Abstract
β-Chitin is an important carbon fixation product of diatoms, and is the most abundant nitrogen-containing polysaccharide in the ocean. It has potential for widespread application, but the characterization of chitin-related enzymes from β-chitin producers has rarely been reported. In this study, a chitin deacetylase (TwCDA) was retrieved from the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) database and was heterologously expressed in vitro for functional analysis. The results showed that both the full-length sequence (TwCDA) and the N-terminal truncated sequence (TwCDA-S) had chitin deacetylase and chitinolytic activities after expression in Escherichia coli. High-performance liquid chromatography (HPLC) and gas chromatography–mass spectrometry (GC-MS) indicated that TwCDA and TwCDA-S could catalyze the deacetylation of oligosaccharide (GlcNAc)5. TwCDA had higher deacetylase activity, and also catalyzed the deacetylation of the β-chitin polymer. A dinitrosalicylic acid (DNS) assay showed that TwCDA-S had high chitinolytic activity for (GlcNAc)5, and the optimal reaction temperature was 35 °C. Liquid chromatography combined with time-of-flight mass spectrometry (LC-coTOF-MS) detected the formation of a N-acetylglucosamine monomer (C8H15NO6) in the reaction mixture. Altogether, we isolated a chitin deacetylase from a marine diatom, which can catalyze the deacetylation and degradation of chitin and chitin oligosaccharides. The relevant results lay a foundation for the internal regulation mechanism of chitin metabolism in diatoms and provide a candidate enzyme for the green industrial preparation of chitosan and chitin oligosaccharides.
1. Introduction
Chitin, β-(1,4)-linked N-acetylglucosamine (GlcNAc) is the most abundant renewable polymer in the ocean [1,2]. It is an important source of carbon and nitrogen in marine environments, where billions of tons are produced each year [3]. Its natural reserves are second only to those of cellulose, widely found in marine mollusks, crustaceans, and diatoms [4]. Chitin has three crystal polymorphs, α, β, and γ [5]. The chain segments of β-chitin are arranged in parallel, which is more accessible to produce amorphous structures. β-Chitin exhibits better reactivity and solubility, and has potential to produce derivatives with higher bioactivities [6,7]. Previous reports showed that β-chitin only exists in marine molluscs (such as squid) and some diatoms [8,9]. Diatoms are the most abundant group of eukaryotic microalgae, which contribute up to 40% of marine primary productivity [10,11,12]. Chitin fibers extend from the theca through the fultoportulae, and have been reported to represent approximately 30% of the fresh weight of diatom cells [13]. Thalassiosira and Cyclotella in centric diatoms are the most important producers of marine chitin [14,15]. Through a cell wall polysaccharide glycosidic linkage analysis, Cheng et al. [16] found that, of the three typical species of Thalassiosira (T. pseudonana, T. rotula, and T. weissflogii), the chitin content in the cell wall of T. weissflogii was the highest, indicating that T. weissflogii might have a strong ability to synthesize chitin and is an excellent material for the exploration of chitin-related enzyme resources and chitin derivatives.
Chitin and its derivatives are used in medicine, pharmaceuticals, cosmetics, the food industry, agriculture, environmental protection, etc. However, due to the strong hydrophobicity of chitin, its application is greatly limited, and its economic value has not been fully appreciated [17]. In contrast, many chitin derivatives, such as chitosan and chitooligosaccharide, have better physicochemical and physiological properties, especially solubility and biocompatibility [18]. Chitosan, also known as soluble chitin, is derived from chitin by the catalyzation of chitin deacetylase [19,20]. Chitooligosaccharides are oligomers of chitosan that are produced by the decomposition of chitosan by chitinase [20,21].
Chitin deacetylase (CDA, E.C.3.5.1.41) belongs to the carbohydrate esterase 4 (CE4) superfamily. It participates in the degradation pathway of chitin and has a wide range of substrate specificity. It can catalyze the deacetylation of both chitin oligosaccharides and insoluble chitin [22]. The initial study of chitin deacetylase focused on screening bacteria and fungal strains producing CDA [23,24]. Subsequently, CDA genes were extensively identified and isolated, facilitating the exploration of their enzymatic activities [25,26]. In insects, CDA has been confirmed to play an important role in morphogenesis and the immune response [27,28,29]. In bacteria and fungi, CDAs may contribute to chitin degradation for nutritional purposes, and most research has focused on cloning the CDA genes and identifying their enzyme activities [30,31,32]. It has also been reported that a few pathogenic fungal CDA can render the fungus less susceptible to host immunosurveillance [33]. Recently, RNA interference (RNAi) and gene knockout have also been applied to study the function of chitin deacetylase. Yang et al. [34] characterized four CDAs from Sogatella furcifera and found that three of them were associated with molting defects and high mortality with nymph–adult molting. Li et al. [35] knocked out the EScDA-l gene in Eriocheir sinensis, which resulted in an abnormal ultrastructure of the cuticle, suggesting that EsCDA-l is indispensable for molting. However, present research is still largely restricted to bacteria, fungi, and crustaceans, and there have been few reports of chitin deacetylase in eukaryotic microalgae. To our knowledge, the only report on functional CDA in diatoms noted the overexpression of two diatom CDA genes (PtCDA and TpCDA) in Phaeodactylum tricornutum to address the ambiguous characteristics of CDAs from P. tricornutum and T. pseudonana [36].
In this study, a putative CDA (TwCDA) was identified from T. weissflogii, and its sequence structure, evolutionary relationships, and enzymatic activities were integrally characterized. The results verified the role of TwCDA in the degradation of chitin in vitro, which lays a foundation for determining the underlying regulation mechanism of chitin metabolism in diatoms, and they provide a new candidate enzyme for the preparation of chitosan and chitooligosaccharides.
2. Materials and Methods
2.1. Sample Culture
T. weissflogii (9021) was acquired from the Microalgae Collection Center at Ningbo University, Ningbo, China, and grown in an optimized f/2 liquid medium provided by Shanghai Guangyu Biological Technology Co., Ltd. (Shanghai, China) (Table S1). Cells were cultured at 20 °C in 12 h:12 h light:dark diurnal cycles (100 µmol m−2 s−1) with shaking at 100× rpm. Cells at the exponential phase were collected by centrifugation, flash-frozen in liquid nitrogen, and stored at −80 °C for further experiments.
