Hecogenin a Plant Derived Small Molecule as an Antagonist to BACE-1: A Potential Target for Neurodegenerative Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction of Bioactive Compounds from Justicia adhatoda and Sida cordifolia
2.3. Antioxidant Profiling of Justicia adhatoda and Sida cordifolia Methanolic Extracts
2.3.1. Total Phenolic Content (TPC)
2.3.2. Total Flavonoid Content
2.3.3. 2, 2-Diphenyl-1-picrylhydrazyl (DPPH) Assay
2.3.4. FRAP Assay
2.4. LC-ESI-MS Analysis of Justicia adhatoda
2.5. Molecular Docking
2.6. ADMET Profiling of Bioactive Compounds
2.7. Molecular Dynamics Simulation
2.8. Binding Free Energy Calculations of Molecules with BACE-1
3. Results
3.1. Antioxidant Profiling of Justicia adhatoda and Sida cordifolia
3.2. LC-MS/MS Analysis of Justicia adhatoda L.
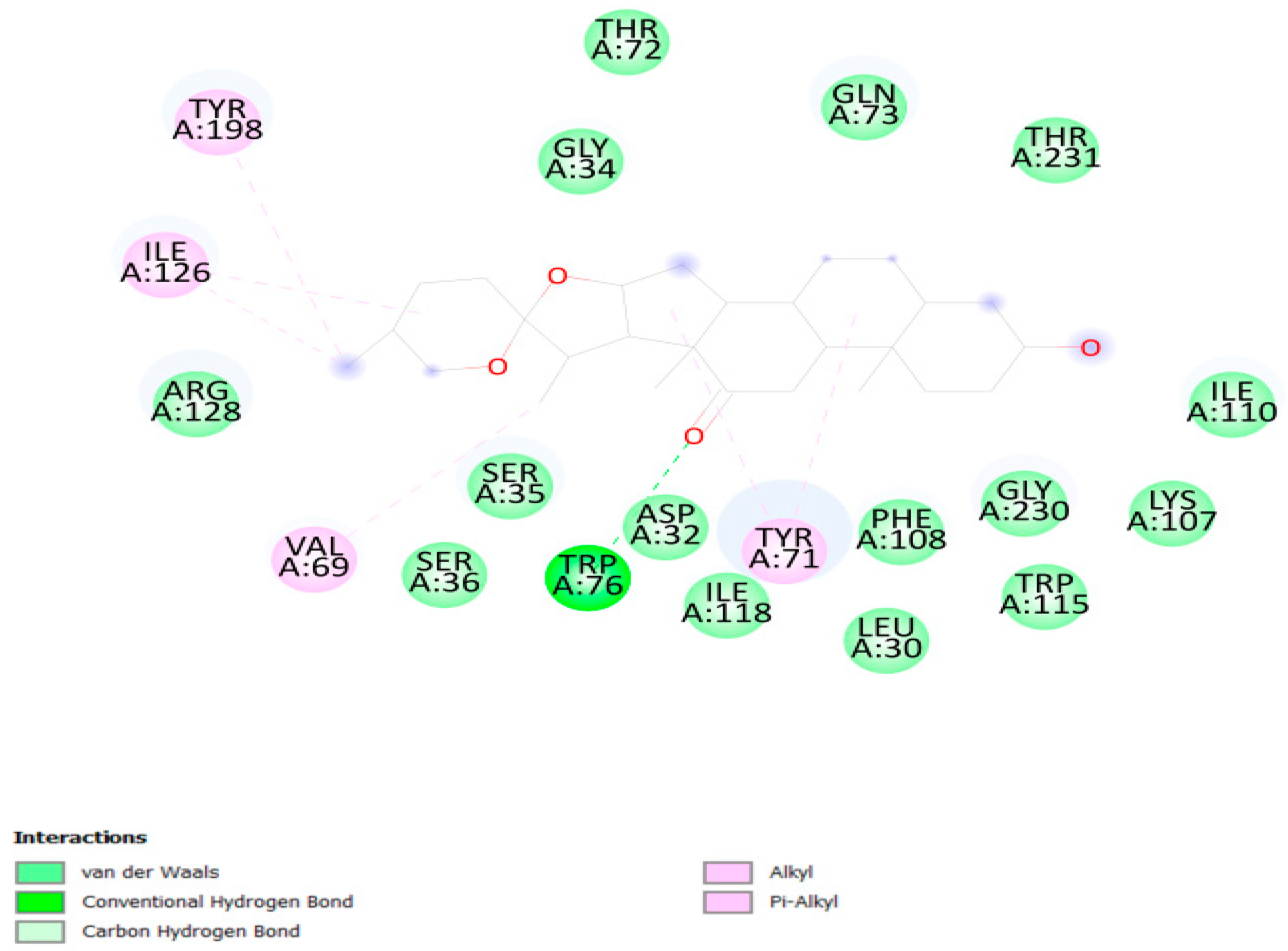
3.3. Molecular Docking
3.4. ADMET Profiling
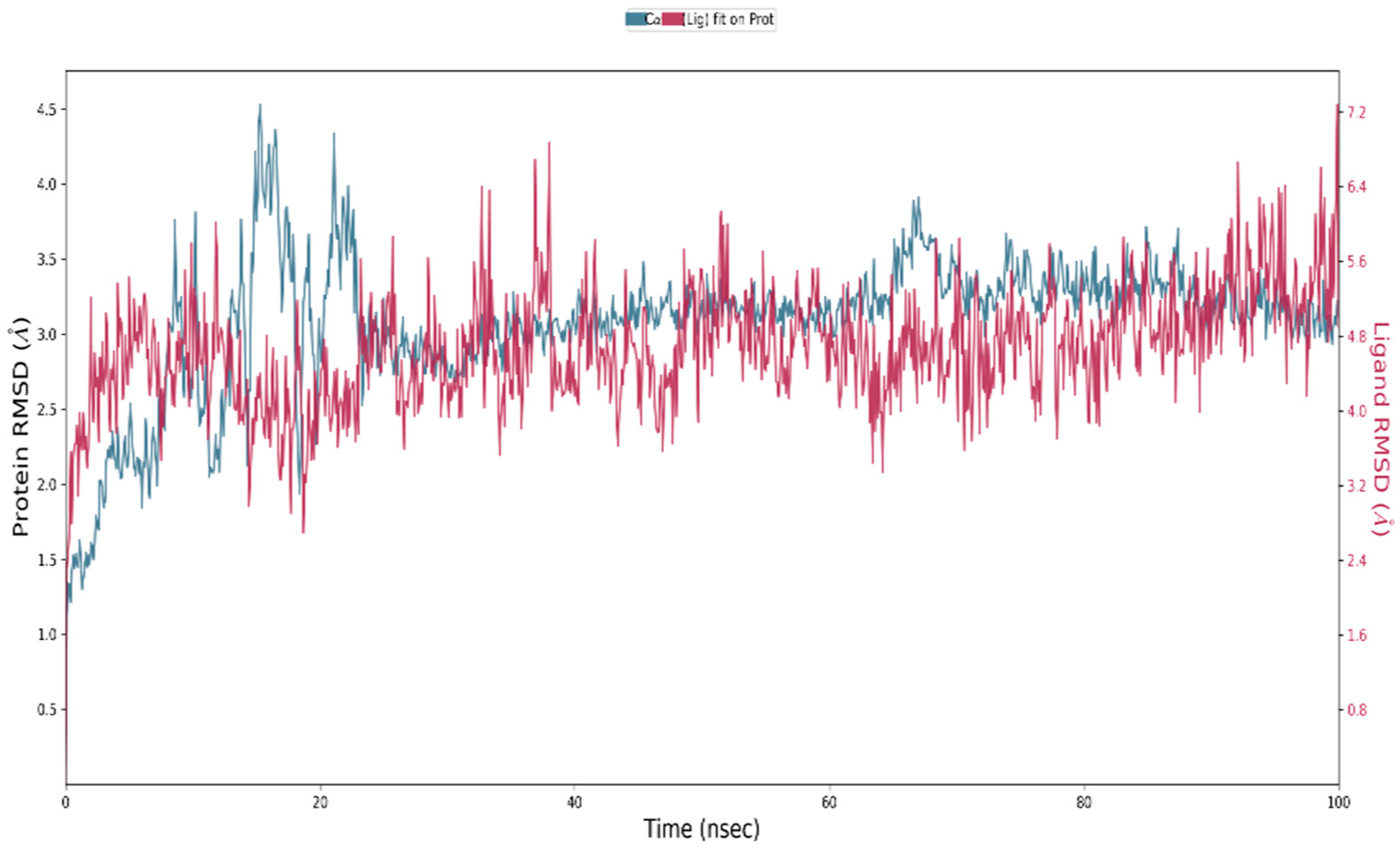
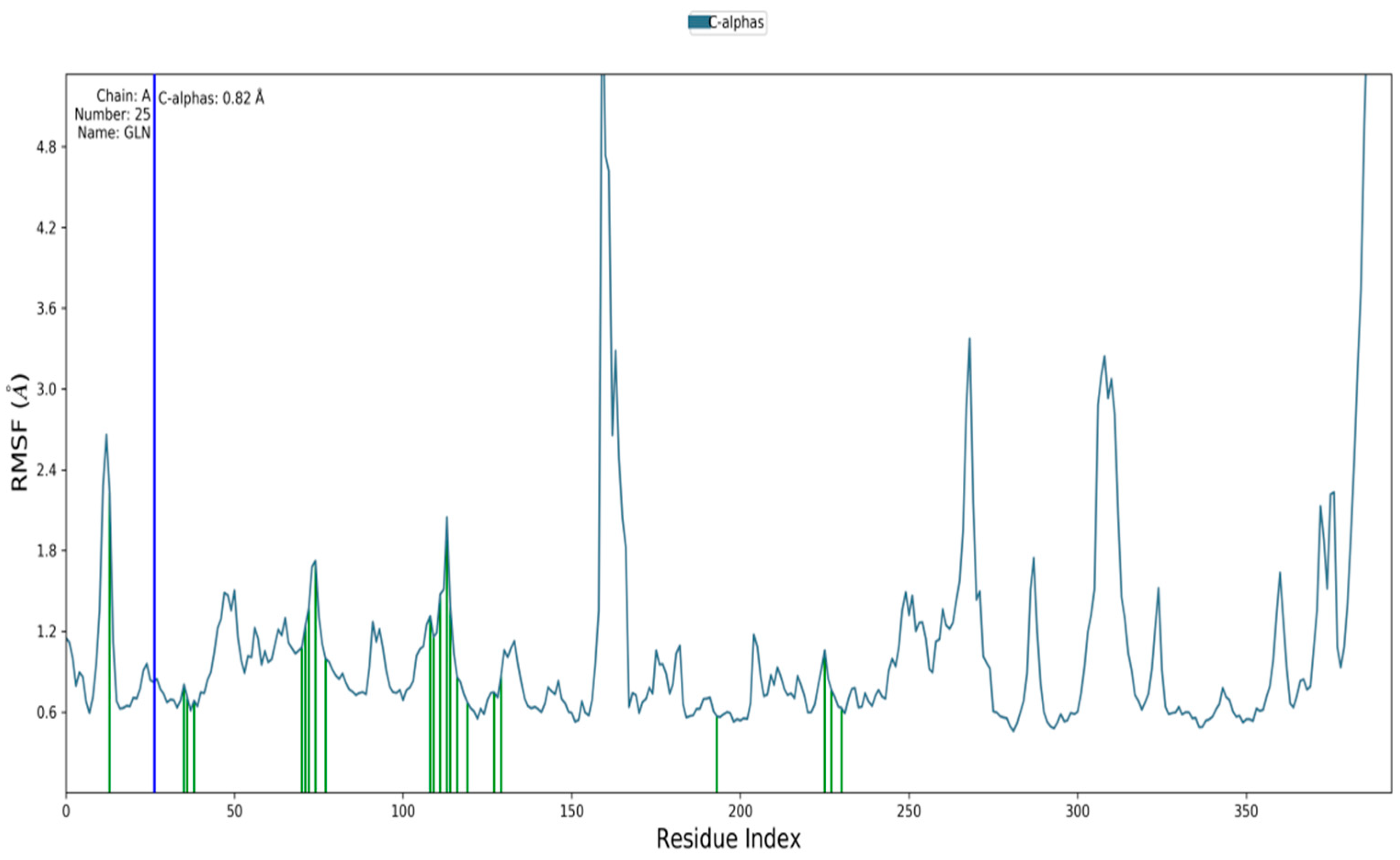
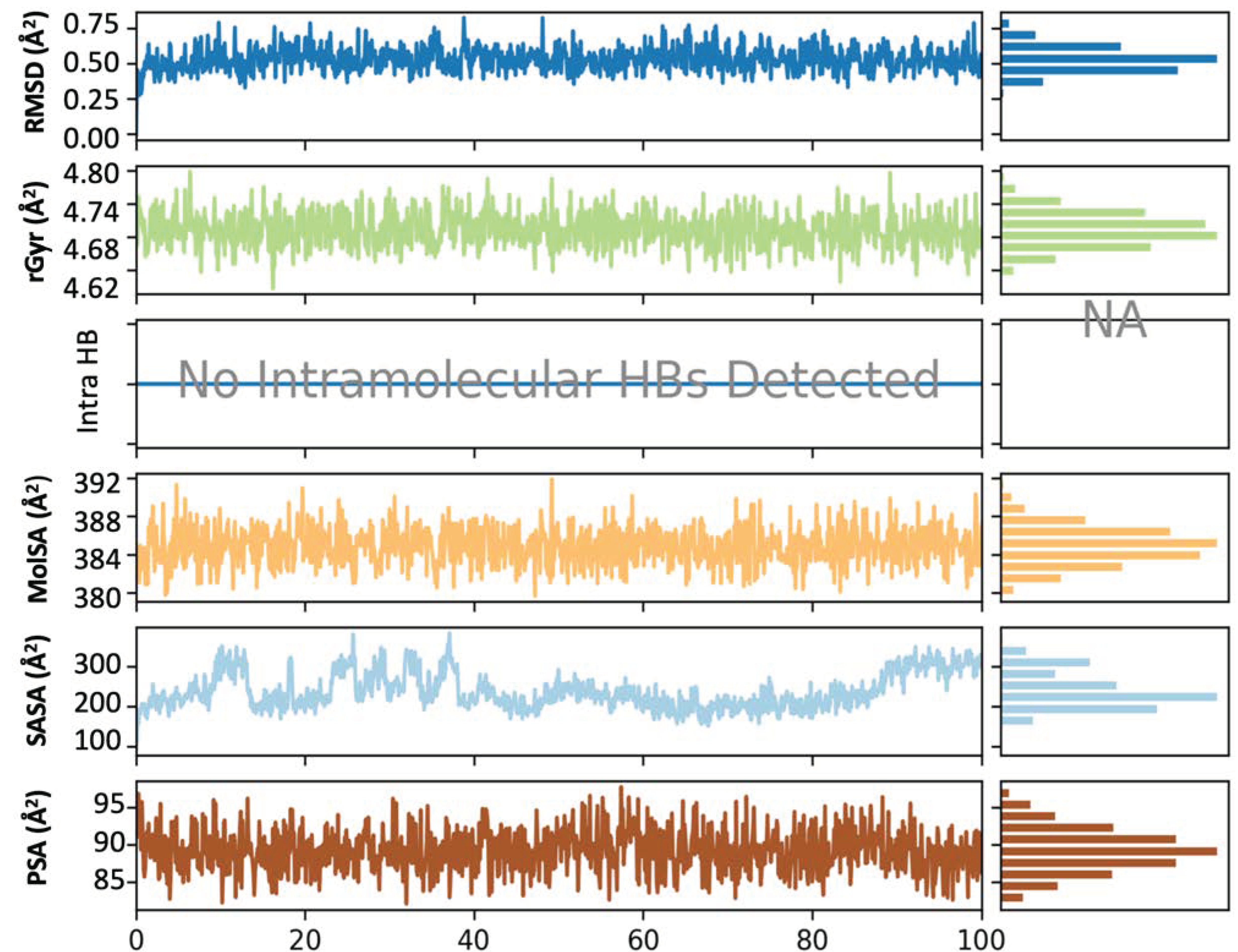
3.5. Molecular Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.K.; Sharma, A. Plant Secondary Metabolites: Physico-Chemical Properties and Therapeutic Applications; Springer Nature: Berlin/Heidelberg, Germany, 2022; ISBN 9789811647796. [Google Scholar]
- Hassan, M.; Bala, S.Z.; Bashir, M.; Waziri, P.M.; Musa Adam, R.; Umar, M.A.; Kini, P. LC-MS and GC-MS Profiling of Different Fractions of Stem Bark Ethanolic Extract. J. Anal. Methods Chem. 2022, 2022, 6349332. [Google Scholar] [CrossRef]
- Basit, A.; Shutian, T.; Khan, A.; Khan, S.M.; Shahzad, R.; Khan, A.; Khan, S.; Khan, M. Anti-Inflammatory and Analgesic Potential of Leaf Extract of Justicia adhatoda L. (Acanthaceae) in Carrageenan and Formalin-Induced Models by Targeting Oxidative Stress. Biomed. Pharmacother. 2022, 153, 113322. [Google Scholar] [CrossRef]
- Chanu, W.S.; Sarangthem, K. Phytochemical constituents of Justicia adhatoda Linn. Found in Manipur. Indian J. Plant Sci. 2014, 3, 19–21. [Google Scholar]
