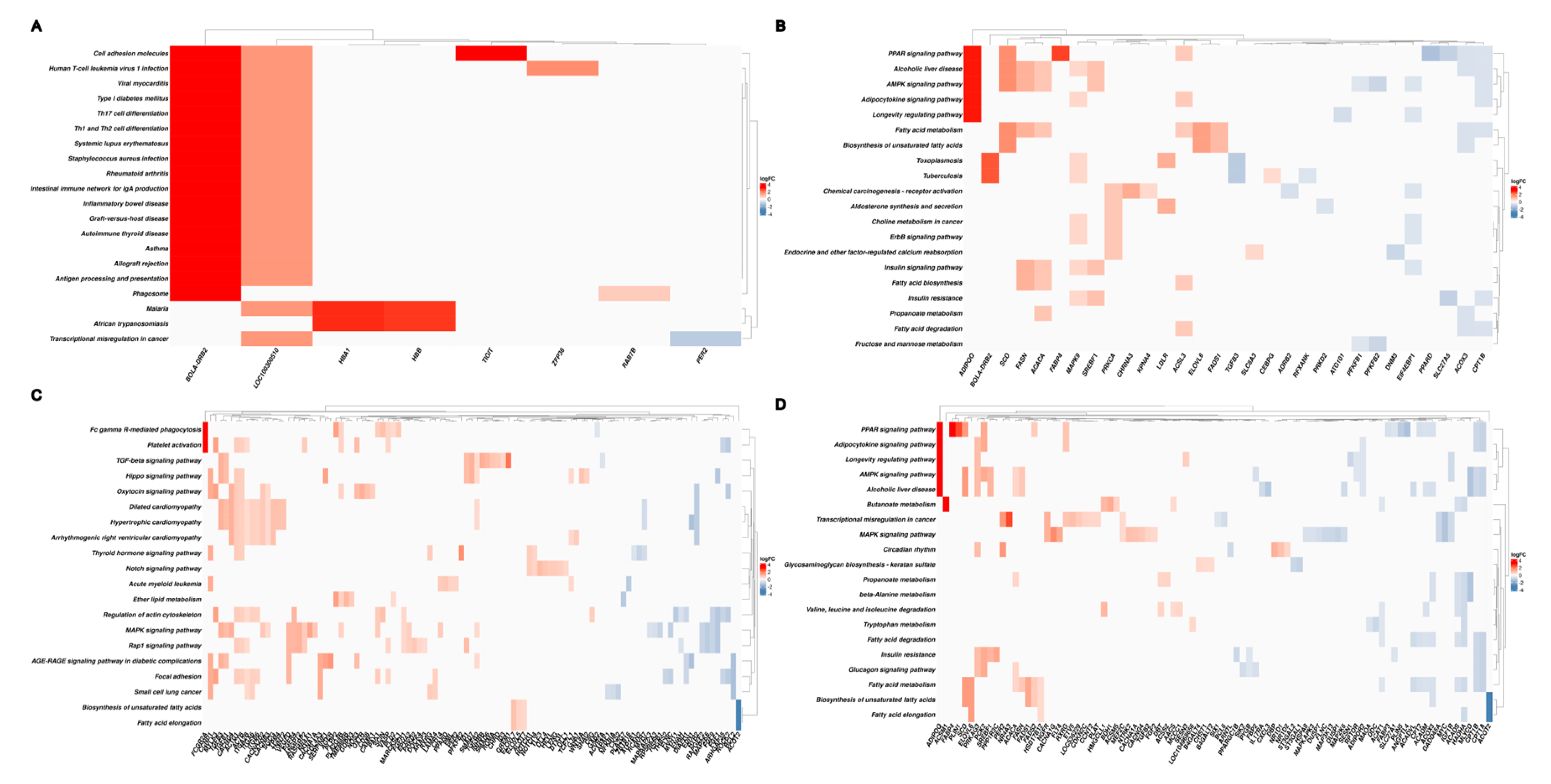
3.5. Functional Enrichment Analysis of Differentially Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beatty, D.T.; Barnes, A.; Taylor, E.; Pethick, D.; McCarthy, M.; Maloney, S.K. Physiological responses of Bos taurus and Bos indicus cattle to prolonged, continuous heat and humidity. J. Anim. Sci. 2006, 84, 972–985. [Google Scholar] [CrossRef]
- Burrow, H.M. Importance of adaptation and genotype × environment interactions in tropical beef breeding systems. Animal 2012, 6, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.E.; Bowman, P.J.; Pryce, J.E.; Hayes, B.J.; Goddard, M.E. Long-term selection strategies for complex traits using high-density genetic markers. J. Dairy Sci. 2012, 95, 4646–4656. [Google Scholar] [CrossRef]
- Mrode, R.; Ojango, J.M.K.; Okeyo, A.M.; Mwacharo, J.M. Genomic selection and use of molecular tools in breeding programs for indigenous and crossbred cattle in developing countries: Current status and future prospects. Front. Genet. 2019, 9, 694. [Google Scholar] [CrossRef]
- Carvalho, V.V.; Paulino, M.F.; Detmann, E.; Valadares Filho, S.C.; Lopes, S.A.; Rennó, L.N.; Sampaio, C.B.; Silva, A.G. A meta-analysis of the effects of creep-feeding supplementation on performance and nutritional characteristics by beef calves grazing on tropical pastures. Livest. Sci. 2019, 227, 175–182. [Google Scholar] [CrossRef]
- Tambara, A.A.C.; Härter, C.J.; Rabelo, C.H.S.; Kozloski, G.V. Effects of supplementation on production of beef cattle grazing tropical pastures in Brazil during the wet and dry seasons: A meta-analysis. Rev. Bras. De Zootec. 2021, 50, e20210020. [Google Scholar] [CrossRef]
- Fordyce, G.; Cooper, N.J.; Kendall, I.E.; O’Leary, B.M.; Faveri, J.D.E.; De Faveri, J. Creep feeding and prepartum supplementation effects on growth and fertility of Brahman-cross cattle in the dry tropics. Aust. J. Exp. Agric. 1996, 36, 389–395. [Google Scholar] [CrossRef]
- Henriques, L.T.; Valadares Filho, S.D.C.; Fonseca, M.A.; Paulino, P.V.R.; Detmann, E.; Valadares, R.F.D. Evaluation of non-linear models and the effects of primiparous cows and calves intake on the lactation curve of Nelore cows. Rev. Bras. De Zootec. 2011, 40, 1287–1295. [Google Scholar] [CrossRef]
- Lusby, K.S.; Wettemann, R.P.; Turman, E.J. Effects of early weaning calves from first-calf heifers on calf and heifer performance. J. Anim. Sci. 1981, 53, 1193–1197. [Google Scholar] [CrossRef]
- Barnes, K.C.; Selk, G.E.; Lusby, K.S. Early weaning fall-born beef calves for improved rebreeding or decreased winter feed. Okla. Agric. Exp. Sta. Res. Rep. 1996, 951, 85–87. [Google Scholar]
- Cattelam, J. Desmame precoce na podução de bovinos de corte. Rev. De Ciências Agroveterinárias 2014, 13, 190–198. [Google Scholar]
- Ferraz, J.B.S.; de Felício, P.E. Production systems–An example from Brazil. Meat Sci. 2010, 84, 238–243. [Google Scholar] [CrossRef]
- Costa e Silva, L.F.; Engle, T.E.; de Valadares Filho, S.; Rotta, P.P.; Villadiego, F.A.C.; Silva, F.A.S.; Martins, E.C.; Silva, L.H.R.; Paulino, M.F. Nellore cows and their calves during the lactation period: Performance, intake, milk composition, and total apparent digestibility. Trop. Anim. Health Prod. 2015, 47, 735–741. [Google Scholar] [CrossRef]