2.2. Substrate Preparation
Thus far, there is no pure beta-chitin standard for sale on the market. β-Chitin was extracted from T. weissflogii by a method optimized from the report by Noishiki et al. [37]. The cells were collected by centrifugation at 2000× g. The supernatant and pellets were decolorized by adding methanol (65 °C, 2 h), deproteinized with 5% KOH (room temperature, overnight), demineralized with 0.34% NaClO2 (70 °C, 6 h) and 0.1 N HCl (boiling, 1 h), and purified with 1% HF (room temperature, overnight). At the end of each step, the sample was centrifuged at 19,000× g to remove the supernatant, and finally it was dehydrated at 80 °C and stored at −80 °C [37]. An α-chitin standard product was purchased from Macklin (Shanghai, China). The crystalline structure of the extracted chitin and α-chitin standard were detected by infrared spectra (Thermo Fisher Scientific, IS50, Waltham, MA, USA). Chitin oligomers (pentamer) were kindly supplied by the laboratory of Prof. Bruno M. Moerschbacher, University of Münster, Germany.
2.3. Cloning and Sequence Analysis of TwCDA
Total RNA was extracted by grinding the T. weissflflogii frozen sample in TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Then, 1.5 mL TRIzol reagent was added to the frozen algal pellet; it was placed at room temperature for 5 min and centrifuged at 10,000× rpm, at 4 °C. The supernatant was added to a new 2 mL tube with 300 μL chloroform and incubated at room temperature for 10 min prior to centrifuging at 10,000× rpm for 5 min. The supernatant was added with 750 μL isopropyl alcohol and centrifuged at 10,000× rpm, 4 °C, for 10 min. Finally, 500 μL 75% ethanol was added and the supernatant was removed by centrifugation at 8500× rpm at 4 °C to obtain RNA. The extracted RNA was reverse-transcribed into cDNA using a reverse transcription kit (SparkJade, Jinan, China), with 200 μL of a reaction mixture of RNA, gDNA eraser, and 2 × SPARK scriptII RT Plus Master Mix. The mixture was incubated in a thermal cycler with the following protocol: 42 °C for 5 min, 50 °C for 30 min, and 80 °C for 5 min. The synthesized cDNA can be directly used for subsequent PCR amplification.
A full-length CDA homologous sequence (CAMPEP_0203574602, TwCDA) was retrieved from the T. weissflogii transcriptome dataset, using a CDA from T. pseudonana (TpCDA) [36] to query against the MMETSP database by BLASTP [38]. The full-length TwCDA sequence was synthesized with codons optimized for E. coli (Sangon Biotech, Shanghai, China). In order to improve the subsequent heterologous expression, we also removed the transmembrane helices to amplify the N-terminal truncated TwCDA-S. TwCDA-S-F (5′-CGGGATCCATGGAAGATACCATGGCAGC-3′) and TwCDA-S-R (5′-CCAAGCTTCTCGAGCAGGATCATAGTCG-3′), containing a BamHI and a HindIII restriction site, respectively, were designed for PCR amplification. A total of 20 μL PCR reaction mixture was used, with 1 μL of each primer, 10 μL of 2 × Phanta Master Mix (Vazyme, Nanjing, China), 2 μL of cDNA template, and 6 μL of ddH2O. The PCR amplification procedure was as follows: 95 °C for 30 s; 35 cycles of 95 °C for 10 s, 55 °C for 5 s, and 72 °C for 10 s; and 72 °C for 1 min. The products were sequenced and analyzed using BLASTN.
2.4. Bioinformatic Analysis of TwCDA
The TwCDA amino acid sequence was aligned with CDA proteins from T. pseudonana and Cyclotella cryptica by CLUSTALW (https://www.genome.jp/tools-bin/clustalw, accessed on 17 September 2022). ProtParam was used to predict the physical and chemical parameters (molecular weight and isoelectric point) of TwCDA [39]. The Motif search tool (http://www.genome.jp/tools/motif/, accessed on 27 February 2022) was applied to analyze the motifs in the Pfam and NCBI-CDD databases. EUK-MPLOC and PSORT were jointly used to predict the TwCDA subcellular localization [40,41], with the SignalP v4.1 server (http://www.cbs.dtu.dk/services/SignalP-4.1/, accessed on 27 February 2022) for signal peptide and signal anchoring sequence prediction [42]. TMHMM Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 27 February 2022) was used to predict transmembrane helices. The secondary structures were illustrated by ESPript [43], and the tertiary structure was predicted by SWISS-MODEL [44]. For phylogenetic analysis, sequences highly homologous to the full-length amino acid sequence of TwCDA were retrieved from three genome resources: MMETSP (https://www.imicrobe.us/#/projects/104, accessed on 10 July 2022), PhycoCosm (https://phycocosm.jgi.doe.gov/phycocosm/home/, accessed on 10 July 2022), and PLAZA (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_diatoms_01/, accessed on 10 July 2022). Homology alignment was conducted using BLASTP. MEGA X was used to construct the neighbor-joining tree based on the Jones–Taylor–Thornton (JTT) model and the Gamma-distributed matrix-based model with 1000 bootstrap replicates [45].
2.5. Expression of Recombinant TwCDA Proteins
The pET system was used to express TwCDA and TwCDA-S in vitro. The codon-optimized full-length sequence of TwCDA was synthesized into pET-32a vectors with a His-tag (pET-TwCDA, Sangon Biotech, Shanghai, China). At the same time, TwCDA-S with a reduced transmembrane helix structure, which was amplified as an BamHI/HindIII fragment by PCR, was cloned into the pET-32a vectors with His-tag, resulting in the fusion plasmid pET-TwCDA-S.
The E. coli cells (BL21 (DE3) pLysS and Transetta (DE3)) were transformed with the recombinant expression plasmids (pET-TwCDA and pET-TwCDA-S). The E. coli cells were then incubated in 1 L of Luria–Bertani (LB) medium with 100 µg/mL of ampicillin and 20 µg/mL of chloramphenicol at 37 °C. When OD600 reached 0.6, 0.1 mM IPTG was supplemented, and the induction conditions were changed to 120× rpm and 15 °C. After 24 h of incubation, the transformed cells were harvested by centrifugation at 4500× rpm at 4 °C for 30 min. Then, the cells were disrupted by sonication for 25 min (3 s on and 5 s off cycle) (Xinzhi, Ningbo, China). One EDTA-free protease inhibitor tablet (Roche, Basel, Switzerland) was added to the 35 mL extracts prior to the centrifugation at 12,000× rpm at 4 °C for 45 min. The supernatant was then filtered through a 0.45 μm filter membrane to obtain a crude enzyme solution. The crude protein concentration was measured using a BCA protein quantification kit (Vazyme, Nanjing, China).