- Iqbal, H.; Wright, C.L.; Jones, S.; da Silva, G.R.; McKillen, J.; Gilmore, B.F.; Kavanagh, O.; Green, B.D. Extracts of Sida Cordifolia Contain Polysaccharides Possessing Immunomodulatory Activity and Rosmarinic Acid Compounds with Antibacterial Activity. BMC Complement. Med. Ther. 2022, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K.; et al. Preclinical Alzheimer’s Disease: Definition, Natural History, and Diagnostic Criteria. Alzheimer’s Dement. 2016, 12, 292–323. [Google Scholar] [CrossRef] [PubMed]
- Zagni, E.; Simoni, L.; Colombo, D. Sex and Gender Differences in Central Nervous System-Related Disorders. Neurosci. J. 2016, 2016, 2827090. [Google Scholar] [CrossRef]
- Bustamante-Barrientos, F.A.; Méndez-Ruette, M.; Ortloff, A.; Luz-Crawford, P.; Rivera, F.J.; Figueroa, C.D.; Molina, L.; Bátiz, L.F. The Impact of Estrogen and Estrogen-Like Molecules in Neurogenesis and Neurodegeneration: Beneficial or Harmful? Front. Cell. Neurosci. 2021, 15, 636176. [Google Scholar] [CrossRef]
- Cole, S.L.; Vassar, R. The Alzheimer’s Disease Beta-Secretase Enzyme, BACE1. Mol. Neurodegener. 2007, 2, 22. [Google Scholar] [CrossRef]
- Nirmalraj, S.; Gayathiri, E.; Sivamurugan, M.; Manivasagaperumal, R.; Jayanthi, J.; Prakash, P.; Selvam, K. Molecular Docking Based Screening Dynamics for Plant Based Identified Potential Compounds of PDE12 Inhibitors. Curr. Res. Green Sustain. Chem. 2021, 4, 100122. [Google Scholar] [CrossRef]
- Alrasheid, A.A.; Babiker, M.Y.; Awad, T.A. Evaluation of Certain Medicinal Plants Compounds as New Potential Inhibitors of Novel Corona Virus (COVID-19) Using Molecular Docking Analysis. Silico Pharmacol. 2021, 9, 10. [Google Scholar] [CrossRef]
- Padmanabhan, D.; Natarajan, P.; Palanisamy, S. Integrated Metabolite and Transcriptome Profiling-Mediated Gene Mining of Sida cordifolia Reveals Medicinally Important Genes. Genes 2022, 13, 1909. [Google Scholar] [CrossRef]
- Kozłowska, M.; Ścibisz, I.; Przybył, J.L.; Laudy, A.E.; Majewska, E.; Tarnowska, K.; Małajowicz, J.; Ziarno, M. Antioxidant and Antibacterial Activity of Extracts from Selected Plant Material. Appl. Sci. 2022, 12, 9871. [Google Scholar] [CrossRef]
- Ralte, L.; Khiangte, L.; Thangjam, N.M.; Kumar, A.; Singh, Y.T. GC–MS and Molecular Docking Analyses of Phytochemicals from the Underutilized Plant, Parkia Timoriana Revealed Candidate Anti-Cancerous and Anti-Inflammatory Agents. Sci. Rep. 2022, 12, 3395. [Google Scholar] [CrossRef]
