Identification of Genetic Markers Linked to The Activity of Indoleamine 2,3-Dioxygenase and Kidney Function
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Metabolite Measurements
2.3. Genotyping and Imputation
2.4. Statistical Analysis
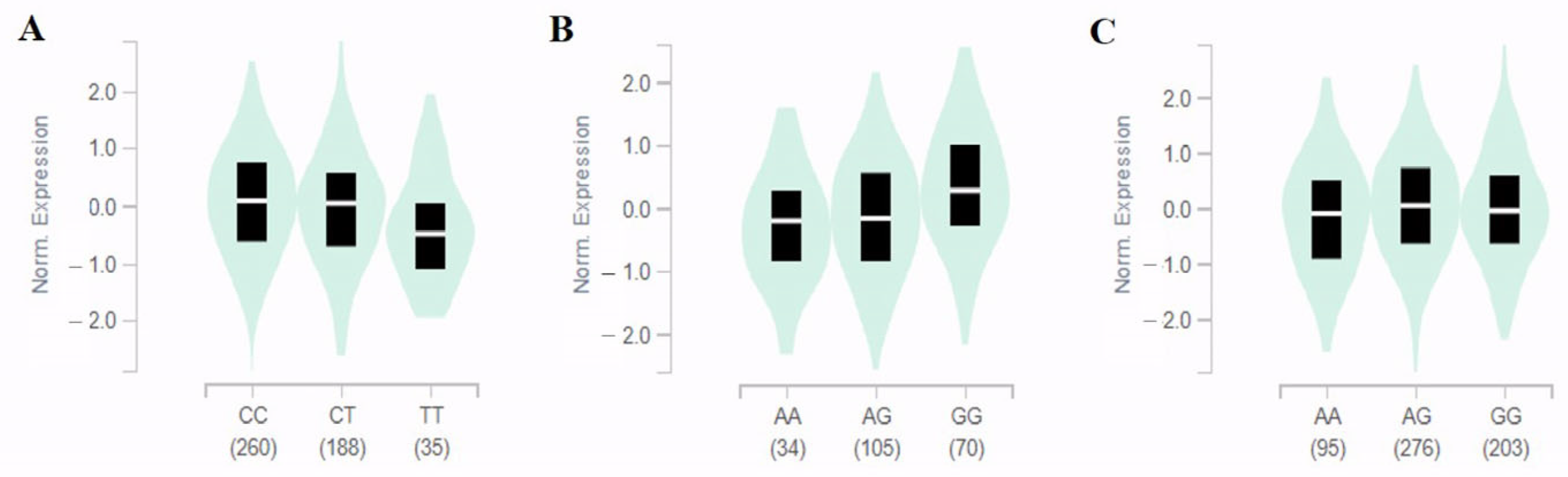
2.5. Genotype-Tissue Expression (GTEx)
2.6. Ethics Statement
3. Results
3.1. Characteristics of Study Participants
3.2. Variants Associated with Both IDO Activity and CKD
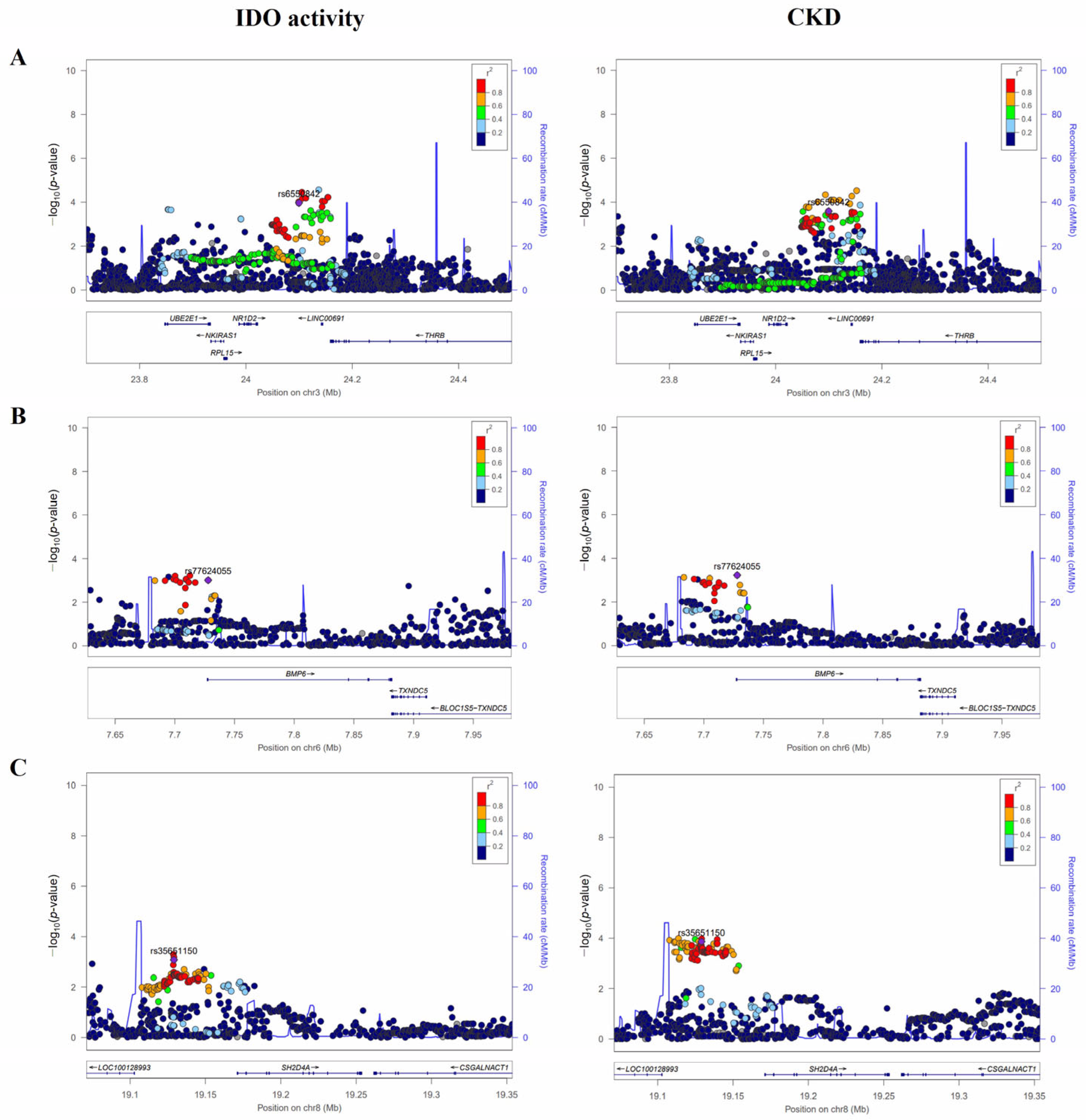
3.3. In Silico Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levey, A.; Atkins, R.; Coresh, J.; Cohen, E.; Collins, A.; Eckardt, K.-U.; Nahas, M.; Jaber, B.; Jadoul, M.; Levin, A. Chronic kidney disease as a global public health problem: Approaches and initiatives—A position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007, 72, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.S. Chronic renal disease. BMJ 2002, 325, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention (CDC). Chronic Kidney Disease in the United States. 2021. Available online: https://www.cdc.gov/kidneydisease/publications-resources/CKD-national-facts.html (accessed on 23 July 2022).