2.6. SDS-PAGE and Western Blotting
The protein concentration in crude protein was determined with a BCA kit (Solarbio, Beijing, China) according to the manufacturer’s instructions. We then boiled the samples for 10 min in loading dye (125 mM Tris/HCl, 10% SDS, 0.25% BPB, 10% 2-mercaptoethanol, 50% glycerol), and loaded 60 μg of the crude extract onto 12% acrylamide gels for separation by SDS-PAGE. Primary (anti his-tag mouse monoclonal antibody at a concentration of 1:500) and secondary (goat anti-mouse IgG, HRP-conjugated at a concentration of 1:2000) antibodies were used for the Western blotting test via the iBind™Flex Western System (Thermo Fisher Scientific, Waltham, MA, USA) at room temperature for 4 h, following the manufacturer’s protocol. After washing with ddH2O solution three times, CDA target bands were visualized by incubating the membranes in ECL reagents with the ChemiDoc XRS Imaging System and the Image Lab 6.0 software (Bio-Rad Lab, Hercules, CA, USA).
2.7. Detection of Chitin Deacetylase Activity
Crude protein extracts of TwCDA and TwCDA-S were prepared with a phosphate buffer containing 20 mM sodium phosphate pH 7.5, 20 mM imidazole, 500 mM sodium chloride, 5% glycerol, and a protease inhibitor. The reaction volume was set to 10 mL, containing 50 mM triethanolamine (TEA) pH 7.0, 2 mg/mL substrate, and 1 mL (c. 3 mg) crude proteins. The reaction was carried out at 250× g, 37 °C for 48 h, and terminated by freezing the mixtures in liquid nitrogen [36]. The inactivated crude enzyme was used as the control group. The substrate specificity was tested towards chitin pentamers (GlcNAc)5 and β-chitin. Each experiment was performed with two replicates. Deacetylation activity was determined by measuring the released acetic acid via high-performance liquid chromatography (HPLC) with an acetic acid content detection kit (Solarbio, Beijing, China). The experimental conditions were as follows: an Ultimate AQ-C18 (Yuexu Technology, Shanghai, China) HPLC column (5 µm, 250 × 4.6 mm) was used for detection at 30 °C with a 210 nm UV filter, and the flow rate was set to 0.4 mL/min. Each acetic acid assay was performed with three replicates. A standard curve generated from 175, 87.5, 17.5, 8.75, 1.75, and 0.875 μmol/mL acetic acid was used to calculate the acetic acid concentration in the sample, using Equation (1). Gas chromatography-mass spectrometry (GC-MS) was used to further detect whether acetic acid was produced in the product.
y = 59506x + 72815 (R2 = 0.9994)
2.8. Detection of Chitinolytic Activity
Some chitin deacetylases also have the ability to degrade chitin sugar chains [46]. The dinitrosalicylic acid (DNS) method was used to detect the presence of reducing sugar in the reaction products to verify whether TwCDA has chitinolytic activity. β-Chitin and (GlcNAc)5 were used as substrates. The inactivated crude enzyme was used as the control group, and the denatured protein precipitate was removed by centrifugation. The conditions of the enzymatic reaction were as follows: 50 μL of crude enzyme was added to 50 μL of substrate and it was incubated at 37 °C for 60 min. Subsequently, 75 μL of DNS reagent was added for the chromogenic reaction, and the mixture was boiled at 98 °C for 5 min to terminate the reaction. OD530 was detected in a 96-well plate using a Biotek EON microplate reader (Biotek, Santa Clara, CA, USA). To obtain the optimal reaction conditions for the TwCDA, we measured the enzyme activity at various temperatures (20, 25, 30, 35, 40, and 45 °C). Each experiment was performed in triplicate, and a statistical analysis was conducted with SPSS 26.0 (SPSS Inc., Chicago, IL, USA). The chemical formula prediction of the reaction products was detected by liquid chromatography combined with time-of-flight mass spectrometry (LC-coTOF-MS), with the inactivated crude enzyme group and blank group (i.e., (GlcNAc)5 aqueous solution) as the control groups.
2.9. Statistical Analysis
The SPSS version 26 software was used to analyze the experimental data statistically, and one-way analysis of variance was used for the data. The results were expressed as mean ± standard deviation (SD). p < 0.05 was considered a significant difference; p < 0.01 was considered an extremely significant difference.
3. Results
3.1. Extraction and Analysis of Chitin from T. weissflogii
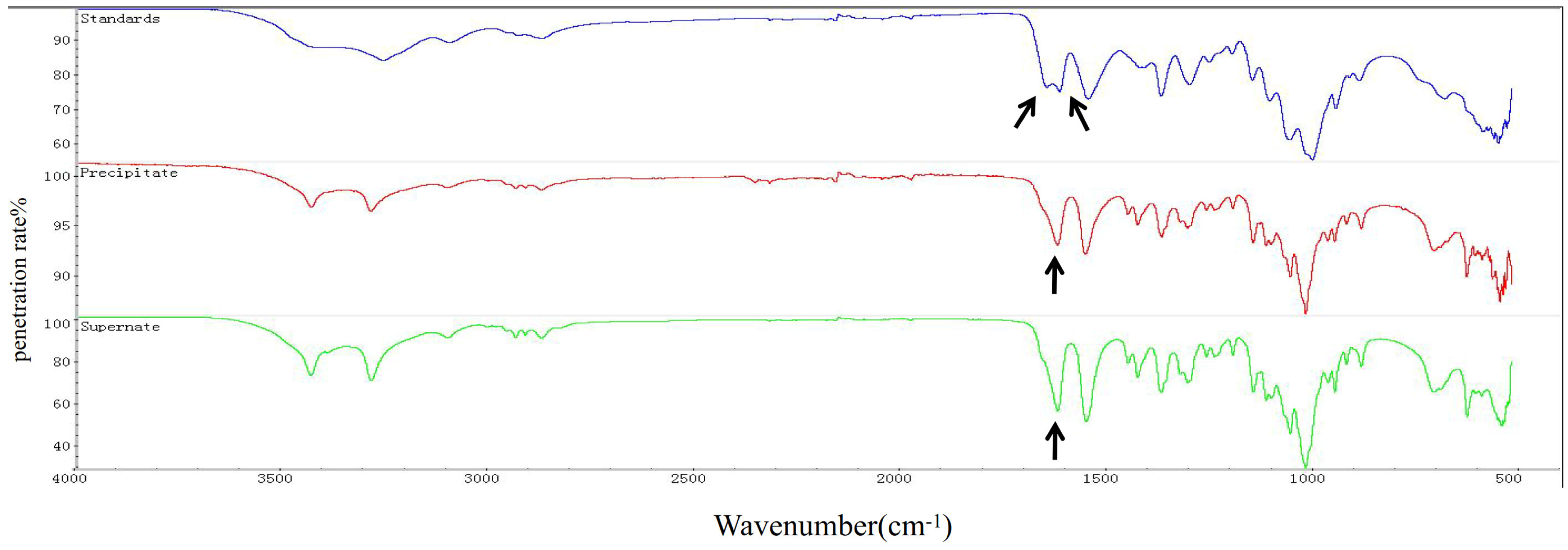
An approximately 600 mL culture of T. weissflogii was used to extract chitin. The content of chitin obtained from the supernatant and cell pellets was 0.047 g and 0.0762 g, respectively. The total yield of chitin was calculated to be 20.53% of the total cell mass (fresh weight). Infrared spectrum detection showed that the chitin extracted from T. weissflogii had a single peak at 1625 cm−1, while α-chitin standard forms a double peak at 1620 cm−1 and 1652 cm−1 (Figure 1). The antiparallel sugar chains in α-chitin make the C=O bond stretch and vibrate, forming a hydrogen bond between C=O and the hydroxyl group and another hydrogen bond between C=O and N–H, whereas β-chitin can only form the former because of the parallel sugar segments [47]. Therefore, it was verified that the crystallographic structure of chitin produced by T. weissflogii is β-chitin.