- Al-Rimawi, F.; Imtara, H.; Khalid, M.; Salah, Z.; Parvez, M.K.; Saleh, A.; Al kamaly, O.; ShawkiDahu, C. Assessment of Antimicrobial, Anticancer, and Antioxidant Activity of Verthimia iphionoides Plant Extract. Processes 2022, 10, 2375. [Google Scholar] [CrossRef]
- Abebe, A.; Hilawea, K.T.; Mekonnen, A.; Tigineh, G.T.; Sitotaw, B.; Liyew, M.; Wubieneh, T.A. Assessment on Antioxidant Activity of the Aqueous Leaf Extracts of Combretum microphyllum and the Effect of Co(II)-Leaf Extract Complex on Antibacterial Activity of Leaf Extracts of the Plant Material. Sci. Afr. 2022, 18, e01432. [Google Scholar] [CrossRef]
- Otari, S.S.; Patel, S.B.; Lekhak, M.M.; Ghane, S.G. Phytochemical Studies on Two Unexplored Endemic Medicinal Plants of India, Barleria terminalis and Calacanthus grandiflorus. Front. Pharmacol. 2021, 12, 817885. [Google Scholar] [CrossRef]
- Aati, H.Y.; Anwar, M.; Al-Qahtani, J.; Al-Taweel, A.; Khan, K.-U.-R.; Aati, S.; Usman, F.; Ghalloo, B.A.; Asif, H.M.; Shirazi, J.H.; et al. Phytochemical Profiling, In Vitro Biological Activities, and In-Silico Studies of: An Unexplored Plant. Antibiotics 2022, 11, 1155. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Banerjee, P.; Dehnbostel, F.O.; Preissner, R. Prediction Is a Balancing Act: Importance of Sampling Methods to Balance Sensitivity and Specificity of Predictive Models Based on Imbalanced Chemical Data Sets. Front. Chem. 2018, 6, 362. [Google Scholar] [CrossRef]
- Bora, H.; Kamle, M.; Hassan, H.; Al-Emam, A.; Chopra, S.; Kirtipal, N.; Bharadwaj, S.; Kumar, P. Exploration of Potent Antiviral Phytomedicines from Lauraceae Family Plants against SARS-CoV-2 Main Protease. Viruses 2022, 14, 2783. [Google Scholar] [CrossRef]
- Bhat, B.A.; Mir, W.R.; Sheikh, B.A.; Alkanani, M.; Mir, M.A. Metabolite Fingerprinting of Phytoconstituents from Fritillaria cirrhosa D. Don and Molecular Docking Analysis of Bioactive Peonidin with Microbial Drug Target Proteins. Sci. Rep. 2022, 12, 7296. [Google Scholar] [CrossRef] [PubMed]