- Zoccali, C.; Kramer, A.; Jager, K.J. Chronic kidney disease and end-stage renal disease—A review produced to contribute to the report ‘the status of health in the European union: Towards a healthier Europe’. NDT Plus 2010, 3, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.; Castro, A.F., III; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.D.; Madias, N.E.; Levey, A.S. Serum creatinine as an index of renal function: New insights into old concepts. Clin. Chem. 1992, 38, 1933–1953. [Google Scholar] [CrossRef]
- Mor, A.; Kalaska, B.; Pawlak, D. Kynurenine pathway in chronic kidney disease: What’s old, what’s new, and what’s next? Int. J. Tryptophan. Res. 2020, 13, 1178646920954882. [Google Scholar] [CrossRef]
- Robinson, C.M.; Hale, P.T.; Carlin, J.M. The role of IFN-γ and TNF-α-responsive regulatory elements in the synergistic induction of indoleamine dioxygenase. J. Interferon. Cytokine Res. 2005, 25, 20–30. [Google Scholar] [CrossRef]
- Hou, W.; Li, S.; Wu, Y.; Du, X.; Yuan, F. Inhibition of indoleamine 2,3-dioxygenase-mediated tryptophan catabolism accelerates crescentic glomerulonephritis. Clin. Exp. Immunol. 2009, 156, 363–372. [Google Scholar] [CrossRef]
- Mohib, K.; Wang, S.; Guan, Q.; Mellor, A.L.; Sun, H.; Du, C.; Jevnikar, A.M. Indoleamine 2,3-dioxygenase expression promotes renal ischemia-reperfusion injury. Am. J. Physiol. Renal. Physiol. 2008, 295, F226–F234. [Google Scholar] [CrossRef]
- Matheus, L.H.G.; Simão, G.M.; Amaral, T.A.; Brito, R.B.O.; Malta, C.S.; Matos, Y.S.T.; Santana, A.C.; Rodrigues, G.G.C.; Albejante, M.C.; Bach, E.E. Indoleamine 2,3-dioxygenase (IDO) increases during renal fibrogenesis and its inhibition potentiates TGF-β 1-induced epithelial to mesenchymal transition. BMC Nephrol. 2017, 18, 287. [Google Scholar] [CrossRef]
- Baban, B.; Liu, J.Y.; Mozaffari, M.S. Endoplasmic reticulum stress response and inflammatory cytokines in type 2 diabetic nephropathy: Role of indoleamine 2,3-dioxygenase and programmed death-1. Exp. Mol. Pathol. 2013, 94, 343–351. [Google Scholar] [CrossRef]
- Schefold, J.C.; Zeden, J.-P.; Fotopoulou, C.; von Haehling, S.; Pschowski, R.; Hasper, D.; Volk, H.-D.; Schuett, C.; Reinke, P. Increased indoleamine 2,3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: A possible link between chronic inflammation and uraemic symptoms. Nephrol. Dial. Transplant. 2009, 24, 1901–1908. [Google Scholar] [CrossRef]
- Bao, Y.-S.; Ji, Y.; Zhao, S.-L.; Ma, L.-L.; Xie, R.-J.; Na, S.-P. Serum levels and activity of indoleamine2,3-dioxygenase and tryptophanyl-tRNA synthetase and their association with disease severity in patients with chronic kidney disease. Biomarkers 2013, 18, 379–385. [Google Scholar] [CrossRef]
- Kim, H.R.; Jin, H.S.; Eom, Y.B. Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease. Genes 2021, 12, 1905. [Google Scholar] [CrossRef]