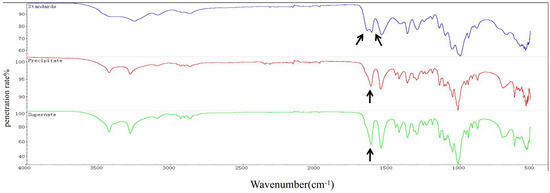
Figure 1.
FTIR spectra of chitin (0–4000 cm−1). α-Chitin standard forms a double peak at 1620 cm−1 and 1652 cm−1 (the blue curve); the chitin extracted from the precipitation of T. weissflogii had a single peak at 1625 cm−1 (the red curve); the chitin extracted from the supernatant of T. weissflogii had a single peak at 1625 cm−1 (the green curve). The arrow indicates the location of the characteristic peaks.
3.2. Sequence Analysis of TwCDA
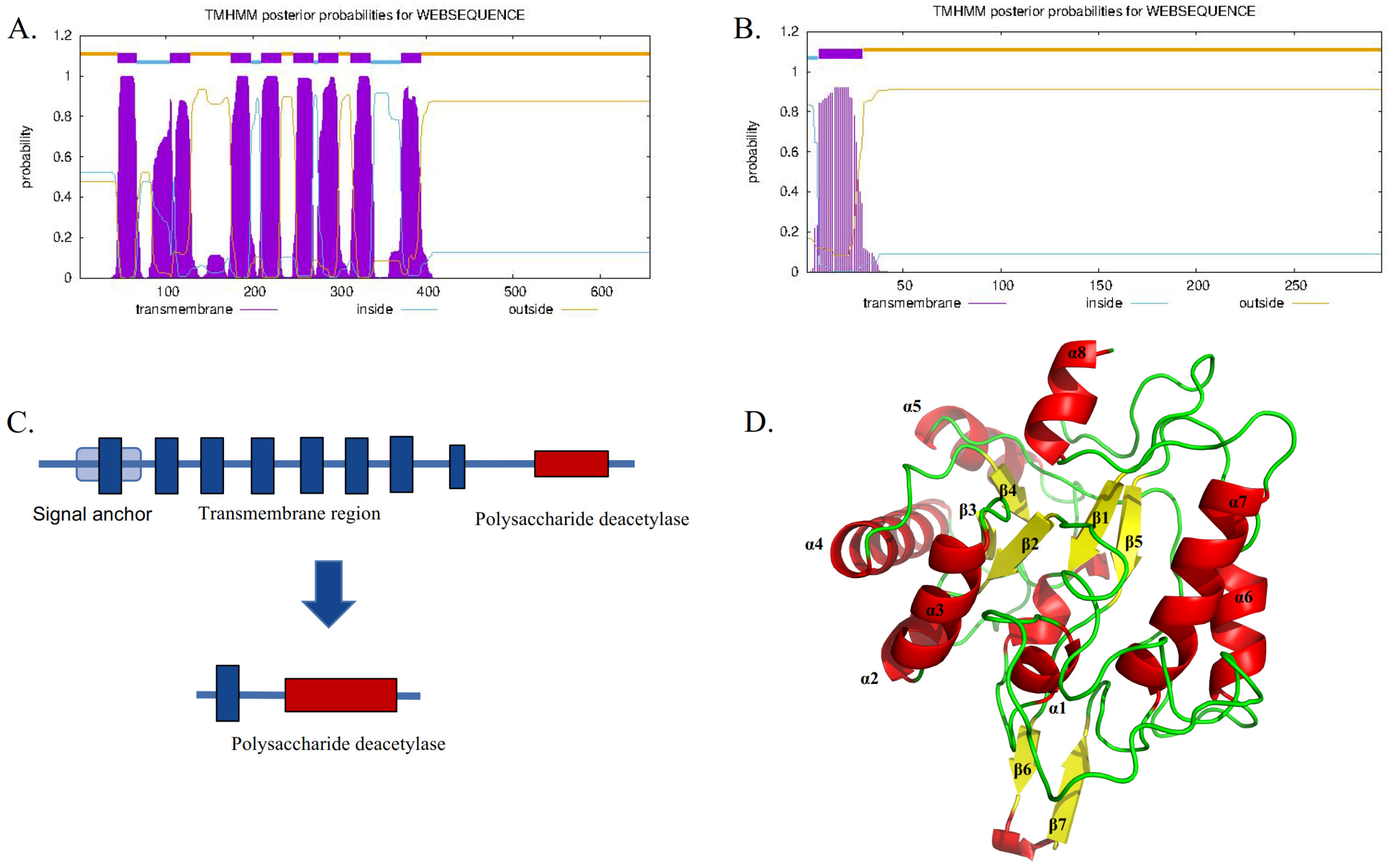
TwCDA-S is the N-terminal truncated sequence of TwCDA with only one transmembrane helical structure but retaining the complete polysaccharide deacetylation domain (Figure 2). The sequence features of TwCDA and TwCDA-S are summarized in Table 1. TwCDA (1974 bp) encoded 658 amino acids, which had a predicted molecular weight (MW) of 71.15 kDa and an isoelectric point (pI) of 6.21. TwCDA-S (885 bp) encoded 295 amino acids, with a predicted MW of 31.89 kDa and a pI of 5.11. TwCDA has a signal anchor, whereas TwCDA-S does not. Secondary structure prediction showed that both CDAs had a similar composition, with a predominance of the α-helix structure (~45%). The TwCDA tertiary structure reveals a single domain (residues 403–651) adopting a fold reminiscent of an (α/β)8 topology (Figure 2D), characteristic of the NodB homology domain present in all CE4 esterases [48].
