The Battle for Survival: The Role of RNA Non-Canonical Tails in the Virus–Host Interaction
Abstract
1. Introduction
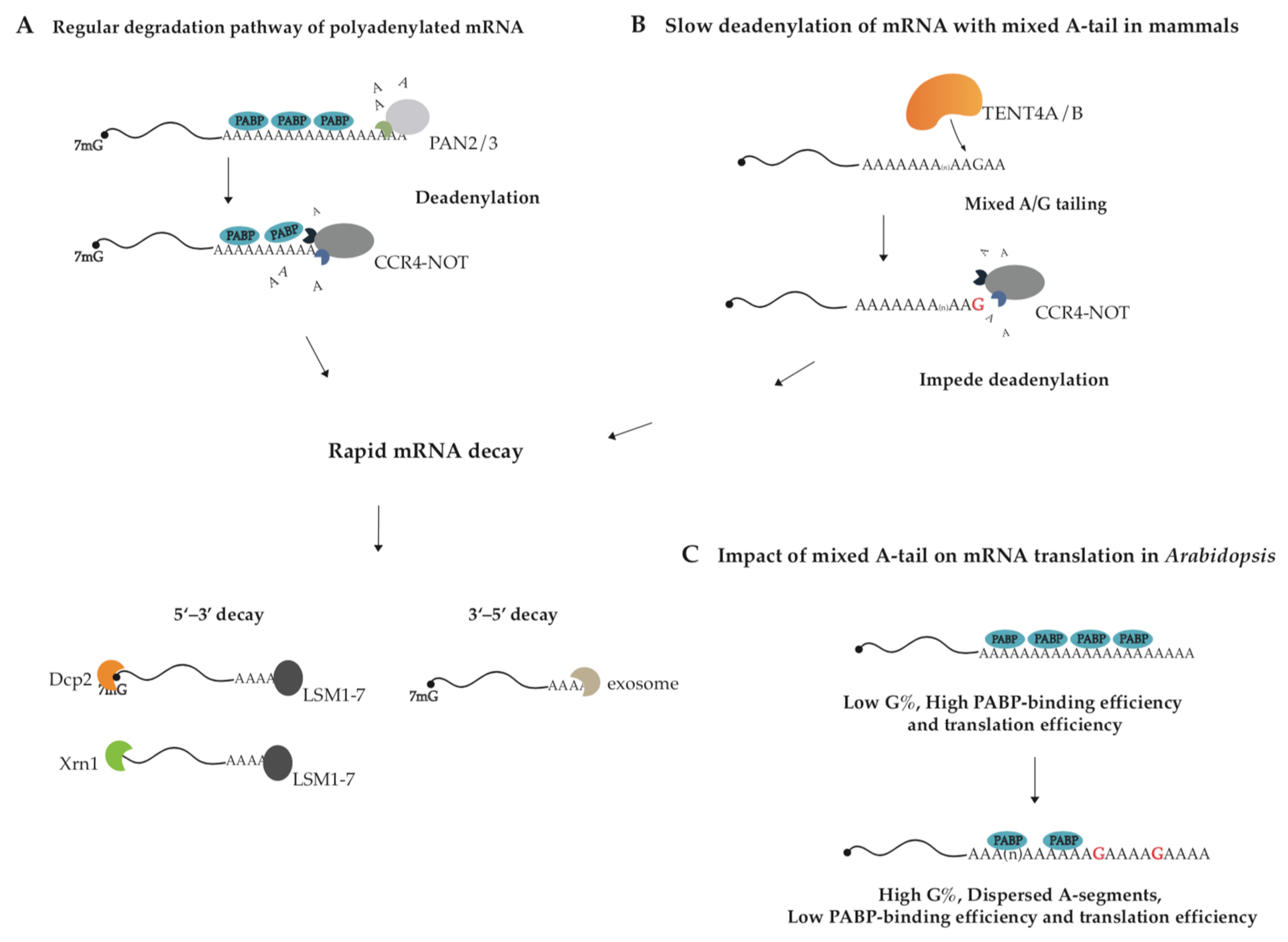
2. Mixed Tail in Viral Infection
2.1. The Cellular Function of Mixed Tail
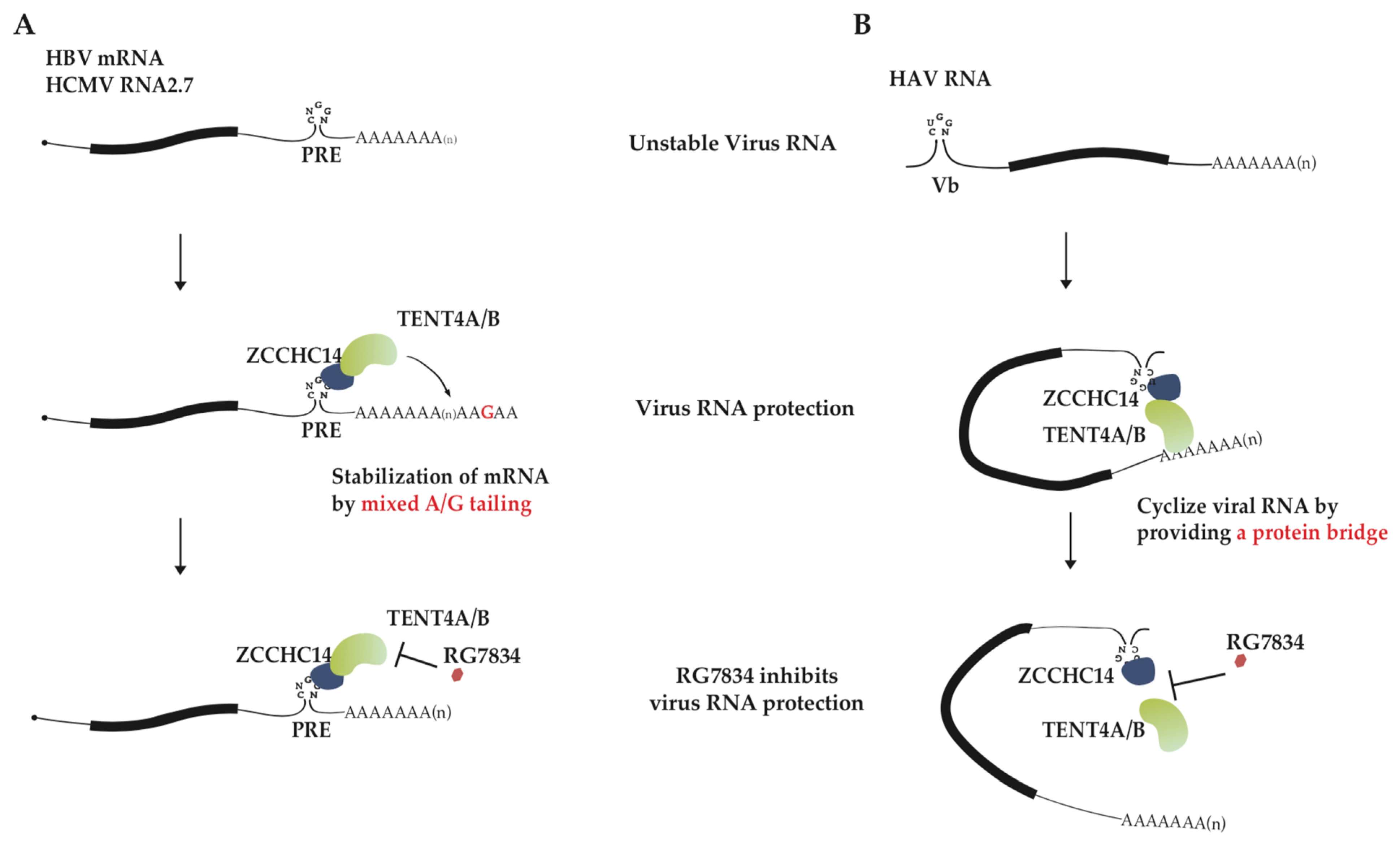
2.2. The Pathological Function of Mixed Tail in Virus Infection
2.3. TENT4-ZCCHC14 and Anti-Hepatitis Virus Therapy
3. U-Rich Tail in Antiviral Innate Immune Response
3.1. The Interferon and RNAi in Antiviral Immune Response
3.2. U-Rich Tail in Antiviral Immune Response
4. Conclusions and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Stewart, M. Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 2019, 294, 2977–2987. [Google Scholar] [CrossRef]
- Weill, L.; Belloc, E.; Bava, F.A.; Méndez, R. Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 2012, 19, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.L.; Pasquinelli, A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Slomovic, S.; Portnoy, V.; Yehudai-Resheff, S.; Bronshtein, E.; Schuster, G. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim. Biophys. Acta 2008, 1779, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid. Res. Mol. Biol. 2002, 71, 285–389. [Google Scholar]
- Norbury, C.J. Cytoplasmic RNA: A case of the tail wagging the dog. Nat. Rev. Mol. Cell Biol. 2013, 14, 643–653. [Google Scholar] [CrossRef]
- Yu, S.; Kim, V.N. A tale of non-canonical tails: Gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol. 2020, 21, 542–556. [Google Scholar] [CrossRef]
- Barnard, D.C.; Ryan, K.; Manley, J.L.; Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 2004, 119, 641–651. [Google Scholar] [CrossRef]
- Suh, N.; Jedamzik, B.; Eckmann, C.R.; Wickens, M.; Kimble, J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. USA 2006, 103, 15108–151123. [Google Scholar] [CrossRef]
- Benoit, P.; Papin, C.; Kwak, J.E.; Wickens, M.; Simonelig, M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 2008, 135, 1969–1979. [Google Scholar] [CrossRef]
- Kwak, J.E.; Drier, E.; Barbee, S.A.; Ramaswami, M.; Yin, J.C.; Wickens, M. GLD2 poly(A) polymerase is required for long-term memory. Proc. Natl. Acad. Sci. USA 2008, 105, 14644–14649. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Wilson, T.L.; Kimble, J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 2010, 107, 17445–17450. [Google Scholar] [CrossRef] [PubMed]
- Sartain, C.V.; Cui, J.; Meisel, R.P.; Wolfner, M.F. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development 2011, 138, 1619–1629. [Google Scholar] [CrossRef]
- Cui, J.; Sartain, C.V.; Pleiss, J.A.; Wolfner, M.F. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 2013, 383, 121–131. [Google Scholar] [CrossRef]
- Menezes, M.R.; Balzeau, J.; Hagan, J.P. 3′ RNA Uridylation in Epitranscriptomics, Gene Regulation, and Disease. Front. Mol. Biosci. 2018, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Rissland, O.S.; Mikulasova, A.; Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007, 27, 3612–3624. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Piao, W.; Jin, H. Uridylation: A vital way for cellular RNA surveillance. Hereditas 2022, 44, 449–465. [Google Scholar] [PubMed]