- Wardrop, I.D. The effects of the plane of nutrition in early post-natal life on the subsequent growth and development of cattle. Aust. J. Agric. Res. 1966, 17, 375–385. [Google Scholar] [CrossRef]
- Restle, J.; Polli, V.A.; Alves Filho, D.C.; Senna, D.B.D.; Vaz, R.Z.; Bernardes, R.A.C.; Silva, J.H.S.D. Desenvolvimento de bovinos de corte de diferentes grupos genéticos desmamados aos 3 ou 7 meses de idade. Rev. Bras. De Zootec. 1999, 28, 1023–1030. [Google Scholar] [CrossRef]
- Arthington, J.D.; Spears, J.W.; Miller, D.C. The effect of early weaning on feedlot performance and measures of stress in beef calves. J. Anim. Sci. 2005, 83, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Vaz, R.Z.; Lobato, J.F.P. Efeito da idade do desmame no desenvolvimento de novilhas de corte até os 14/15 meses de idade. Rev. Bras. De Zootec. 2010, 39, 289–298. [Google Scholar] [CrossRef]
- Neville, W.E., Jr.; McCormick, W.C. Performance of early-and normal-weaned beef calves and their dams. J. Anim. Sci. 1981, 52, 715–724. [Google Scholar] [CrossRef]
- Fluharty, F.L.; Loerch, S.C.; Turner, T.B.; Moeller, S.J.; Lowe, G.D. Effects of weaning age and diet on growth and carcass characteristics in steers. J. Anim. Sci. 2000, 78, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.E.; Faulkner, D.B.; Ireland, F.A.; Parrett, D.F. Comparison of three weaning ages on cow-calf performance and steer carcass traits. J. Anim. Sci. 1999, 77, 323–329. [Google Scholar] [CrossRef]
- Reddy, K.E.; Jeong, J.; Lee, S.D.; Baek, Y.C.; Oh, Y.; Kim, M.; So, K.M.; Kim, D.W.; Kim, J.H.; Park, S.; et al. Effect of different early weaning regimens for calves on adipogenic gene expression in Hanwoo loin at the fattening stage. Livest. Sci. 2017, 195, 87–98. [Google Scholar] [CrossRef]
- Schoonmaker, J.P.; Cecava, M.J.; Faulkner, D.B.; Fluharty, F.L.; Zerby, H.N.; Loerch, S.C. Effect of source of energy and rate of growth on performance, carcass characteristics, ruminal fermentation, and serum glucose and insulin of early-weaned steers. J. Anim. Sci. 2003, 81, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Sithyphone, K.; Yabe, M.; Horita, H.; Hayashi, K.; Fumita, T.; Shiotsuka, Y.; Etoh, T.; Ebara, F.; Samadmanivong, O.; Wegner, J.; et al. Comparison of feeding systems: Feed cost, palatability and environmental impact among hay-fattened beef, consistent grass-only-fed beef and conventional marbled beef in Wagyu (Japanese Black cattle). J. Anim. Sci. 2011, 82, 352–359. [Google Scholar] [CrossRef]
- Scheffler, J.M.; McCann, M.A.; Greiner, S.P.; Jiang, H.; Hanigan, M.D.; Bridges, G.A.; Gerrard, D.E. Early metabolic imprinting events increase marbling scores in fed cattle. J. Anim. Sci. 2014, 92, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Rasby, R. Early weaning beef calves. Vet. Clin. N. Am. Food Anim. Pract. 2007, 23, 29–40. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 June 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- R Core Team. R: A Language Land Environment for Statistical Computing. R Foundation for Statistical Computing. 2022. Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 28 September 2022).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0. 