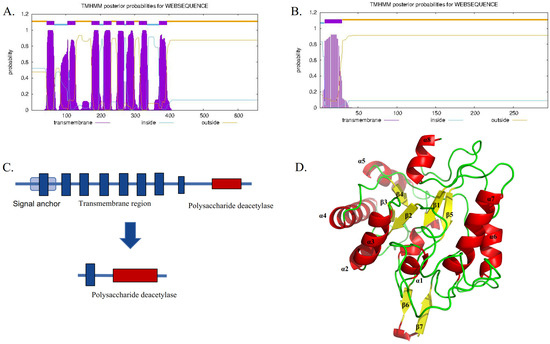
Figure 2.
Sequence comparison between TwCDA and TwCDA-S. (A) TwCDA has 8 transmembrane helices at amino acids 44 to 393 and a signal anchor. (B) TwCDA-S has 1 transmembrane helix at amino acids 7 to 29. (C) Both TwCDA and TwCDA-S have a complete polysaccharide deacetylation motif at their C-terminal. (D) TwCDA’s tertiary structure. Stereo images of TwCDA and the canonical TIM barre (α/β)8 fold as observed in Tiny-TIM.

Table 1.
Comparisons of TwCDA and TwCDA-S in terms of gene/protein structure, subcellular localization, and activities. CDS, coding sequences; ER, endoplasmic reticulum.
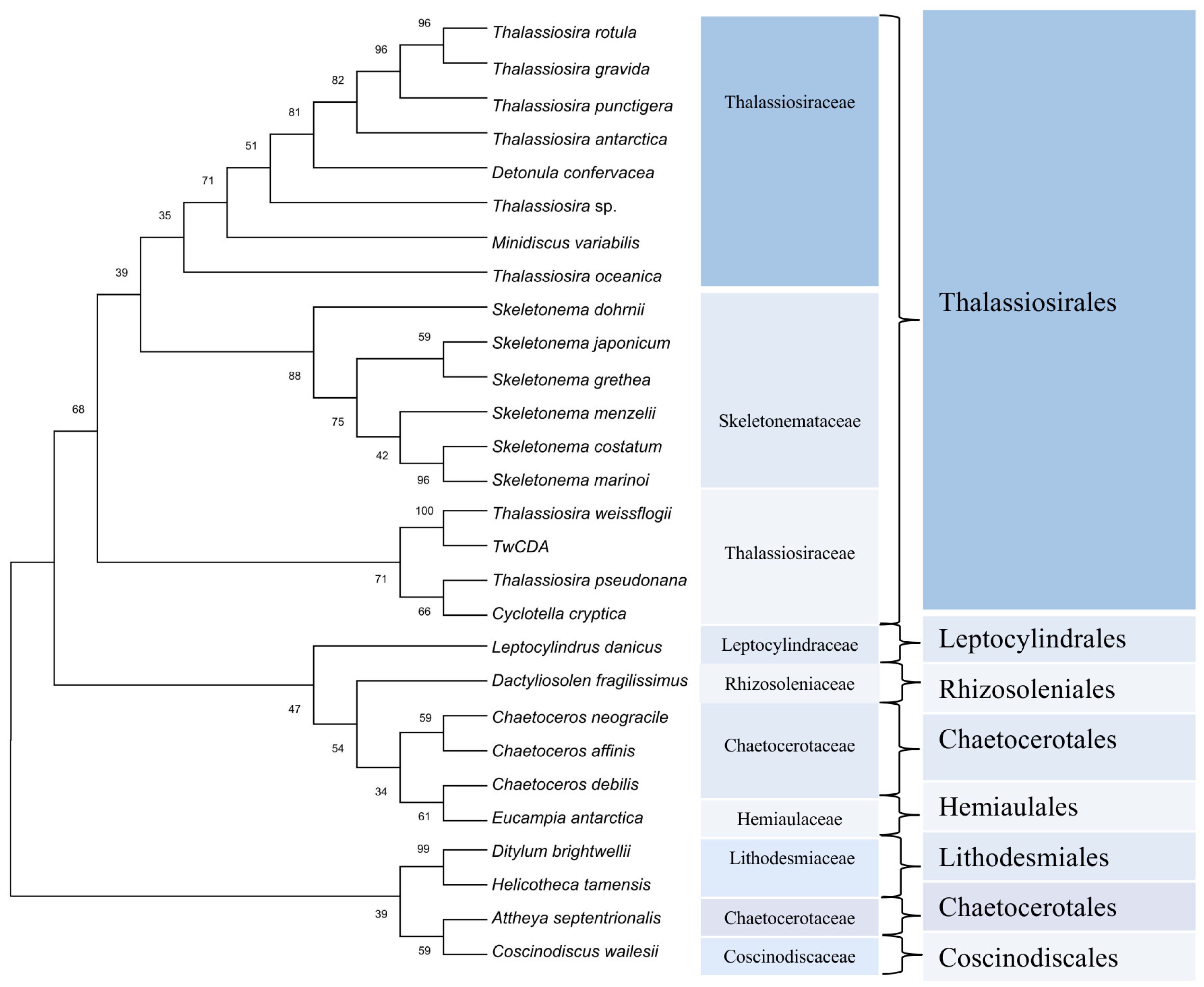
For the phylogenetic analysis, 93, 37, and 6 diatom sequences highly homologous to the full-length amino acid sequence of TwCDA were retrieved from the three available genome resources of the MMETSP [38], PhycoCosm [49], and PLAZA databases, respectively (Tables S2–S4). These sequences were highly enriched in Thalassiosirales (74.3%). In the 101 Thalassiosirales sequences, 81 (80.2%) were from Thalassiosiraceae and 17 (16.8%) from Skeletonemataceae. We arranged these 136 sequences according to sequence identity with TwCDA after BLASTP homologous alignment, and selected 28 CDA amino acid sequences with the highest similarity from each homologous species to illustrate the evolutionary history (Figure 3). The CDA sequences of these 28 CDAs are listed in Tables S5–S7. Among them, all the species of Thalassiosirales were clustered into one clade, and the CDAs from the genus Skeletonema were the most conserved, forming an independent group. Members of the genus Thalassiosira were distributed across different lineages within the Thalassiosirales. The CDA of genus Chaetoceros is more closely related to Eucampia, and Leptocylindrus and Dactyliosolen form individual branches. Figure 3 also shows that TwCDA was in the same node as the CDA from diatom T. pseudonana (NCBI Reference Sequence: XP_002293781.1) and C. cryptica (PLAZA Reference Sequence: CC00G25130). Multiple-sequence alignments indicated that TwCDA has 95% and 96% query coverage and 38.61% and 39.31% identity with T. pseudonana CDA and C. cryptica CDA, respectively (Figure S1). We also retrieved two CDA genes from a pennate diatom Synedra acus, with the amino acid sequences listed in Table S7.