- Ustianenko, D.; Chiu, H.S.; Treiber, T.; Weyn-Vanhentenryck, S.M.; Treiber, N.; Meister, G.; Sumazin, P.; Zhang, C. LIN28 Selectively Modulates a Subclass of Let-7 MicroRNAs. Mol. Cell 2018, 71, 271–283.e5. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, Q.; Vrettos, N.; Maragkakis, M.; Alexiou, P.; Gregory, B.D.; Mourelatos, Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Mol. Cell 2014, 55, 868–879. [Google Scholar] [CrossRef]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.J.; Park, J.E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef]
- Liu, M.; Sun, Z.; Tang, Y.; Zhang, S.; Luo, J. The Regulation of Exosome-Mediated miR-132-3p/miR-132-3p-UUU on Radiation-Induced Esophageal Injury. Radiat. Res. 2023, 23, 231–241. [Google Scholar] [CrossRef]
- Liu, L.; Wang, X.; Zhao, W.; Li, Q.; Li, J.; Chen, H.; Shan, G. Systematic characterization of small RNAs associated with C. elegans Argonautes. Sci. China Life Sci. 2023, 66, 1303–1322. [Google Scholar] [CrossRef] [PubMed]
- Lipińska-Zubrycka, L.; Grochowski, M.; Bähler, J.; Małecki, M. Pervasive mRNA uridylation in fission yeast is catalysed by both Cid1 and Cid16 terminal uridyltransferases. PLoS ONE 2023, 18, e0285576. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, D.; Lee, Y.S.; Ha, M.; Lee, M.; Yeo, J.; Chang, H.; Song, J.; Ahn, K.; Kim, V.N. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 2018, 361, 701–704. [Google Scholar] [CrossRef]
- Hyrina, A.; Jones, C.; Chen, D.; Clarkson, S.; Cochran, N.; Feucht, P.; Hoffman, G.; Lindeman, A.; Russ, C.; Sigoillot, F.; et al. A Genome-wide CRISPR Screen Identifies ZCCHC14 as a Host Factor Required for Hepatitis B Surface Antigen Production. Cell Rep. 2019, 29, 2970–2978. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Makino, S. Interplay between viruses and host mRNA degradation. Biochim. Biophys. Acta 2013, 1829, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Park, J.; Ha, M.; Lim, J.; Chang, H.; Kim, V.N. PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol. Cell 2018, 70, 1081–1088. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.H.; Stowell, J.A.W.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef]
- Passmore, L.A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef]
- Zhao, T.; Huan, Q.; Sun, J.; Liu, C.; Hou, X.; Yu, X.; Silverman, I.M.; Zhang, Y.; Gregory, B.D.; Liu, C.M.; et al. Impact of poly(A)-tail G-content on Arabidopsis PAB binding and their role in enhancing translational efficiency. Genome Biol. 2019, 20, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Misumi, I.; Shiota, T.; Sun, L.; Lenarcic, E.M.; Kim, H.; Shirasaki, T.; Hertel-Wulff, A.; Tibbs, T.; Mitchell, J.E.; et al. The ZCCHC14/TENT4 complex is required for hepatitis A virus RNA synthesis. Proc. Natl. Acad. Sci. USA 2022, 119, 22045–22061. [Google Scholar] [CrossRef] [PubMed]