7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 3 October 2022).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Int. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Myers, S.E.; Faulkner, D.B.; Nash, T.G.; Berger, L.L.; Parrett, D.F.; McKeith, F.K. Performance and carcass traits of early-weaned steers receiving either a pasture growing period or a finishing diet at weaning. J. Anim. Sci. 1999, 77, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, L.E.; Moisá, S.J. Stress, immunity, and the management of calves. J. Dairy Sci. 2016, 99, 3199–3216. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.L.; Kerley, M.S.; Walker, E.L.; Keisler, D.H.; Pierce, V.L.; Schmidt, T.B.; Stahl, C.A.; Linville, M.L.; Berg, E.P. Growth rate, body composition, and meat tenderness in early vs. traditionally weaned beef calves. J. Anim. Sci. 2005, 83, 2752–2761. [Google Scholar] [CrossRef] [PubMed]
- Bonny, S.; Gardner, G.; Pethick, D.; Legrand, I.; Polkinghorne, R.; Hocquette, J. Biochemical measurements of beef are a good predictor of untrained consumer sensory scores across muscles. Animal 2015, 9, 179–190. [Google Scholar] [CrossRef]
- Fonseca, L.F.S.; dos Santos Silva, D.B.; Gimenez, D.F.J.; Baldi, F.; Ferro, J.A.; Chardulo, L.A.L.; de Albuquerque, L.G. Gene expression profiling and identification of hub genes in Nellore cattle with different marbling score levels. Genomics 2020, 112, 873–879. [Google Scholar] [CrossRef]
- Ladeira, M.M.; Schoonmaker, J.P.; Swanson, K.C.; Duckett, S.K.; Gionbelli, M.P.; Rodrigues, L.M.; Teixeira, P.D. Nutrigenomics of marbling and fatty acid profile in ruminant meat. Animal 2018, 12, s282–s294. [Google Scholar] [CrossRef]
- Reggio, A.; Rosina, M.; Palma, A.; Cerquone Perpetuini, A.; Petrilli, L.L.; Gargioli, C.; Fuoco, C.; Micarelli, E.; Giuliani, G.; Cerretani, M.; et al. Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/β-catenin axis. Cell Death Differ. 2020, 10, 2921–2941. [Google Scholar] [CrossRef]
- Cianzo, D.S.; Topel, D.G.; Whitehurst, G.B.; Beitz, D.C.; Self, H.L. Adipose tissue growth and cellularity: Changes in bovine adipocyte and number. J. Anim. Sci. 1985, 60, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, Q.; Harris, C.L.; Nelson, M.L.; Busboom, J.R.; Zhu, M.J.; Du, M. Nutrigenomic regulation of adipose tissue development—Role of retinoic acid: A review. Meat Sci. 2016, 120, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Moisá, S.J.; Shike, D.W.; Faulkner, D.B.; Meteer, W.T.; Keisler, D.; Loor, J.J. Central role of the PPARγ gene network in coordinating beef cattle intramuscular adipogenesis in response to weaning age and nutrition. Gene Regul. Syst. Biol. 2014, 8, GRSB-S11782. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, N.; Klein, R.L.; Garvey, W.T. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J. Lipid Res. 2005, 46, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Bower, N.I.; Reverter, A.; Tan, S.H.; De Jager, N.; Wang, R.; McWilliam, S.M.; Cafe, L.M.; Greenwood, P.L.; Lehnert, S.A.; et al. Gene expression patterns during intramuscular fat development in cattle. J. Anim. Sci. 2009, 87, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, J.; Wang, X.; Ma, Y. Transcription factors regulate adipocyte differentiation in beef cattle. Anim. Genet. 2020, 51, 351–357. [Google Scholar] [CrossRef]