- Mulakala, C.; Viswanadhan, V.N. Could MM-GBSA be accurate enough for calculation of absolute protein/ligand binding free energies? J. Mol. Graph. Model. 2013, 46, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.K.; Singh, S.K.; Singh, P.; Chellaperumal, P.; Reddy, K.K.; Selvaraj, C. Exploring the selectivity of a ligand complex with CDK2/CDK1: A molecular dynamics simulation approach: Exploring the selectivity of a ligand complex with CDK2 and CDK1. J. Mol. Recognit. 2012, 25, 504–512. [Google Scholar] [CrossRef]
- Noor, F.; Tahir ul Qamar, M.; Ashfaq, U.A.; Albutti, A.; Alwashmi, A.S.S.; Aljasir, M.A. Network Pharmacology Approach for Medicinal Plants: Review and Assessment. Pharmaceuticals 2022, 15, 572. [Google Scholar] [CrossRef] [PubMed]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef]
- Ouédraogo, M.; Konaté, K.; Lepengué, A.N.; Souza, A.; M’Batchi, B.; Sawadogo, L.L. Free Radical Scavenging Capacity, Anticandicidal Effect of Bioactive Compounds from Sida cordifolia L., in Combination with Nystatin and Clotrimazole and Their Effect on Specific Immune Response in Rats. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 33. [Google Scholar] [CrossRef]
- Ahmad, M.; Prawez, S.; Sultana, M.; Raina, R.; Pankaj, N.K.; Verma, P.K.; Rahman, S. Anti-Hyperglycemic, Anti-Hyperlipidemic and Antioxidant Potential of Alcoholic-Extract of Sida cordifolia (Areal Part) in Streptozotocin-Induced-Diabetes in Wistar-Rats. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2014, 84, 397–405. [Google Scholar] [CrossRef]
- Subramanya, M.D.; Pai, S.R.; Upadhya, V.; Ankad, G.M.; Bhagwat, S.S.; Hegde, H.V. Total polyphenolic contents and in vitro antioxidant properties of eight Sida species from Western Ghats, India. J. Ayurveda Integr. Med. 2015, 6, 24–28. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, R.; Dutta, D.; Chandel, S.; Bhattacharya, A.; Ravichandiran, V.; Sukla, S. In Vitro Anticancer Activity of Methanolic Extract of Justicia adhatoda Leaves with Special Emphasis on Human Breast Cancer Cell Line. Molecules 2022, 27, 8222. [Google Scholar] [CrossRef]
- Das, B.; Yan, R. Role of BACE1 in Alzheimer’s Synaptic Function. Transl. Neurodegener. 2017, 6, 23. [Google Scholar] [CrossRef]
- Ahmed, R.R.; Holler, C.J.; Webb, R.L.; Li, F.; Beckett, T.L.; Murphy, M.P. BACE1 and BACE2 Enzymatic Activities in Alzheimer’s Disease. J. Neurochem. 2010, 112, 1045–1053. [Google Scholar] [CrossRef]