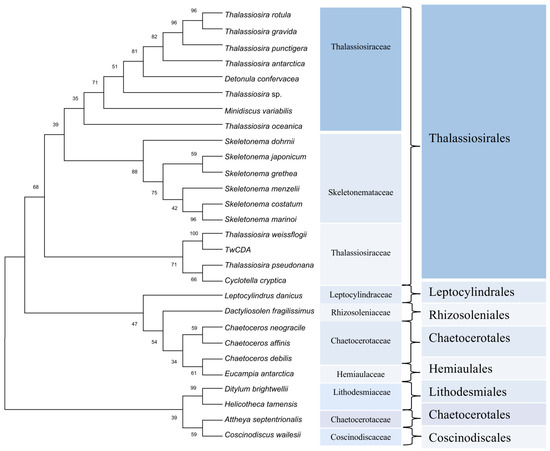
Figure 3.
Phylogenetic tree constructed based on 28 putative chitin deacetylase amino acid sequences in diatoms. Proteins are from the MMETSP, PhycoCosm, and PLAZA databases. The tree was constructed using the neighbor-joining algorithm with 1000 bootstrap replicates. The number at each branch node represents a bootstrap value.
3.3. Heterologous Expression and Deacetyl Activity of TwCDAs
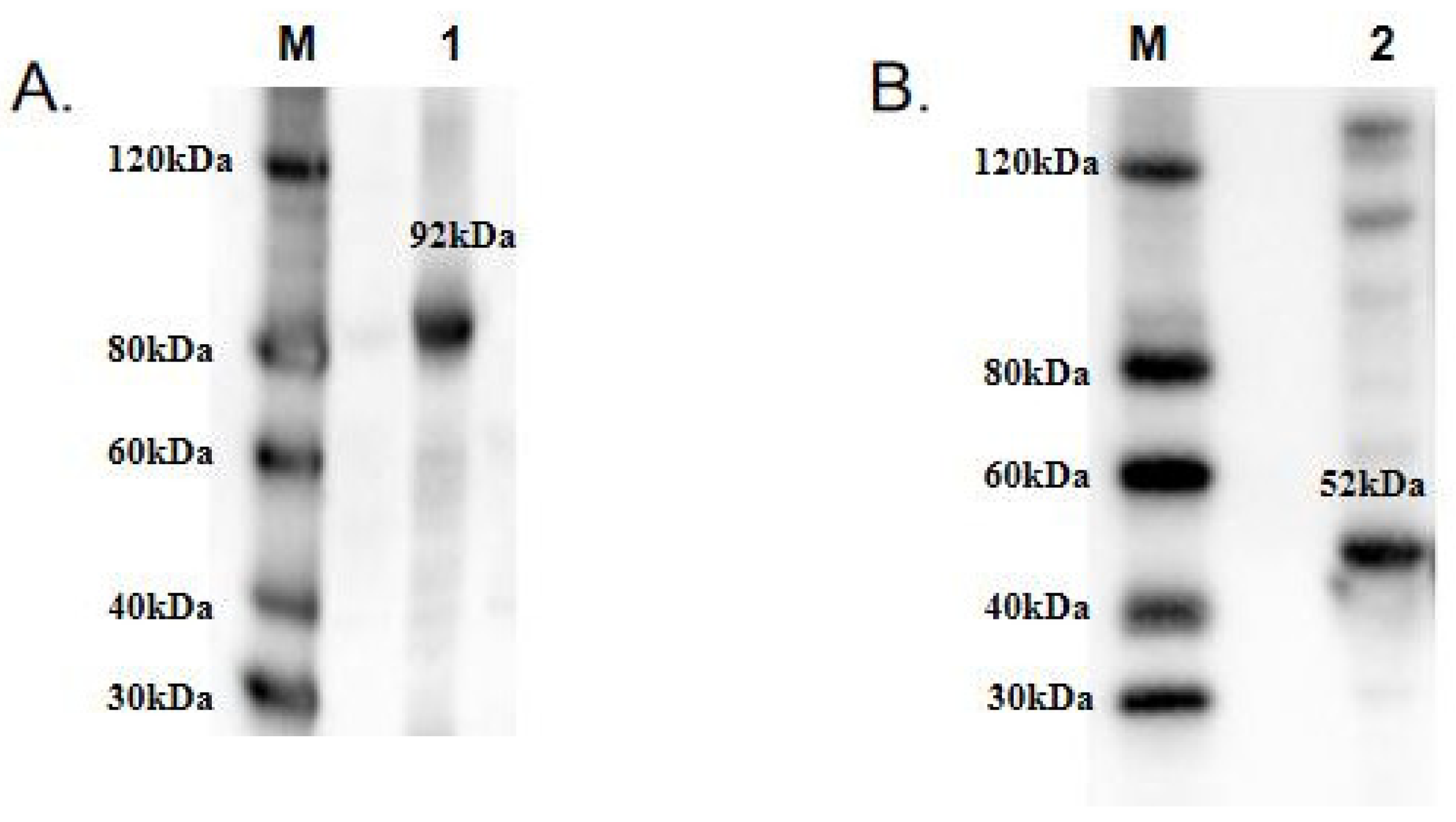
To functionally characterize TwCDAs and compare their enzyme characteristics, we induced large amounts of CDA expression. The protein concentrations of TwCDA and TwCDA-S were 3.1171 mg/mL and 3.374 mg/mL, respectively. The MWs of TwCDA and TwCDA-S fusion proteins were approximately 92 kDa and 52 kDa, respectively. The Western blot analysis confirmed the presence of proteins with the expected sizes (Figure 4 and Figure S2).
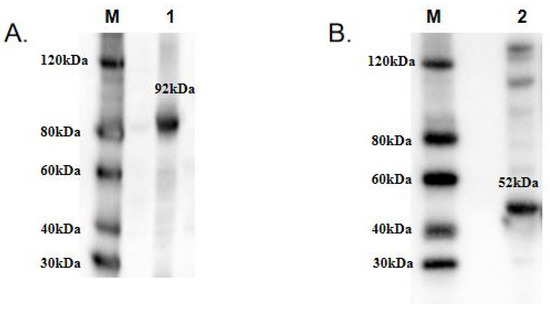
Figure 4.
Western blot analysis of recombinant (A) TwCDA; (B) TwCDA-S. M: protein ladder; 1–2: TwCDA and TwCDA-S fusion proteins. To improve the clarity and conciseness of the presentation, these two gels have been cropped. Original Western blots are shown in Figure S3 (Supplementary Materials).