- Graugnard, D.E.; Piantoni, P.; Bionaz, M.; Berger, L.L.; Faulkner, D.B.; Loor, J.J. Adipogenic and energy metabolism gene networks in longissimus lumborum during rapid post-weaning growth in Angus and Angus × Simmental cattle fed high-starch or low-starch diets. BMC Genom. 2009, 10, 142. [Google Scholar] [CrossRef]
- Du, M.; Wang, B.; Fu, X.; Yang, Q.; Zhu, M.J. Fetal programming in meat production. Meat Sci. 2015, 109, 40–47. [Google Scholar] [CrossRef]
- Smith, S.B.; Gill, C.A.; Lunt, D.K.; Brooks, M.A. Regulation of fat and fatty acid composition in beef cattle. Asian-Australas. J. Anim. Sci. 2009, 22, 1225–1233. [Google Scholar] [CrossRef]
- Smith, S.B.; Blackmon, T.L.; Sawyer, J.E.; Miller, R.K.; Baber, J.R.; Morrill, J.C.; Wickersham, T.A. Glucose and acetate metabolism in bovine intramuscular and subcutaneous adipose tissues from steers infused with glucose, propionate, or acetate. J. Anim. Sci. 2018, 96, 921–929. [Google Scholar] [CrossRef]
- Shijun, L.; Khan, R.; Raza SH, A.; Jieyun, H.; Chugang, M.; Kaster, N.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef]
- Hudson, N.J.; Reverter, A.; Greenwood, P.L.; Guo, B.; Cafe, L.M.; Dalrymple, B.P. Longitudinal muscle gene expression patterns associated with differential intramuscular fat in cattle. Animal 2015, 9, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yin, J.; Zhu, M.J. Cellular signaling pathways regulating the initial stage of adipogenesis and marbling of skeletal muscle. Meat Sci. 2010, 86, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Hossner, K.L. Hormonal Regulation of Farm Animal Growth; CABI: Wallingford, UK, 2005. [Google Scholar]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.A.; Partridge, T.A.; Beauchamp, J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef] [PubMed]
- Ustanina, S.; Carvajal, J.; Rigby, P.; Braun, T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells 2007, 25, 2006–2016. [Google Scholar] [CrossRef]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. J. Biol. 2005, 132, 2685–2695. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Guo, T.; Jou, W.; Chanturiya, T.; Portas, J.; Gavrilova, O.; McPherron, A.C. Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS ONE 2009, 4, e4937. [Google Scholar] [CrossRef]
- Kim, H.S.; Liang, L.; Dean, R.G.; Hausman, D.B.; Hartzell, D.L.; Baile, C. Inhibition of preadipocyte differentiation by myostatin treatment in 3T3-L1 cultures. Biochem. Biophys. Res. Commun. 2001, 281, 902–906. [Google Scholar] [CrossRef]
- Obregon, M.J. Thyroid hormone and adipocyte differentiation. Thyroid 2008, 18, 185–195. [Google Scholar] [CrossRef]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S.; et al. Board-invited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef]