- Kulsuptrakul, J.; Wang, R.; Meyers, N.L.; Ott, M.; Puschnik, A.S. A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection. Cell Rep. 2021, 34, 108859–108867. [Google Scholar] [CrossRef] [PubMed]
- Ashe, A.; Sarkies, P.; Le Pen, J.; Tanguy, M.; Miska, E.A. Antiviral RNA Interference against Orsay Virus Is neither Systemic nor Transgenerational in Caenorhabditis elegans. J. Virol. 2015, 89, 12035–12046. [Google Scholar] [CrossRef]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- MacKay, C.R.; Wang, J.P.; Kurt-Jones, E.A. Dicer’s role as an antiviral: Still an enigma. Curr. Opin. Immunol. 2014, 26, 49–55. [Google Scholar] [CrossRef]
- Koralewska, N.; Ciechanowska, K.; Pokornowska, M.; Figlerowicz, M.; Kurzyńska-Kokorniak, A. Human ribonuclease Dicer—Structure and functions. Postep. Biochem. 2019, 65, 173–182. [Google Scholar] [CrossRef][Green Version]
- Le Pen, J.; Jiang, H.; Di Domenico, T.; Kneuss, E.; Kosałka, J.; Leung, C.; Morgan, M.; Much, C.; Rudolph, K.L.M.; Enright, A.J.; et al. Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nat. Struct. Mol. Biol. 2018, 25, 778–786. [Google Scholar] [CrossRef]
- Huo, Y.; Shen, J.; Wu, H.; Zhang, C.; Guo, L.; Yang, J.; Li, W. Widespread 3′-end uridylation in eukaryotic RNA viruses. Sci. Rep. 2016, 6, 25454–25461. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Lim, J.; Ha, M.; Kim, V.N. TAIL-seq: Genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 2014, 53, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Shen, Y.R.; Chang, C.C.; Guo, X.Y.; Young, Y.Y.; Lai, T.Y.; Yu, I.S.; Lee, C.Y.; Chuang, T.H.; Tsai, H.Y.; et al. Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation. Nat. Commun. 2021, 12, 3878–3885. [Google Scholar] [CrossRef]
- Gupta, A.; Li, Y.; Chen, S.H.; Papas, B.N.; Martin, N.P.; Morgan, M. TUT4/7-mediated uridylation of a coronavirus subgenomic RNAs delays viral replication. Commun. Biol. 2023, 6, 438–444. [Google Scholar] [CrossRef]
- Kim, D.; Lee, Y.S.; Jung, S.J.; Yeo, J.; Seo, J.J.; Lee, Y.Y.; Lim, J.; Chang, H.; Song, J.; Yang, J.; et al. Viral hijacking of the TENT4-ZCCHC14 complex protects viral RNAs via mixed tailing. Nat. Struct. Mol. Biol. 2020, 27, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Nozaki, A.; Uno, H.; Ishida, Y.; Nagahama, M. Interaction properties of human TRAMP-like proteins and their role in pre-rRNA 5′ETS turnover. FEBS Lett. 2016, 590, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Leung, S.W.; Banerjee, A.; Kodani, M.O.; Chavez, R.; Bowman, E.A.; Purohit, M.K.; Rubinson, M.E.; Rubinson, E.H.; Corbett, A.H. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J. Biol. Chem. 2011, 286, 37429–37445. [Google Scholar] [CrossRef]
- Tang, T.T.L.; Passmore, L.A. Recognition of Poly(A) RNA through Its Intrinsic Helical Structure. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Huch, S.; Nissan, T. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA 2014, 5, 747–763. [Google Scholar] [CrossRef]