- Benjannet, S.; Elagoz, A.; Wickham, L.; Mamarbachi, M.; Munzer, J.S.; Basak, A.; Lazure, C.; Cromlish, J.A.; Sisodia, S.; Checler, F.; et al. Post-Translational Processing of Beta-Secretase (beta-Amyloid-Converting Enzyme) and Its Ectodomain Shedding. The pro- and Transmembrane/cytosolic Domains Affect Its Cellular Activity and Amyloid-Beta Production. J. Biol. Chem. 2001, 276, 10879–10887. [Google Scholar] [CrossRef]
- Mukerjee, N.; Das, A.; Jawarkar, R.D.; Maitra, S.; Das, P.; Castrosanto, M.A.; Paul, S.; Samad, A.; Zaki, M.E.A.; Al-Hussain, S.A.; et al. Repurposing Food Molecules as a Potential BACE1 Inhibitor for Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 878276. [Google Scholar] [CrossRef]
- Mirza, F.J.; Zahid, S.; Amber, S.; Sumera; Jabeen, H.; Asim, N.; Ali Shah, S.A. Multitargeted Molecular Docking and Dynamic Simulation Studies of Bioactive Compounds from Rosmarinus officinalis against Alzheimer’s Disease. Molecules 2022, 27, 7241. [Google Scholar] [CrossRef]
- Quintans, J.S.S.; Barreto, R.S.S.; De Lucca, W.; Villarreal, C.F.; Kaneto, C.M.; Soares, M.B.P.; Branco, A.; Almeida, J.R.G.S.; Taranto, A.G.; Antoniolli, A.R.; et al. Evidence for the Involvement of Spinal Cord-Inhibitory and Cytokines-Modulatory Mechanisms in the Anti-Hyperalgesic Effect of Hecogenin Acetate, a Steroidal Sapogenin-Acetylated, in Mice. Molecules 2014, 19, 8303–8316. [Google Scholar] [CrossRef]
- Cerqueira, G.S.; dos Santos, G.; Vasconcelos, E.R.; de Freitas, A.P.F.; Moura, B.A.; Macedo, D.S.; Souto, A.L.; Barbosa Filho, J.M.; de Almeida Leal, L.K.; de Castro Brito, G.A.; et al. Effects of Hecogenin and Its Possible Mechanism of Action on Experimental Models of Gastric Ulcer in Mice. Eur. J. Pharmacol. 2012, 683, 260–269. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Sivalingam, K.; Shibu, M.A.; Liao, P.-H.; Ho, T.-J.; Kuo, W.-W.; Chen, R.-J.; Day, C.-H.; Viswanadha, V.P.; Ju, D.-T. Induction of Autophagy by Vasicinone Protects Neural Cells from Mitochondrial Dysfunction and Attenuates Paraquat-Mediated Parkinson’s Disease Associated α-Synuclein Levels. Nutrients 2020, 12, 1707. [Google Scholar] [CrossRef]
- Semwal, D.K.; Semwal, R.B.; Combrinck, S.; Viljoen, A. Myricetin: A Dietary Molecule with Diverse Biological Activities. Nutrients 2016, 8, 90. [Google Scholar] [CrossRef]
