A Fecal-Microbial-Extracellular-Vesicles-Based Metabolomics Machine Learning Framework and Biomarker Discovery for Predicting Colorectal Cancer Patients
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design, Data, and Compliance with Ethical Standards
2.2. Microbe-Derived Extracellular Vesicles’ Isolation and Gas Chromatography Time-of-Flight Mass Spectrometry Analysis
2.3. Data Analysis
3. Results
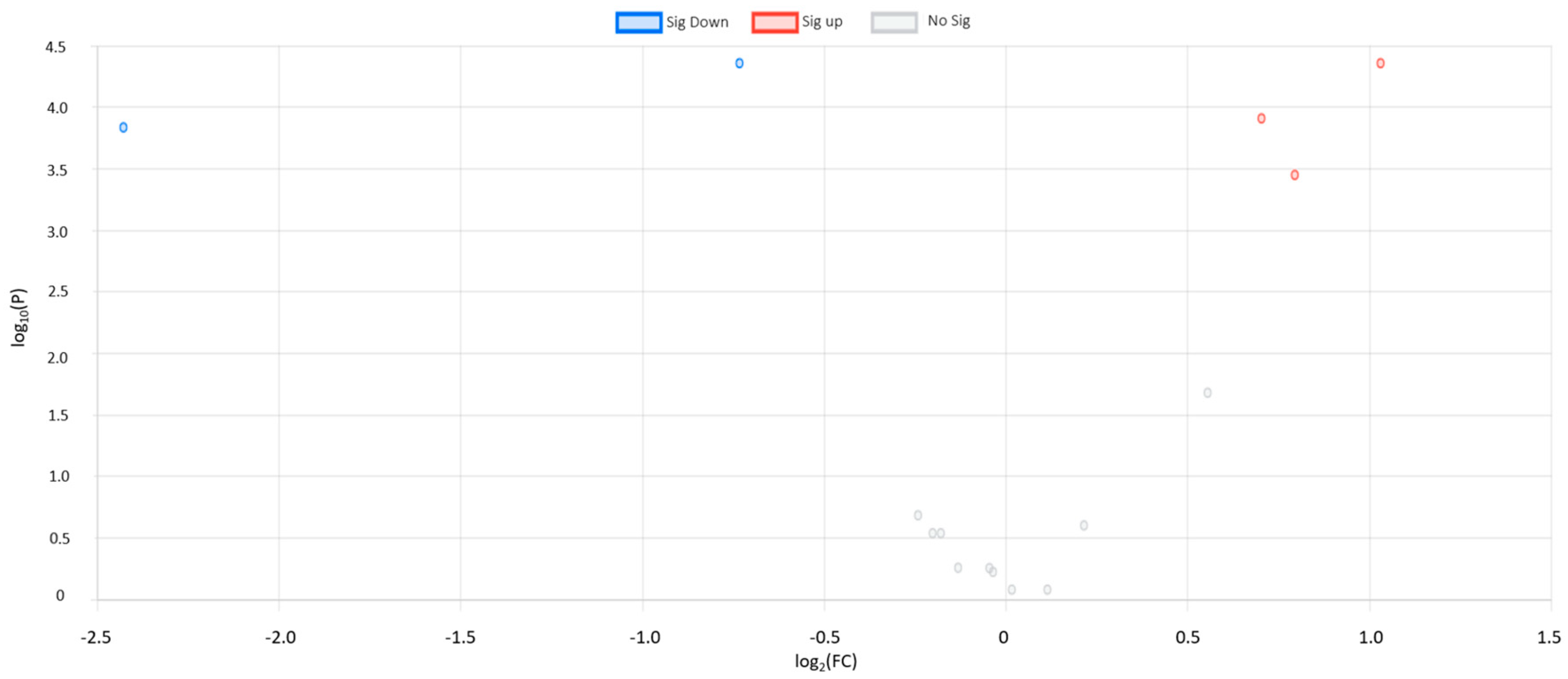
3.1. Univariate Statistical Analysis
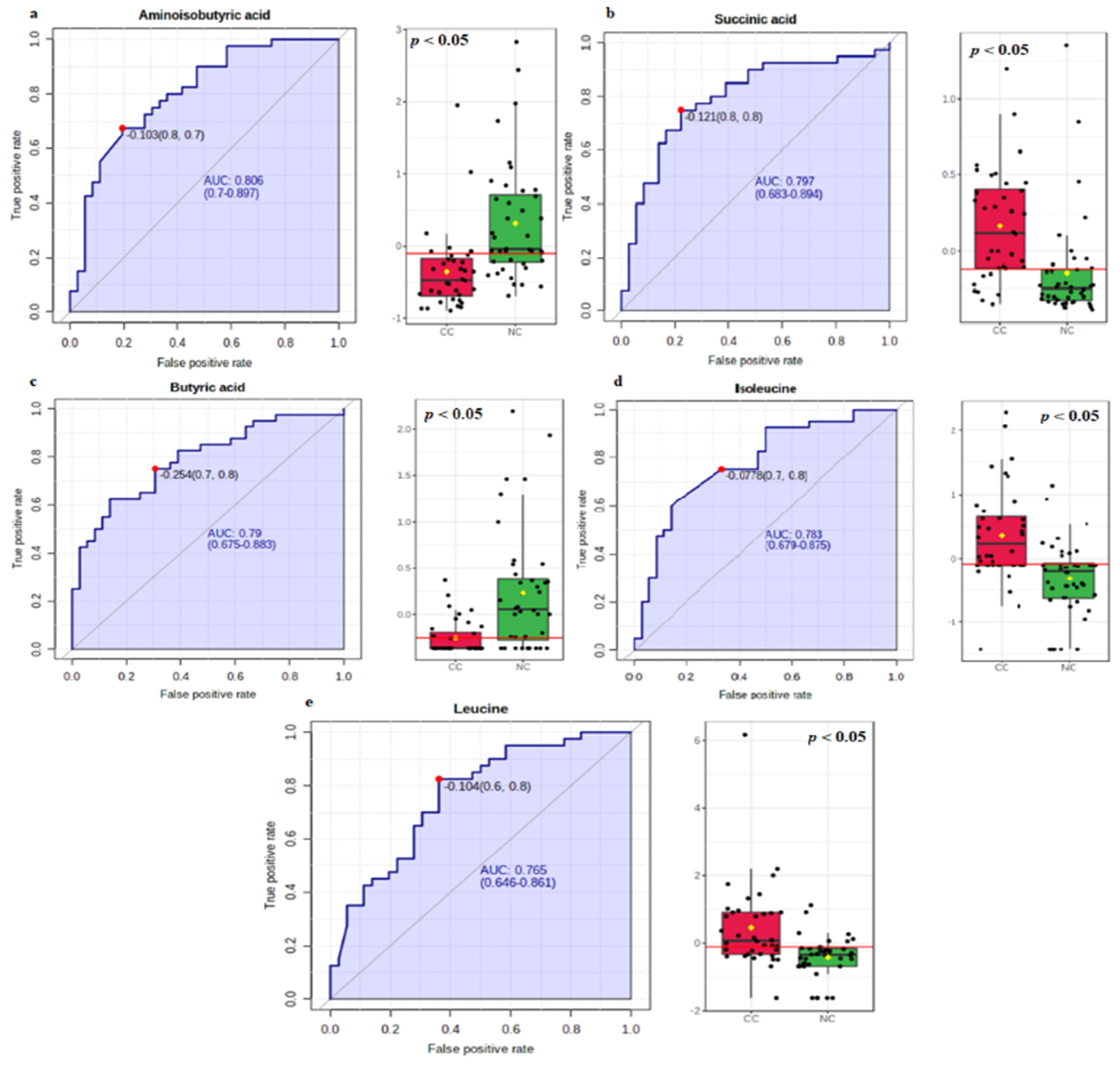
3.2. Biomarker Analysis
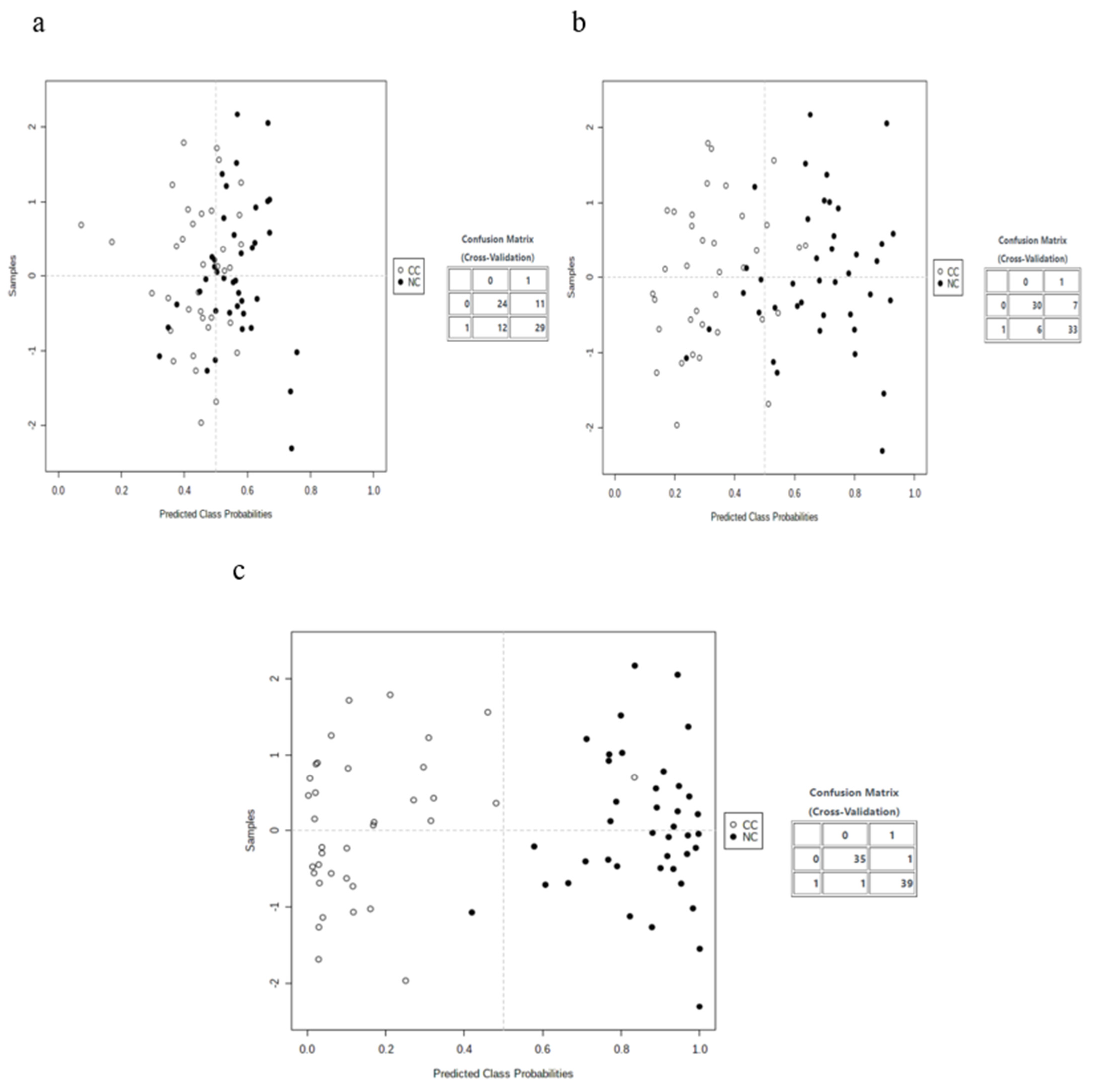
3.3. Multivariate Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 63–70. [Google Scholar]
- Atikukke, G.; Alkhateeb, A.; Porter, L.; Fifield, B.; Cavallo-Medved, D.; Facca, J.; Elfiki, T.; Elkeilani, A.; Rueda, L.; Misra, S. P-370 Comprehensive targeted genomic profiling and comparative genomic analysis to identify molecular mechanisms driving cancer progression in young-onset sporadic colorectal cancer. Ann. Oncol. 2020, 31, S209–S210. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Han, Y.; Wang, X. Metabolomics in diagnosis and biomarker discovery of colorectal cancer. Cancer Lett. 2014, 345, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.E.; Short, S.P.; Williams, C.S. Colorectal cancer and metabolism. Curr. Color. Cancer Rep. 2018, 14, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.I.; Tam, S.Y.; Azam, Z.; Law, H.K.W. Proteomic profiling of metabolic proteins as potential biomarkers of radioresponsiveness for colorectal cancer. J. Proteom. 2022, 262, 104600. [Google Scholar] [CrossRef]
- Kim, D.J.; Yang, J.; Seo, H.; Lee, W.H.; Ho Lee, D.; Kym, S.; Park, Y.S.; Kim, J.G.; Jang, I.-J.; Kim, Y.-K. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci. Rep. 2020, 10, 2860. [Google Scholar] [CrossRef]