- Kajjo, S.; Sharma, S.; Chen, S.; Brothers, W.R.; Cott, M.; Hasaj, B.; Jovanovic, P.; Larsson, O.; Fabian, M.R. PABP prevents the untimely decay of select mRNA populations in human cells. EMBO J. 2022, 41, e108650–e108661. [Google Scholar] [CrossRef]
- Yeo, J.; Kim, V.N. U-tail as a guardian against invading RNAs. Nat. Struct. Mol. Biol. 2018, 25, 903–905. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Young, J.A.T.; Javanbakht, H.; Sofia, M.J.; Zhou, T. Host RNA quality control as a hepatitis B antiviral target. Antiviral Res. 2021, 186, 104972–104980. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lee, A.C.H.; Guo, F.; Kondratowicz, A.S.; Micolochick Steuer, H.M.; Miller, A.; Bailey, L.D.; Wang, X.; Chen, S.; Kultgen, S.G.; et al. Host Poly(A) Polymerases PAPD5 and PAPD7 Provide Two Layers of Protection That Ensure the Integrity and Stability of Hepatitis B Virus RNA. J. Virol. 2021, 95, e0057421–e0057429. [Google Scholar] [CrossRef] [PubMed]
- Mueller, H.; Lopez, A.; Tropberger, P.; Wildum, S.; Schmaler, J.; Pedersen, L.; Han, X.; Wang, Y.; Ottosen, S.; Yang, S.; et al. PAPD5/7 Are Host Factors That Are Required for Hepatitis B Virus RNA Stabilization. Hepatology 2019, 69, 1398–1411. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.J.; Jung, S.J.; Yang, J.; Choi, D.E.; Kim, V.N. Functional viromic screens uncover regulatory RNA elements. Cell 2023, 186, 3291–3306. [Google Scholar] [CrossRef]
- McKnight, K.L.; Lemon, S.M. Hepatitis A Virus Genome Organization and Replication Strategy. Cold Spring Harb. Perspect. Med. 2018, 8, 23–41. [Google Scholar] [CrossRef]
- Das, A.; Barrientos, R.; Shiota, T.; Madigan, V.; Misumi, I.; McKnight, K.L.; Sun, L.; Li, Z.; Meganck, R.M.; Li, Y.; et al. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat. Microbiol. 2020, 5, 1069–1078. [Google Scholar] [CrossRef]
- Abutaleb, A.; Kottilil, S. Hepatitis A: Epidemiology, Natural History, Unusual Clinical Manifestations, and Prevention. Gastroenterol. Clin. N. Am. 2020, 49, 191–199. [Google Scholar] [CrossRef]
- De Clercq, E.; Férir, G.; Kaptein, S.; Neyts, J. Antiviral treatment of chronic hepatitis B virus (HBV) infections. Viruses 2010, 2, 1279–1305. [Google Scholar] [CrossRef]
- Ganem, D.; Prince, A.M. Hepatitis B virus infection—Natural history and clinical consequences. N. Engl. J. Med. 2004, 350, 1118–1129. [Google Scholar] [CrossRef]
- Yuen, M.F.; Chen, D.S.; Dusheiko, G.M.; Janssen, H.L.A.; Lau, D.T.Y.; Locarnini, S.A.; Peters, M.G.; Lai, C.L. Hepatitis B virus infection. Nat. Rev. Dis. Primers 2018, 4, 18035–18041. [Google Scholar] [CrossRef] [PubMed]
- Trépo, C.; Chan, H.L.; Lok, A. Hepatitis B virus infection. Lancet 2014, 384, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Maynard, J.E. Hepatitis A. Yale J. Biol. Med. 1976, 49, 227–233. [Google Scholar]
- Mathiesen, L.R. The hepatitis A virus infection. Liver 1981, 1, 81–109. [Google Scholar] [CrossRef]