- Meng, X.M. Inflammatory Mediators and Renal Fibrosis. Adv. Exp. Med. Biol. 2019, 1165, 381–406. [Google Scholar]
- Akiyama, M. Multi-omics study for interpretation of genome-wide association study. J. Hum. Genet. 2021, 66, 3–10. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef]
- Cañadas-Garre, M.; Anderson, K.; Cappa, R.; Skelly, R.; Smyth, L.J.; McKnight, A.J.; Maxwell, A.P. Genetic susceptibility to chronic kidney disease–some more pieces for the heritability puzzle. Front. Genet. 2019, 10, 453. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G.; KoGES Group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, 1350. [Google Scholar] [CrossRef]
- Levey, A.S.; Coresh, J.; Greene, T.; Marsh, J.; Stevens, L.A.; Kusek, J.W.; Van Lente, F.; Collaboration, C.K.D.E. Expressing the Modification of Diet in Renal Disease Study equation for estimating glomerular filtration rate with standardized serum creatinine values. Clin. Chem. 2007, 53, 766–772. [Google Scholar] [CrossRef]
- Levey, A.S.; Eckardt, K.-U.; Tsukamoto, Y.; Levin, A.; Coresh, J.; Rossert, J.; Zeeuw, D.D.; Hostetter, T.H.; Lameire, N.; Eknoyan, G. Definition and classification of chronic kidney disease: A position statement from Kidney Disease: Improving Global Outcomes (KDIGO). Kidney Int. 2005, 67, 2089–2100. [Google Scholar] [CrossRef]
- Lee, H.-S.; Xu, T.; Lee, Y.; Kim, N.-H.; Kim, Y.-J.; Kim, J.-M.; Cho, S.Y.; Kim, K.-Y.; Nam, M.; Adamski, J. Identification of putative biomarkers for type 2 diabetes using metabolomics in the Korea Association REsource (KARE) cohort. Metabolomics 2016, 12, 178. [Google Scholar] [CrossRef]
- Cho, Y.S.; Go, M.J.; Kim, Y.J.; Heo, J.Y.; Oh, J.H.; Ban, H.-J.; Yoon, D.; Lee, M.H.; Kim, D.-J.; Park, M. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009, 41, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Howie, B.N.; Donnelly, P.; Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Lee, H.; Jang, H.B.; Yoo, M.-G.; Park, S.I.; Lee, H.-J. Amino acid metabolites associated with chronic kidney disease: An eight-year follow-up Korean epidemiology study. Biomedicines 2020, 8, 222. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; De Geus, E.J.; Boomsma, D.I.; Wright, F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Postler, T.S.; Rao, P.; Schmitt, H.; Schmitt, V.; Grinberg-Bleyer, Y.; Kühn, L.I.; Gruber, C.W.; Lienhard, G.E.; Ghosh, S. κB-Ras proteins regulate both NF-κB-dependent inflammation and Ral-dependent proliferation. Cell Rep. 2014, 8, 1793–1807. [Google Scholar] [CrossRef]
- Esteban, V.; Lorenzo, O.; Rupérez, M.; Suzuki, Y.; Mezzano, S.; Blanco, J.; Kretzler, M.; Sugaya, T.; Egido, J.; Ruiz-Ortega, M. Angiotensin II, via AT1 and AT2 receptors and NF-κB pathway, regulates the inflammatory response in unilateral ureteral obstruction. J. Am. Soc. Nephrol. 2004, 15, 1514–1529. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, S.-C. NF-κB in inflammation and renal diseases. Cell Biosci. 2015, 5, 63. [Google Scholar] [CrossRef]