- Hossain, M.J.; Chowdhury, U.N.; Islam, M.B.; Uddin, S.; Ahmed, M.B.; Quinn, J.M.; Moni, M.A. Machine learning and network-based models to identify genetic risk factors to the progression and survival of colorectal cancer. Comput. Biol. Med. 2021, 135, 104539. [Google Scholar] [CrossRef]
- Ahmadi Badi, S.; Moshiri, A.; Fateh, A.; Rahimi Jamnani, F.; Sarshar, M.; Vaziri, F.; Siadat, S.D. Microbiota-derived extracellular vesicles as new systemic regulators. Front. Microbiol. 2017, 8, 1610. [Google Scholar] [CrossRef]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Uronis, J.M.; Mühlbauer, M.; Herfarth, H.H.; Rubinas, T.C.; Jones, G.S.; Jobin, C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 2009, 4, e6026. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Gormley, I.C.; Fan, Y.; Gallagher, W.M.; Brennan, L. MetSizeR: Selecting the optimal sample size for metabolomic studies using an analysis based approach. BMC Bioinform. 2013, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-C.; Chen, K.; Chiu, C.-Y.; Lu, K.-Y.; Lu, H.-Y.; Chiang, M.-H.; Tsai, C.-K.; Lo, C.-J.; Cheng, M.-L.; Chang, T.-C. Metabolomic biomarkers in cervicovaginal fluid for detecting endometrial cancer through nuclear magnetic resonance spectroscopy. Metabolomics 2019, 15, 146. [Google Scholar] [CrossRef]
- Yilmaz, R.; Yağin, F.H. Early detection of coronary heart disease based on machine learning methods. Med. Rec. 2022, 4, 1–6. [Google Scholar] [CrossRef]
- Yang, Z.; Luo, F.; Liu, G.; Luo, Z.; Ma, S.; Gao, H.; He, H.; Tao, J. Plasma Metabolomic Analysis Reveals the Relationship between Immune Function and Metabolic Changes in Holstein Peripartum Dairy Cows. Metabolites 2022, 12, 953. [Google Scholar] [CrossRef]