- Nassal, M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015, 64, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Karatayli, E.; Karatayli, S.C.; Cinar, K.; Gokahmetoglu, S.; Güven, K.; Idilman, R.; Yurdaydin, C.; Bozdayi, A.M. Molecular characterization of a novel entecavir mutation pattern isolated from a multi-drug refractory patient with chronic hepatitis B infection. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2012, 53, 130–134. [Google Scholar] [CrossRef]
- Seto, W.K.; Lo, Y.R.; Pawlotsky, J.M.; Yuen, M.F. Chronic hepatitis B virus infection. Lancet 2018, 392, 2313–2324. [Google Scholar] [CrossRef]
- Férir, G.; Kaptein, S.; Neyts, J.; De Clercq, E. Antiviral treatment of chronic hepatitis B virus infections: The past, the present and the future. Rev. Med. Virol. 2008, 18, 19–34. [Google Scholar] [CrossRef]
- Warner, N.; Locarnini, S.; Xu, H. The role of hepatitis B surface antibodies in HBV infection, disease and clearance. Future Virol. 2020, 8, 22–34. [Google Scholar] [CrossRef]
- Kuhns, M.C.; Holzmayer, V.; Anderson, M.C.; McNamara, A.L.; Sauleda, S.; Mbanya, D.; Duong, P.T.; Dung, N.T.T.; Cloherty, G.A. Molecular and Serological Characterization of Hepatitis B Virus (HBV)-Positive Samples with Very Low or Undetectable Levels of HBV Surface Antigen. Viruses 2021, 13, 2053. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, J.; Chow, H.Y.; Wang, J.; Zhang, Y.; Fung, Y.M.E.; Ren, Q.; Li, X. Development of DHQ-based chemical biology probe to profile cellular targets for HBV. Bioorg. Med. Chem. Lett. 2020, 30, 127615–127621. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Block, T.; Liu, F.; Kondratowicz, A.S.; Sun, L.; Rawat, S.; Branson, J.; Guo, F.; Steuer, H.M.; Liang, H.; et al. HBsAg mRNA degradation induced by a dihydroquinolizinone compound depends on the HBV posttranscriptional regulatory element. Antivir. Res. 2018, 149, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhou, C.; Jiang, M.; Wang, Y.; Wang, J.; Cheng, Z.; Wang, M.; Liu, Y.; Liang, C.; Wang, J.; et al. Discovery of RG7834: The First-in-Class Selective and Orally Available Small Molecule Hepatitis B Virus Expression Inhibitor with Novel Mechanism of Action. J. Med. Chem. 2018, 61, 10619–10634. [Google Scholar] [CrossRef] [PubMed]
- Mueller, H.; Wildum, S.; Luangsay, S.; Walther, J.; Lopez, A.; Tropberger, P.; Ottaviani, G.; Lu, W.; Parrott, N.J.; Zhang, J.D.; et al. A novel orally available small molecule that inhibits hepatitis B virus expression. J. Hepatol. 2018, 68, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Bopst, M.; Dinklo, T.; Funk, J.; Greiter-Wilke, A.; Lenz, B.; Kustermann, S.; Jiang, T.; Xie, J. Unexpected neurotoxicity in chronic toxicity studies with a HBV viral expression inhibitor. Regul. Toxicol. Pharmacol. 2023, 141, 105407. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Yu, J.; Zhou, L.; Xu, B.; Dai, Y.; Wang, H.; Zhou, W.; Zhao, H. Prevalence of antibody to hepatitis B surface antigen among qualified blood donors in Nanjing, China. Hum. Vaccin. Immunother. 2023, 19, 2206774–2206778. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, F.; Yuan, Q.; Du, J.; Hu, L.; Gu, Z.; Zhou, Q.; Du, X.; He, S.; Sun, Y.; et al. Discovery and preclinical evaluations of GST-HG131, a novel HBV antigen inhibitor for the treatment of chronic hepatitis B infection. Bioorg. Med. Chem. Lett. 2022, 75, 128977–128987. [Google Scholar] [CrossRef]