- Gerashchenko, G.; Bogatyrova, O.; Rudenko, E.; Kondratov, A.; Gordiyuk, V.; Zgonnyk, Y.; Vozianov, O.; Pavlova, T.; Zabarovsky, E.; Rynditch, A. Genetic and epigenetic changes of NKIRAS1 gene in human renal cell carcinomas. Exp. Oncol. 2010, 32, 71–75. [Google Scholar]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-beta: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Yoshioka, K.; Takemura, T.; Murakami, K.; Okada, M.; Hino, S.; Miyamoto, H.; Maki, S. Transforming growth factor-beta protein and mRNA in glomeruli in normal and diseased human kidneys. Lab. Investig. 1993, 68, 154–163. [Google Scholar]
- Yamamoto, T.; Nakamura, T.; Noble, N.A.; Ruoslahti, E.; Border, W.A. Expression of transforming growth factor beta is elevated in human and experimental diabetic nephropathy. Proc. Natl. Acad. Sci. USA 1993, 90, 1814–1818. [Google Scholar] [CrossRef]
- Chen, W. IDO: More than an enzyme. Nat. Immunol. 2011, 12, 809–811. [Google Scholar] [CrossRef]
- Dendooven, A.; van Oostrom, O.; van der Giezen, D.M.; Leeuwis, J.W.; Snijckers, C.; Joles, J.A.; Robertson, E.J.; Verhaar, M.C.; Nguyen, T.Q.; Goldschmeding, R. Loss of endogenous bone morphogenetic protein-6 aggravates renal fibrosis. Am. J. Pathol. 2011, 178, 1069–1079. [Google Scholar] [CrossRef]
- Jenkins, R.H.; Fraser, D.J. BMP-6 emerges as a potential major regulator of fibrosis in the kidney. Am. J. Pathol. 2011, 178, 964–965. [Google Scholar] [CrossRef]
- Long, J.; Badal, S.S.; Wang, Y.; Chang, B.H.; Rodriguez, A.; Danesh, F.R. MicroRNA-22 is a master regulator of bone morphogenetic protein-7/6 homeostasis in the kidney. J. Biol. Chem. 2013, 288, 36202–36214. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.-D.; Yang, S.; Zhang, J.; Zhu, T.-H. BMP6 reverses TGF-β1-induced changes in HK-2 cells: Implications for the treatment of renal fibrosis. Acta Pharmacol. Sin. 2009, 30, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Paterni, I.; Granchi, C.; Katzenellenbogen, J.A.; Minutolo, F. Estrogen receptors alpha (ERα) and beta (ERβ): Subtype-selective ligands and clinical potential. Steroids 2014, 90, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.-Y.; Chen, S.; Du, Y. Estrogen and estrogen receptors in kidney diseases. Ren. Fail. 2021, 43, 619–642. [Google Scholar] [CrossRef]
- Carrero, J.J.; Hecking, M.; Chesnaye, N.C.; Jager, K.J. Sex and gender disparities in the epidemiology and outcomes of chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 151–164. [Google Scholar] [CrossRef]
- Li, T.; Li, W.; Lu, J.; Liu, H.; Li, Y.; Zhao, Y. SH2D4A regulates cell proliferation via the ERα/PLC-γ/PKC pathway. BMB Rep. 2009, 42, 516–522. [Google Scholar] [CrossRef]