- Paksoy, N.; Yağin, F.H. Artificial Intelligence-based Colon Cancer Prediction by Identifying Genomic Biomarkers. Med. Rec. 2022, 4, 196–202. [Google Scholar] [CrossRef]
- Yang, M.; Yang, H.; Ji, L.; Hu, X.; Tian, G.; Wang, B.; Yang, J. A multi-omics machine learning framework in predicting the survival of colorectal cancer patients. Comput. Biol. Med. 2022, 146, 105516. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, H.; Liu, J.; Long, J. Post-translational modifications on mitochondrial metabolic enzymes in cancer. Free Radic. Biol. Med. 2022, 179, 11–23. [Google Scholar] [CrossRef]
- Terasaki, M.; Mima, M.; Kudoh, S.; Endo, T.; Maeda, H.; Hamada, J.; Osada, K.; Miyashita, K.; Mutoh, M. Glycine and succinic acid are effective indicators of the suppression of epithelial-mesenchymal transition by fucoxanthinol in colorectal cancer stem-like cells. Oncol. Rep. 2018, 40, 414–424. [Google Scholar] [CrossRef]
- Long, L.; Yang, W.; Liu, L.; Tobias, D.K.; Katagiri, R.; Wu, K.; Jin, L.; Zhang, F.F.; Luo, X.; Liu, X. Dietary intake of branched-chain amino acids and survival after colorectal cancer diagnosis. Int. J. Cancer 2021, 148, 2471–2480. [Google Scholar] [CrossRef]
- Bordon, Y. Leucine restriction limits pro-cancer B cell responses. Nat. Rev. Immunol. 2022, 22, 408–409. [Google Scholar] [CrossRef]
- Lamy, C.; Mansard, C.; Blondel, L.; Mercier, L.; Paci, A.; Broutin, S. Quantification of succinic acid levels, linked to succinate dehydrogenase (SDH) dysfunctions, by an automated and fully validated liquid chromatography tandem mass spectrometry method suitable for multi-matrix applications. J. Chromatogr. B 2022, 1189, 123085. [Google Scholar] [CrossRef]
- Holz, M.K. The role of S6K1 in ER-positive breast cancer. Cell Cycle 2012, 11, 3159–3165. [Google Scholar] [CrossRef]