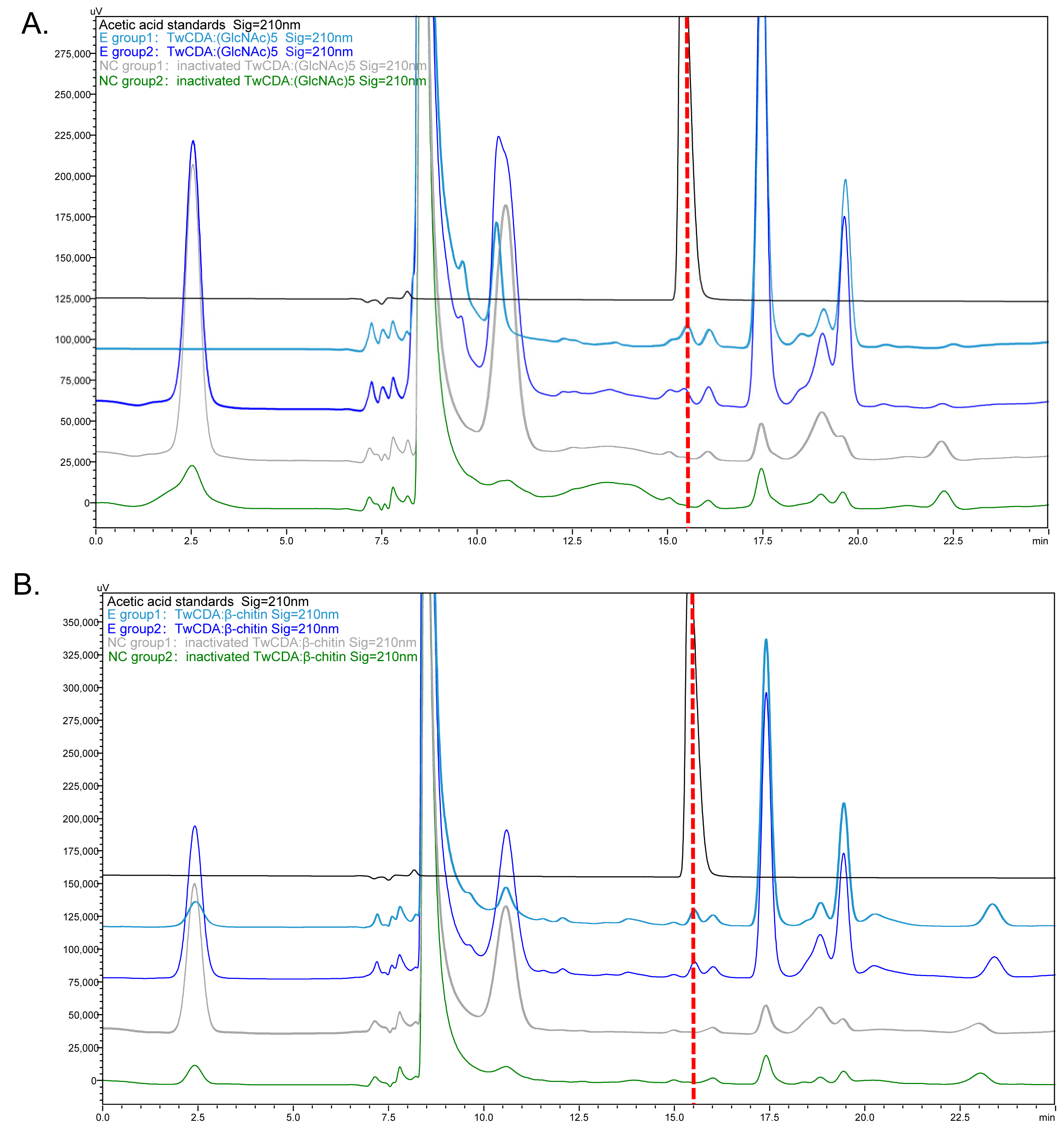
The deacetylation activities of TwCDA and TwCDA-S on (GlcNAc)5 and β-chitin were detected by HPLC (Figure 5 and Figure S3). The concentration of acetic acid produced in the reaction mixtures was calculated (Figure S3C). The HPLC analysis showed that TwCDA could catalyze the deacetylation of both (GlcNAc)5 (Figure 5A) and β-chitin (Figure 5B), and the catalyzation efficiency on the pentamer was 1.20 times higher than that on the polymer (Figure S3C). In contrast, TwCDA-S could not catalyze β-chitin deacetylation (Figure S3A,B), and the deacetylase activity against the oligosaccharide of (GlcNAc)5 was only 62.89% of that of TwCDA (Figure S3C). To summarize, TwCDA had higher deacetylase activity than TwCDA-S, and the activity was stronger when (GlcNAc)5 was used as a substrate. To further confirm whether acetic acid was produced in the reaction mixture, the products of TwCDA against the (GlcNAc)5 group were detected by GC-MS, and it was verified from the molecular weight of the characteristic peak that acetic acid (C2H4O2, MW = 60.02) was indeed produced in the mixture (Figure S4).
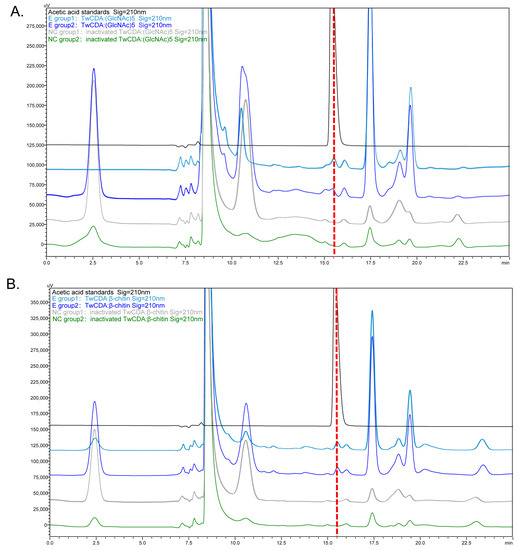
Figure 5.
HPLC chromatograms presenting the deacetylation activity of TwCDA. (A) Chromatogram of TwCDA with (GlcNAc)5 as substrate. (B) Chromatogram of TwCDA with β-chitin as substrate. The retention time of acetic acid standard was 15.389 min (black curves). Acetic acid was produced in both experimental groups (dark blue and light blue curves) but not in the control groups (gray and green curves).