| Characteristics | Quantitative Trait Analysis | Case-Control Analysis for CKD | ||
|---|---|---|---|---|
| Controls | Cases | p-Value 1 | ||
| Number of participants | 2579 | 1550 | 264 | |
| Gender [men/women] | 1218/1361 | 789/761 | 81/183 | <0.001 |
| Age (M years ± SD) | 57.10 ± 9.05 | 54.98 ± 8.64 | 65.72 ± 6.53 | <0.001 |
| Height (M cm ± SD) | 159.55 ± 9.16 | 160.55 ± 8.98 | 155.42 ± 8.28 | <0.001 |
| Weight (M kg ± SD) | 62.63 ± 10.36 | 62.30 ± 10.41 | 60.88 ± 9.53 | 0.042 |
| BMI (M kg/m2 ± SD) | 24.56 ± 3.23 | 24.11 ± 3.09 | 25.20 ± 3.47 | <0.001 |
| eGFR (mL/min/1.73 m2) | 75.58 ± 11.92 | 78.68 ± 9.69 | 55.24 ± 9.21 | <0.001 |
| Creatinine (mg/dL) | 0.98 ± 0.20 | 0.96 ± 0.14 | 1.18 ± 0.42 | <0.001 |
| BUN (mg/dL) | 15.69 ± 4.26 | 15.33 ± 3.92 | 17.91 ± 5.45 | <0.001 |
| SNP | Nearest Gene | Chromosome Position | Minor Allele | MAF | Function | IDO Activity | CKD | eGFR | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± S.E | p-Value | OR (95% CI) | p-Value | β ± S.E | p-Value | ||||||
| rs189138212 | ITPKB | 1:226951229 | C | 0.017 | - | 0.44 ± 0.11 | 7.19 × 10−5 | 3.55 (1.71–7.37) | 6.78 × 10–4 | −3.34 ± 1.13 | 3.28 × 10–3 |
| rs6550842 | NKIRAS1-NR1D2-THRB | 3:24099794 | T | 0.107 | - | 0.18 ± 0.05 | 1.07 × 10−4 | 1.89 (1.34–2.65) | 2.52 × 10–4 | −1.77 ± 0.47 | 1.76 × 10–4 |
| rs145478425 | FHIT | 3:59803424 | T | 0.015 | Intron | 0.41 ± 0.12 | 4.04 × 10−4 | 3.63 (1.78–7.40) | 3.95 × 10–4 | −1.26 ± 1.20 | 0.293 |
| rs7679032 | LINC00616 | 4:138949061 | T | 0.031 | Intron | 0.32 ± 0.08 | 1.26 × 10−4 | 2.65 (1.53–4.60) | 5.24 × 10–4 | −1.32 ± 0.85 | 0.120 |
| rs77624055 | BMP6 | 6:7727931 | A | 0.167 | Intron | 0.13 ± 0.04 | 9.70 × 10−4 | 1.67 (1.25–2.23) | 5.89 × 10–4 | −0.93 ± 0.39 | 0.017 |
| rs35651150 | SH2D4A | 8:19129080 | A | 0.256 | - | −0.11 ± 0.03 | 8.30 × 10−4 | 0.58 (0.44–0.77) | 1.37 × 10–4 | 1.16 ± 0.33 | 4.48 × 10–4 |
| rs149583220 | ADAM7 | 8:24307595 | C | 0.011 | Intron | 0.48 ± 0.13 | 2.99 × 10−4 | 4.64 (2.06–10.44) | 2.16 × 10–4 | −4.26 ± 1.36 | 1.75 × 10–3 |
| rs145951089 | LOC101927318 | 12:50355316 | T | 0.026 | Intron | 0.34 ± 0.09 | 1.40 × 10−4 | 3.13 (1.71–5.73) | 2.15 × 10–4 | −2.43 ± 0.91 | 7.80 × 10–3 |
| rs9533960 | LINC00330 | 13:45333274 | C | 0.320 | - | 0.11 ± 0.03 | 5.04 × 10−4 | 1.51 (1.19–1.92) | 7.31 × 10–4 | −0.63 ± 0.31 | 0.041 |
| rs72898186 | MIR924HG | 18:37071519 | C | 0.163 | Intron | 0.13 ± 0.04 | 9.80 × 10−4 | 1.72 (1.29–2.29) | 2.49 × 10–4 | −0.75 ± 0.39 | 0.056 |
| SNP | Chromosome Position | Minor Allele | MAF | Function | IDO Activity | CKD | eGFR | |||
|---|---|---|---|---|---|---|---|---|---|---|
| β ± S.E | p-Value | OR (95% CI) | p-Value | β ± S.E | p-Value | |||||