- Song, X.; Tan, L.; Wang, M.; Ren, C.; Guo, C.; Yang, B.; Ren, Y.; Cao, Z.; Li, Y.; Pei, J. Myricetin: A review of the most recent research. Biomed. Pharmacother. 2021, 134, 111017. [Google Scholar] [CrossRef]
- Khaldan, A.; Bouamrane, S.; En-Nahli, F.; El-Mernissi, R.; Hmamouchi, R.; Maghat, H.; Ajana, M.A.; Sbai, A.; Bouachrine, M.; Lakhlifi, T. Prediction of potential inhibitors of SARS-CoV-2 using 3D-QSAR, molecular docking modeling and ADMET properties. Heliyon 2021, 7, e06603. [Google Scholar] [CrossRef]
- Jayaraman, S.; Veeraraghavan, V.; Sreekandan, R.N.; Krishna Mohan, S.; Deiva Suga, S.S.; Kamaraj, D.; Mohandoss, S.; Rajagopal, P. Molecular Docking Analysis of Compounds from JusticaAdhatoda L with Glycogen Synthase Kinase-3 β. Bioinformation 2020, 16, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Okimoto, N.; Futatsugi, N.; Fuji, H.; Suenaga, A.; Morimoto, G.; Yanai, R.; Ohno, Y.; Narumi, T.; Taiji, M. High-Performance Drug Discovery: Computational Screening by Combining Docking and Molecular Dynamics Simulations. PLoS Comput. Biol. 2009, 5, e1000528. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.H.S.; Ferreira, R.S.; Caffarena, E.R. Integrating Molecular Docking and Molecular Dynamics Simulations. Methods Mol. Biol. 2019, 2053, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Cerón-Carrasco, J.P. When Virtual Screening Yields Inactive Drugs: Dealing with False Theoretical Friends. ChemMedChem 2022, 17, e202200278. [Google Scholar] [CrossRef]

| S.No. | RT | Compound | Mode | Molecular Formula | m/z | Adduct |
|---|---|---|---|---|---|---|
| 1 | 13.43 | Verapamil | Negative | C27H38O4 | 455.291, 456.298, 303.206 | [M − H]− |
| 2 | 46.84 | Cynarine | Negative | C25H24O12 | 515.123, 353.10, 191.021 | [M − H]− |
| 3 | 53.5 | Coumaroyl Hexoside | Negative | C15H18O8 | 163.038, 145.012, 119.047 | [M − H]− |
| 4 | 88.7 | Teniposide | Negative | C32H32O13S | 701.100, 113.123, 655.126 | [M − H]− |
| 5 | 93.59 | Shingomyelin | Negative | C39H79N2O6P | 687.544, 301.218, 762.510 | [M + Hac-H]− |
| 6 | 8.2 | 2’-Deoxyadenosine | Positive | C10H13N15O3 | 252.399, 113.324, 123.238 | [M + H]+ |