| Metabolite Name | FC | Log2 FC | −Log10 (p-Value) | FDR Adjusted p-Value | Regulation | Main Class | Subclass |
|---|---|---|---|---|---|---|---|
| Succinic acid | 2.04 | 1.03 | 4.35 | 8.64 × | UP | TCA acids | TCA acids |
| Aminoisobutyric acid | 0.60 | 0.73 | 4.35 | 4.95 × | DOWN | Fatty acids | Amino fatty acids |
| Butyric acid | 0.18 | −2.43 | 4.17 | 4.60 × | DOWN | Fatty acids | Saturated fatty acids |
| Isoleucine | 1.63 | 0.70 | 4.03 | 4.60 × | UP | Amino acids and peptides | Amino acids |
| Leucine | 1.73 | 0.79 | 3.52 | 8.19 × | UP | Amino acids and peptides | Amino acids |
| Oxalic acid | 1.55 | 0.07 | Fatty acids | Dicarboxylic acids | |||
| Alanine | 1.02 | 0.20 | Amino acids and peptides | Amino acids | |||
| Ethanolamine | 0.97 | 0.20 | Amines | 1,2-Aminoalcohols | |||
| Caproic acid | 0.62 | 0.39 | Fatty acids | Saturated fatty acids | |||
| Oleic acid | 0.58 | 0.39 | Fatty acids | Unsaturated fatty acids | |||
| Lysine | 0.39 | 0.55 | Amino acids and peptides | Amino acids | |||
| Phenol | 0.33 | 0.58 | Phenolic acids | Phenolic acids | |||
| 2-Furoic acid | 0.12 | 0.86 | Furoic acids | Furoic acid derivatives | |||
| Palmitic acid | 0.09 | 0.86 | Fatty acids | Saturated fatty acids | |||
| Tyramine | 0.04 | 0.91 | Tyrosine alkaloids | Phenylethylamines |
| Metabolite Name | Cut-Off Point | AUC | 95% CI | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Aminoisobutyric acid | −0.103 | 0.806 | 0.700–0.897 | 0.675 | 0.805 |
| Succinic acid | −0.121 | 0.797 | 0.683–0.894 | 0.750 | 0.770 |
| Butyric acid | −0.254 | 0.790 | 0.675–0.883 | 0.750 | 0.694 |
| Isoleucine | −0.078 | 0.783 | 0.679–0.875 | 0.750 | 0.666 |
| Leucine | −0.104 | 0.765 | 0.646–0.861 | 0.820 | 0.638 |
| Oxalic acid | −0.171 | 0.675 | 0.552–0.805 | 0.675 | 0.611 |
| Ethanolamine | −0.149 | 0.609 | 0.492–0.734 | 0.550 | 0.666 |