- Smith, S. The animal fatty acid synthase: One gene, one polypeptide, seven enzymes. FASEB J. 1994, 8, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.D.; Oliveira, D.M.; Chizzotti, M.L.; Chalfun-Junior, A.; Coelho, T.C.; Gionbelli, M.; Paiva, L.V.; Carvalho, J.R.R.; Ladeira, M.M. Subspecies and diet affect the expression of genes involved in lipid metabolism and chemical composition of muscle in beef cattle. Meat Sci. 2017, 133, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Michal, J.J.; Tobey, D.J.; Daniels, T.F.; Rule, D.C.; Macneil, M.D. Significant associations of stearoyl-CoA desaturase (SCD1) gene with fat deposition and composition in skeletal muscle. Int. J. Biol. Sci. 2008, 4, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, Y.; Wang, X.; Qiu, J.; Liang, C.; Cheng, G.; Wang, H.; Zhao, C.; Yang, W.; Zan, L.; et al. Bovine Stearoyl-CoA desaturase 1 promotes adipogenesis by activating the PPARγ receptor. J. Agric. Food Chem. 2020, 68, 12058–12066. [Google Scholar] [CrossRef] [PubMed]
- Jurie, C.; Cassar-Malek, I.; Bonnet, M.; Leroux, C.; Bauchart, D.; Boulesteix, P.; Pethick, D.W.; Hocquette, J.F. Adipocyte fatty acid-binding protein and mitochondrial enzyme activities in muscles as relevant indicators of marbling in cattle. J. Anim. Sci. 2007, 85, 2660–2669. [Google Scholar] [CrossRef]
- Vasconcelos, J.T.; Sawyer, J.E.; Tedeschi, L.O.; McCollum, F.T.; Greene, L.W. Effects of different growing diets on performance, carcass characteristics, insulin sensitivity, and accretion of intramuscular and subcutaneous adipose tissue of feedlot cattle. J. Anim. Sci. 2009, 87, 1540–1547. [Google Scholar] [CrossRef]
| Categories | Group | Days | n | Min. | Max. | Median | Mean 1 | SD 2 |
|---|---|---|---|---|---|---|---|---|
| Live weight (kg) | CW | 120 | 20 | 103 | 155 | 122 | 122.50 ± 2.961 b | 13.241 |
| EW | 120 | 20 | 96 | 141 | 125 | 123.15 ± 2.606 b | 11.654 | |
| CW | 205 | 20 | 169 | 216 | 198 | 194.39 ± 3.023 a | 12.825 | |
| EW | 205 | 20 | 156 | 221 | 191 | 190.32 ± 4.375 a | 19.070 | |
| ADG (kg) | CW | 205–120 | 20 | 0.553 | 1.047 | 0.853 | 0.845 ± 0.028 a | 0.127 |
| EW | 205–120 | 20 | 0.588 | 1.024 | 0.788 | 0.786 ± 0.030 a | 0.133 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedro, A.E.; Torrecilhas, J.A.; Torres, R.N.S.; Ramírez-Zamudio, G.D.; Baldassini, W.A.; Chardulo, L.A.L.; Curi, R.A.; Russo, G.H.; Napolitano, J.A.; Bezerra Tinoco, G.L.; et al. Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves. Metabolites 2023, 13, 1028. https://doi.org/10.3390/metabo13091028
Pedro AE, Torrecilhas JA, Torres RNS, Ramírez-Zamudio GD, Baldassini WA, Chardulo LAL, Curi RA, Russo GH, Napolitano JA, Bezerra Tinoco GL, et al. Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves. Metabolites. 2023; 13(9):1028. https://doi.org/10.3390/metabo13091028
Chicago/Turabian StylePedro, Ariane Enara, Juliana Akamine Torrecilhas, Rodrigo Nazaré Santos Torres, Germán Darío Ramírez-Zamudio, Welder Angelo Baldassini, Luis Artur Loyola Chardulo, Rogério Abdallah Curi, Gustavo Henrique Russo, Juliane Arielly Napolitano, Gustavo Lucas Bezerra Tinoco, and et al. 2023. "Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves" Metabolites 13, no. 9: 1028. https://doi.org/10.3390/metabo13091028
APA StylePedro, A. E., Torrecilhas, J. A., Torres, R. N. S., Ramírez-Zamudio, G. D., Baldassini, W. A., Chardulo, L. A. L., Curi, R. A., Russo, G. H., Napolitano, J. A., Bezerra Tinoco, G. L., Mariano, T. B., Caixeta, J. L., Moriel, P., & Pereira, G. L. (2023). Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves. Metabolites, 13(9), 1028. https://doi.org/10.3390/metabo13091028