| 7 | 13.19 | Atenolol acid | Positive | C14H21NO4 | 269.156, 270.1558, 226.105 | [M + H]+ |
| 8 | 14.06 | Glutamylphenylalanine | Positive | C14H18N2O5 | 278.178, 277.120 | [M + H]+ |
| 9 | 26.83 | 9,10-EODE | Positive | - | - | |
| 10 | 30.91 | trans-4-hydroxycinnamic acid | Positive | C9H8O3 | 147.043, 165.054, 91.053 | [M + H]+ |
| 11 | 39.84 | Chlorogenate | Positive | C16H18O9 | 135.043, 163.037, 145.027 | [M + H]+ |
| 12 | 56.87 | Dinitrocresol | Positive | C7H6N2O5 | 197.020, 137.0244, 167.022 | [M + H]+ |
| 13 | 61.82 | Ritonavir | Positive | C37H48N6O5S2 | 721.325, 296.144, 426.187 | [M + H]+ |
| 14 | 63.24 | Quercetin-3-O-arabinoglucoside | Positive | C26H28O16 | 303.049, 304.052,145.0474 | [M + H]+ |
| 15 | 66.51 | 2-Napthalenesulfonic acid | Positive | C10H9NO3S | 223.001 | [M + H]+ |
| 16 | 69.1 | Rutin | Positive | C27H30O16 | 609.148, 610.151 | [M + H]+ |
| 17 | 79.1 | Trimethoprim | Positive | C14H18N4O3 | 291.146, 123.066 | [M + H]+ |
| Macromolecule | Ligand | Binding Affinity (kcal/mol) |
|---|---|---|
| Human BACE-1 protein | Hecogenin | −11.3 |
| Naringin 6″-malonate | −10.2 | |
| Procyanidin B2 | −10 | |
| Kaempferol-dirhamnoside | −9.9 |
| Compound | Molecular Weight (g/mol) | Num. H-Bond Acceptor | Num. H-Bond Donor | TPSA (Å2) | GI Absorption | BBB Permeant | Lipinski |
|---|---|---|---|---|---|---|---|
| 3-p-Coumaroylquinic acid | 338.31 g/mol | 8 | 5 | 144.52 | Low | No | Yes; 0 violation |
| Acacetin | 284.26 g/mol | 5 | 2 | 79.9 | High | No | Yes; 0 violation |
| Apigenin | 270.24 g/mol | 5 | 3 | 90.9 | high | No | Yes; 0 violation |
| Caffeic acid | 180.16 g/mol | 4 | 3 | 77.76 | High | No | Yes; 0 violation |
| Cinnamaldehyde | 132.16 g/mol | 1 | 0 | 17.07 | High | yes | Yes; 0 violation |
| Cinnamic acid | 148.16 g/mol | 2 | 1 | 37.3 | High | yes | Yes; 0 violation |
| Catechin | 290.27 g/mol | 6 | 5 | 110.38 | High | no | Yes: 0 violation |
| Ferulic acid O-glucoside | 356.32 g/mol | 9 | 5 | 145.91 | Low | no | Yes; 0 violation |
| Hecogenin | 430.62 g/mol | 4 | 1 | 55.76 | High | yes | Yes; 0 violation |
| Loliolide | 196.24 g/mol | 3 | 1 | 46.53 | High | yes | Yes; 0 violation |
| Malic acid | 134.09 g/mol | 5 | 3 | 94.83 | High | no | Yes; 0 violation |