| rs77624055 | 7727931 | A | 0.167 | Intron | 0.125 ± 0.038 | 9.70 × 10–4 | 1.67 (1.25–2.23) | 5.89 × 10−4 | −0.93 ± 0.39 | 0.017 |
| rs7753111 | 7730944 | G | 0.299 | Intron | 0.085 ± 0.031 | 5.56 × 10–3 | 1.31 (1.04–1.66) | 0.024 | −0.74 ± 0.31 | 0.019 |
| rs2224564 | 7737187 | C | 0.297 | Intron | 0.082 ± 0.031 | 8.99 × 10–3 | 0.93 (0.73–1.19) | 0.564 | −0.57 ± 0.32 | 0.079 |
| rs111588693 | 7727271 | A | 0.179 | Non-synonymous (R28Q) | −0.085 ± 0.038 | 0.023 | 0.89 (0.65–1.23) | 0.484 | 0.06 ± 0.38 | 0.875 |
| rs76295967 | 7733744 | A | 0.009 | Intron | −0.305 ± 0.151 | 0.043 | 0.71 (0.14–3.52) | 0.676 | 0.34 ± 1.54 | 0.826 |
| rs7766858 | 7694908 | G | 0.478 | - | 0.043 ± 0.029 | 0.132 | 1.32 (1.05–1.65) | 0.015 | −0.60 ± 0.29 | 0.041 |
| rs270407 | 7738392 | C | 0.478 | Intron | −0.043 ± 0.028 | 0.132 | 1.21 (0.97–1.52) | 0.090 | 0.13 ± 0.29 | 0.657 |
| rs962279 | 7708132 | G | 0.324 | - | −0.043 ± 0.030 | 0.150 | 0.95 (0.74–1.22) | 0.690 | −0.05 ± 0.31 | 0.859 |
| rs1923409 | 7728212 | A | 0.370 | Intron | 0.031 ± 0.029 | 0.297 | 1.25 (0.99–1.58) | 0.057 | −0.59 ± 0.30 | 0.048 |
| rs270417 | 7729614 | C | 0.025 | Intron | −0.095 ± 0.091 | 0.297 | 1.68 (0.84–3.36) | 0.145 | −0.63 ± 0.93 | 0.498 |
| rs9505263 | 7698011 | T | 0.017 | - | −0.106 ± 0.109 | 0.329 | 0.65 (0.26–1.65) | 0.364 | 0.64 ± 1.11 | 0.565 |
| rs79982308 | 7691593 | A | 0.016 | - | 0.068 ± 0.110 | 0.535 | 1.29 (0.60–2.75) | 0.513 | −0.16 ± 1.12 | 0.886 |
| rs75686372 | 7737224 | T | 0.016 | Intron | 0.060 ± 0.115 | 0.603 | 1.84 (0.88–3.85) | 0.105 | −1.05 ± 1.18 | 0.371 |
| rs118168182 | 7735363 | T | 0.028 | Intron | 0.027 ± 0.088 | 0.759 | 0.95 (0.49–1.86) | 0.888 | 1.52 ± 0.89 | 0.090 |
| rs79560447 | 7690928 | T | 0.142 | - | −0.005 ± 0.042 | 0.902 | 0.77 (0.56–1.07) | 0.115 | 0.27 ± 0.42 | 0.526 |
| rs2876117 | 7738931 | T | 0.138 | Intron | 0.000 ± 0.040 | 0.994 | 0.91 (0.67–1.25) | 0.573 | 0.27 ± 0.41 | 0.507 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-R.; Jin, H.-S.; Eom, Y.-B. Identification of Genetic Markers Linked to The Activity of Indoleamine 2,3-Dioxygenase and Kidney Function. Metabolites 2023, 13, 541. https://doi.org/10.3390/metabo13040541
Kim H-R, Jin H-S, Eom Y-B. Identification of Genetic Markers Linked to The Activity of Indoleamine 2,3-Dioxygenase and Kidney Function. Metabolites. 2023; 13(4):541. https://doi.org/10.3390/metabo13040541
Chicago/Turabian StyleKim, Hye-Rim, Hyun-Seok Jin, and Yong-Bin Eom. 2023. "Identification of Genetic Markers Linked to The Activity of Indoleamine 2,3-Dioxygenase and Kidney Function" Metabolites 13, no. 4: 541. https://doi.org/10.3390/metabo13040541
APA StyleKim, H.-R., Jin, H.-S., & Eom, Y.-B. (2023). Identification of Genetic Markers Linked to The Activity of Indoleamine 2,3-Dioxygenase and Kidney Function. Metabolites, 13(4), 541. https://doi.org/10.3390/metabo13040541