- Qin, X.; Yang, L.; Ma, X.; Jiang, B.; Wu, S.; Wang, A.; Xu, S.; Wu, W.; Song, H.; Du, N.; et al. Identification of dihydroquinolizinone derivatives with cyclic ether moieties as new anti-HBV agents. Eur. J. Med. Chem. 2022, 238, 114518–114521. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, X.; Jin, H.; Lu, D.; Chen, S.; Zhang, Y.; Wang, X.; Xu, H.; Ao, W.; Zhang, Y. Discovery, optimization and biological evaluation of novel HBsAg production inhibitors. Eur. J. Med. Chem. 2023, 256, 115387. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, A.C.; Stempel, M.; Chan, B.; Brinkmann, M.M. One Step Ahead: Herpesviruses Light the Way to Understanding Interferon-Stimulated Genes (ISGs). Front. Microbiol. 2020, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, C.; Dishongh, R.; Moore, S.C.; Whitt, M.A.; Chow, M.; Machaca, K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 2005, 436, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, W.X.; Ding, S.W. Induction and Suppression of RNA Silencing by an Animal Virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA Interference in Mammalian Cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef]
- Berkhout, B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 2018, 32, 9–14. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Han, Y.; Fan, X.; Ding, S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef]
- Rissland, O.S.; Norbury, C.J. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 2009, 16, 616–623. [Google Scholar] [CrossRef]
- Wickens, M.; Kwak, J.E. Molecular biology. A tail tale for U. Science 2008, 319, 1344–1345. [Google Scholar] [CrossRef][Green Version]
- Joly, A.C.; Garcia, S.; Hily, J.M.; Koechler, S.; Demangeat, G.; Garcia, D.; Vigne, E.; Lemaire, O.; Zuber, H.; Gagliardi, D. An extensive survey of phytoviral RNA 3′ uridylation identifies extreme variations and virus-specific patterns. Plant Physiol. 2023, 193, 271–290. [Google Scholar] [CrossRef]
- Warkocki, Z.; Krawczyk, P.S.; Adamska, D.; Bijata, K.; Garcia-Perez, J.L.; Dziembowski, A. Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s. Cell 2018, 174, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Strzyz, P. TUT-TUTting retrotransposons. Nat. Rev. Mol. Cell Biol. 2018, 19, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Jupin, I.; Bouzoubaa, S.; Richards, K.; Jonard, G.; Guilley, H. Multiplication of beet necrotic yellow vein virus RNA 3 lacking a 3′ poly(A) tail is accompanied by reappearance of the poly(A) tail and a novel short U-rich tract preceding it. Virology 1990, 178, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Pyle, J.D.; Mandadi, K.K.; Scholthof, K.B.G. Panicum Mosaic Virus and Its Satellites Acquire RNA Modifications Associated with Host-Mediated Antiviral Degradation. mBio 2019, 10, 01900–01913. [Google Scholar] [CrossRef] [PubMed]