| Luteolin | 286.24 g/mol | 6 | 4 | 111.13 | High | no | Yes; 0 violation |
| Quercetin | 302.24 g/mol | 7 | 5 | 131.36 | High | no | Yes; 0 violation |
| Quinic acid | 192.17 g/mol | 6 | 5 | 118.22 | Low | no | Yes; 0 violation |
| Rosmarinic acid | 360.31 g/mol | 8 | 5 | 144.52 | Low | no | Yes; 0 violation |
| 3’,4’,7-trihydroxyisoflavanone | 272.25 g/mol | 5 | 3 | 86.99 | High | no | Yes; 0 violation |
| Verapamil | 454.60 g/mol | 6 | 0 | 63.95 | High | yes | Yes; 0 violation |
| Atenolol | 266.34 g/mol | 4 | 3 | 84.58 | High | no | Yes; 0 violation |
| 4-Hydroxycinnamic acid | 164.16 g/mol | 3 | 2 | 57.53 | High | yes | Yes; 0 violation |
| Dinitrocresol | 198.13 g/mol | 5 | 1 | 111.87 | High | no | Yes; 0 violation |
| Trimethoprim | 290.32 g/mol | 5 | 2 | 105.51 | High | no | Yes; 0 violation |
| Compound | MMGBSA-dG-Binding Energy (kcal/mol) | MMGBSA-dG-Bind in Coulomb (kcal/mol) | MMGBSA-dG-Bind(NS) (kcal/mol) | MMGBSA-dG-Bind(NS)-Coulomb (kcal/mol) |
|---|---|---|---|---|
| Hecogenin | −57.7393 | −7.53358 | −60.3922 | −7.43166 |
| Naringin6″-malonate | −50.3731 | −15.0102 | −52.1919 | −15.2455 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padmanabhan, D.; Siddiqui, M.H.; Natarajan, P.; Palanisamy, S. Hecogenin a Plant Derived Small Molecule as an Antagonist to BACE-1: A Potential Target for Neurodegenerative Disorders. Metabolites 2023, 13, 758. https://doi.org/10.3390/metabo13060758
Padmanabhan D, Siddiqui MH, Natarajan P, Palanisamy S. Hecogenin a Plant Derived Small Molecule as an Antagonist to BACE-1: A Potential Target for Neurodegenerative Disorders. Metabolites. 2023; 13(6):758. https://doi.org/10.3390/metabo13060758
Chicago/Turabian StylePadmanabhan, Deepthi, Manzer H. Siddiqui, Purushothaman Natarajan, and Senthilkumar Palanisamy. 2023. "Hecogenin a Plant Derived Small Molecule as an Antagonist to BACE-1: A Potential Target for Neurodegenerative Disorders" Metabolites 13, no. 6: 758. https://doi.org/10.3390/metabo13060758
APA StylePadmanabhan, D., Siddiqui, M. H., Natarajan, P., & Palanisamy, S. (2023). Hecogenin a Plant Derived Small Molecule as an Antagonist to BACE-1: A Potential Target for Neurodegenerative Disorders. Metabolites, 13(6), 758. https://doi.org/10.3390/metabo13060758