| Alanine | −1.130 | 0.601 | 0.471–0.725 | 0.425 | 0.805 |
| Caproic acid | −0.089 | 0.588 | 0.465–0.705 | 0.550 | 0.583 |
| Oleic acid | 0.003 | 0.587 | 0.448–0.720 | 0.500 | 0.722 |
| Lysine | −0.225 | 0.556 | 0.435–0.687 | 0.600 | 0.611 |
| 2-Furoic acid | −0.207 | 0.551 | 0.412–0.673 | 0.650 | 0.472 |
| Palmitic acid | −4.170 | 0.544 | 0.413–0.682 | 0.675 | 0.472 |
| Tyramine | −0.220 | 0.514 | 0.384–0.640 | 0.525 | 0.527 |
| Phenol | −0.465 | 0.514 | 0.377–0.640 | 0.475 | 0.583 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yagin, F.H.; Alkhateeb, A.; Colak, C.; Azzeh, M.; Yagin, B.; Rueda, L. A Fecal-Microbial-Extracellular-Vesicles-Based Metabolomics Machine Learning Framework and Biomarker Discovery for Predicting Colorectal Cancer Patients. Metabolites 2023, 13, 589. https://doi.org/10.3390/metabo13050589
Yagin FH, Alkhateeb A, Colak C, Azzeh M, Yagin B, Rueda L. A Fecal-Microbial-Extracellular-Vesicles-Based Metabolomics Machine Learning Framework and Biomarker Discovery for Predicting Colorectal Cancer Patients. Metabolites. 2023; 13(5):589. https://doi.org/10.3390/metabo13050589
Chicago/Turabian StyleYagin, Fatma Hilal, Abedalrhman Alkhateeb, Cemil Colak, Mohammad Azzeh, Burak Yagin, and Luis Rueda. 2023. "A Fecal-Microbial-Extracellular-Vesicles-Based Metabolomics Machine Learning Framework and Biomarker Discovery for Predicting Colorectal Cancer Patients" Metabolites 13, no. 5: 589. https://doi.org/10.3390/metabo13050589
APA StyleYagin, F. H., Alkhateeb, A., Colak, C., Azzeh, M., Yagin, B., & Rueda, L. (2023). A Fecal-Microbial-Extracellular-Vesicles-Based Metabolomics Machine Learning Framework and Biomarker Discovery for Predicting Colorectal Cancer Patients. Metabolites, 13(5), 589. https://doi.org/10.3390/metabo13050589