- Viegas, S.C.; Silva, I.J.; Apura, P.; Matos, R.G.; Arraiano, C.M. Surprises in the 3′-end: ‘U’ can decide too! FEBS J. 2015, 282, 3489–3499. [Google Scholar] [CrossRef]
- Rehwinkel, J. Is anti-viral defence the evolutionary origin of mRNA turnover? Bioessays 2016, 38, 817–824. [Google Scholar] [CrossRef]
- Berman-Booty, L.D.; Sargeant, A.M.; Rosol, T.J.; Rengel, R.C.; Clinton, S.K.; Chen, C.S.; Kulp, S.K. A review of the existing grading schemes and a proposal for a modified grading scheme for prostatic lesions in TRAMP mice. Toxicol. Pathol. 2012, 40, 5–17. [Google Scholar] [CrossRef]
- Molleston, J.M.; Sabin, L.R.; Moy, R.H.; Menghani, S.V.; Rausch, K.; Gordesky-Gold, B.; Hopkins, K.C.; Zhou, R.; Jensen, T.H.; Wilusz, J.E.; et al. A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation. Genes Dev. 2016, 30, 1658–1670. [Google Scholar] [CrossRef]
- Schmidt, K.; Butler, J.S. Nuclear RNA surveillance: Role of TRAMP in controlling exosome specificity. Wiley Interdiscip. Rev. RNA 2013, 4, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Takeuchi, R.; Takata, K.; Shimanouchi, K.; Abe, Y.; Kanai, Y.; Ruike, T.; Ihara, A.; Sakaguchi, K. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol. Cell. Biol. 2008, 28, 6620–6631. [Google Scholar] [CrossRef] [PubMed]
- Rammelt, C.; Bilen, B.; Zavolan, M.; Keller, W. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 2011, 17, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Lubas, M.; Christensen, M.S.; Kristiansen, M.S.; Domanski, M.; Falkenby, L.G.; Lykke-Andersen, S.; Andersen, J.S.; Dziembowski, A.; Jensen, T.H. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 2011, 43, 624–637. [Google Scholar] [CrossRef]
- Manokaran, G.; Finol, E.; Wang, C.; Gunaratne, J.; Bahl, J.; Ong, E.Z.; Tan, H.C.; Sessions, O.M.; Ward, A.M.; Gubler, D.J.; et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015, 350, 217–221. [Google Scholar] [CrossRef]
- Brinton, M.A.; Basu, M. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res. 2015, 206, 108–119. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Qin, C.F. Structure and function of cis-acting RNA elements of flavivirus. Rev. Med. Virol. 2020, 30, e2092–e2099. [Google Scholar] [CrossRef]
- Preston, M.A.; Porter, D.F.; Chen, F.; Buter, N.; Lapointe, C.P.; Keles, S.; Kimble, J.; Wickens, M. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 2019, 16, 437–445. [Google Scholar] [CrossRef]
- Shukla, A.; Yan, J.; Pagano, D.J.; Dodson, A.E.; Fei, Y.; Gorham, J.; Seidman, J.G.; Wickens, M.; Kennedy, S. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 2020, 582, 283–2888, reprinted in Nature 2021, 592, E27–E34. [Google Scholar] [CrossRef]
- Lee, M.; Kim, B.; Kim, V.N. Emerging roles of RNA modification: M(6)A and U-tail. Cell 2014, 158, 980–987. [Google Scholar] [CrossRef]
- Lu, M.; Xue, M.; Wang, H.T.; Kairis, E.L.; Ahmad, S.; Wei, J.; Zhang, Z.; Liu, Q.; Zhang, Y.; Gao, Y.; et al. Nonsegmented Negative-Sense RNA Viruses Utilize N(6)-Methyladenosine (m(6)A) as a Common Strategy To Evade Host Innate Immunity. J. Virol. 2021, 95, 01939–01947. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Cheng, W.; Zhao, F.; Tang, M.; Diao, Y.; Xu, R. Association of N6-methyladenosine with viruses and related diseases. Virol. J. 2019, 16, 133–136. [Google Scholar] [CrossRef] [PubMed]

| Subfamily | Enzyme (Synonyms) | RNA Substrate | Activity | Localization |
|---|---|---|---|---|
| TENT1 | TUT1 (U6 TUTase, PAPD2, RBM21, URLC6, STARPAP) | U6 snRNA Pre-mRNA | oligouridylation polyadenylation | nucleolus nuclear speckle nucleoplasm cytosol mitochondrion |
| TENT2 | TENT2 (GLD-2, PAPD4, TUT2, APD4) | mRNA miRNA | monoadenylation oligoadenylation polyadenylation | part of nuclear RNA-directed RNA polymerase complex cytoplasm |
| TENT3 | TUT4 (PAPD3, TENT3A, ZCCHC11) | mRNA Histone mRNA LINE-1 mRNA Pre-miRNA miRNA Viral RNA Pre-rRNA Pol III-ncRNA TSS RNA | monouridylation oligouridylation | nucleolus cytosol cytoplasm cytoplasmic ribonucleoprotein granule extracellular space extracellular exosome |
| TUT7 (PAPD6, TENT3B, ZCCHC6) | nucleoplasm cytosol cytoplasm | |||
| TENT4 | TENT4A (PAPD7, TUT5, TRF4-1, LAK1, POLK, POLS) | mRNA Viral RNA | polyadenylation mixed tailing | nucleus nucleoplasm nuclear membrane nucleolus Golgi apparatus part of TRAMP complex |
| TENT4B (PAPD5, TUT3, TRF4-2) | mRNA Viral RNA miRNA Pre-rRNA rRNA snoRNA scaRNA Y RNA hTR | monoadenylation oligoadenylation polyadenylation mixed tailing | nucleolus plasma membrane cytosol cytoplasm part of TRAMP complex | |
| TENT5 | TENT5A (OI18, XTP11, FAM46A, C6orf37) | mRNA | polyadenylation | nucleus cytoplasm |
| TENT5B (FAM46B) | nucleus cytoplasm | |||
| TENT5C (FAM46C) | nucleus nucleoplasm cytoplasm centrosome | |||
| TENT5D (CT112, CT1.26, FAM46D) | nucleus cytoplasm | |||
| TENT6 | MTPAP (PAPD1, TENT6, SPAX4) | MT-mRNA MT-tRNA | oligoadenylation polyadenylation | nucleoplasm mitochondrion intracellular membrane-bounded organelle |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, X.; Irshad, A.; Jin, H. The Battle for Survival: The Role of RNA Non-Canonical Tails in the Virus–Host Interaction. Metabolites 2023, 13, 1009. https://doi.org/10.3390/metabo13091009
Wen X, Irshad A, Jin H. The Battle for Survival: The Role of RNA Non-Canonical Tails in the Virus–Host Interaction. Metabolites. 2023; 13(9):1009. https://doi.org/10.3390/metabo13091009
Chicago/Turabian StyleWen, Xianghui, Ahsan Irshad, and Hua Jin. 2023. "The Battle for Survival: The Role of RNA Non-Canonical Tails in the Virus–Host Interaction" Metabolites 13, no. 9: 1009. https://doi.org/10.3390/metabo13091009
APA StyleWen, X., Irshad, A., & Jin, H. (2023). The Battle for Survival: The Role of RNA Non-Canonical Tails in the Virus–Host Interaction. Metabolites, 13(9), 1009. https://doi.org/10.3390/metabo13091009