3.4. Chitinolytic Activity of TwCDA
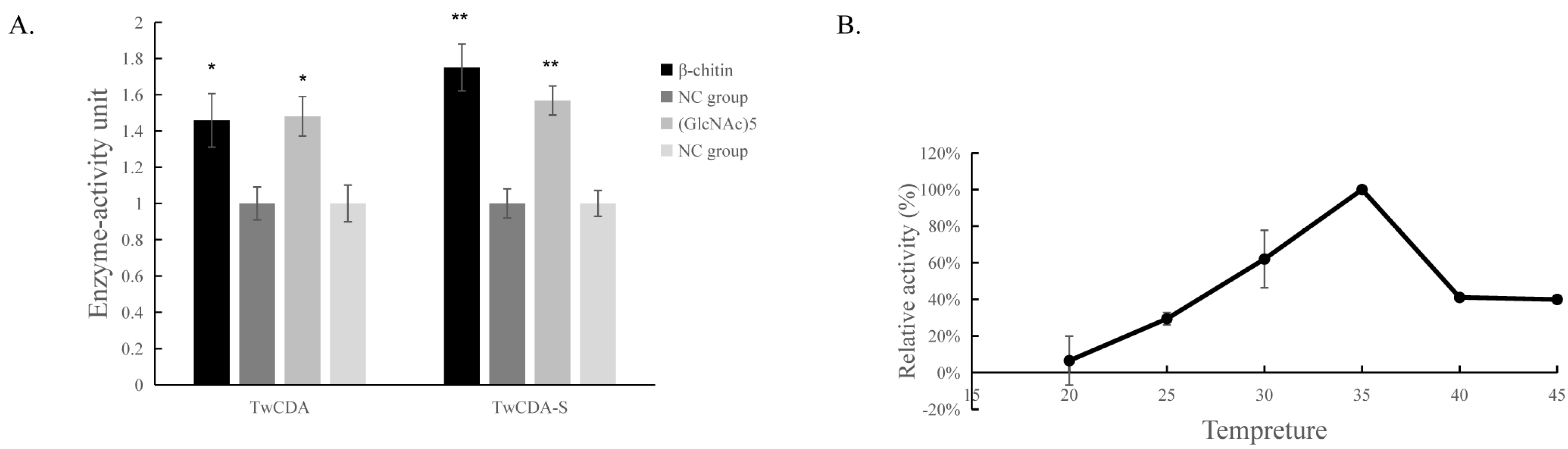
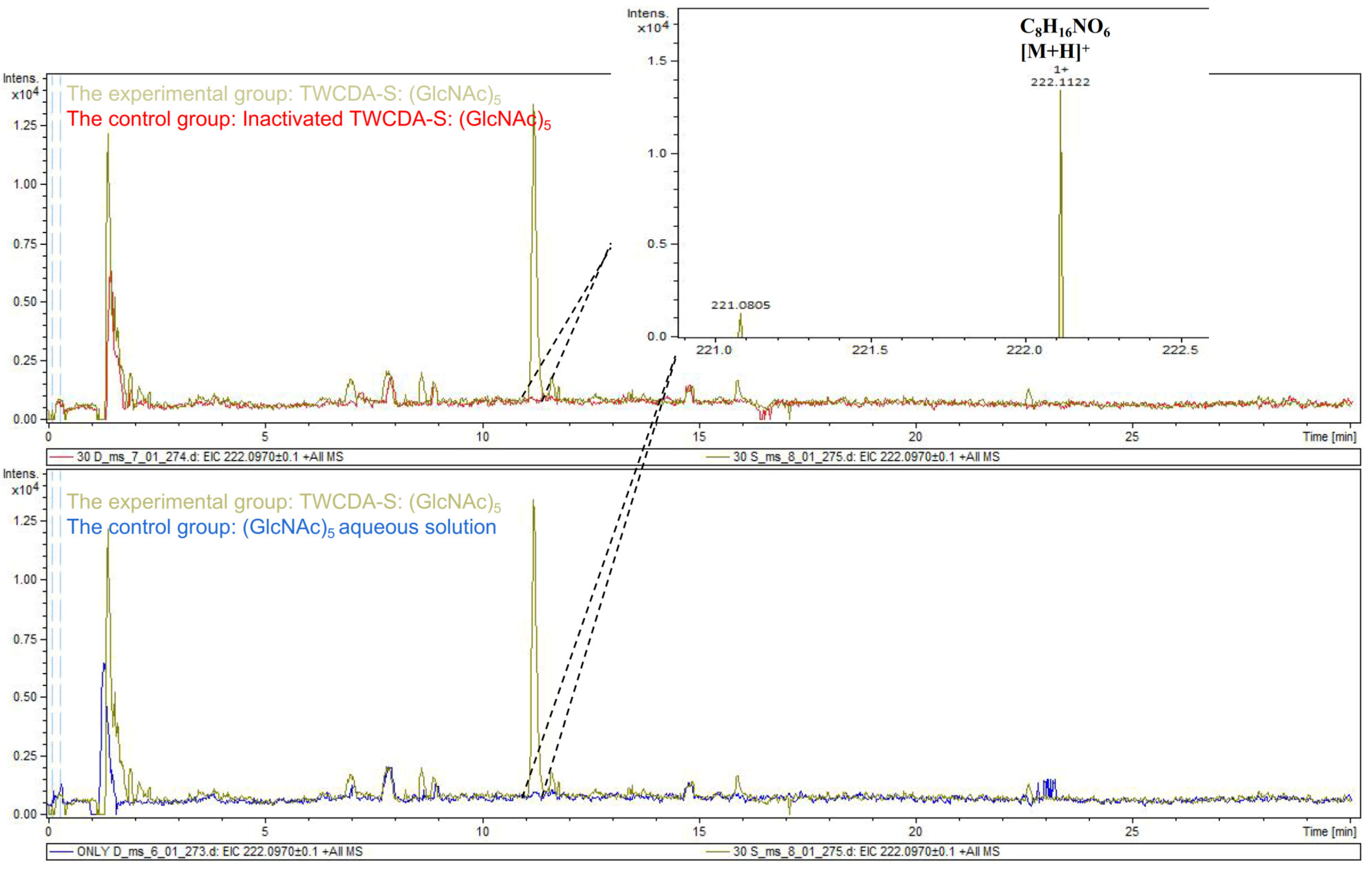
The DNS assay showed that reducing sugars were produced in the products of the four experimental groups (TwCDA and TwCDA-S against pentamer and β-chitin). Taking the amount of enzyme required to produce 0.1 μmol NAG in 1 h as the unit of enzyme activity, we drew the specific activity bar chart with the number of units of enzyme activity per milligram of enzyme (Figure S5). We compared the difference in OD530 between the experimental group and the control group. The results indicated that regardless of whether β-chitin or (GlcNAc)5 was used as the substrate, TwCDA showed significantly higher chitinolytic activity than the control groups, with activity 1.48-fold (against pentamer) and 1.46-fold (against β-chitin) higher than the control groups (p < 0.05), respectively (Figure 6A). Compared with TwCDA, TwCDA-S presented higher chitinolytic activity on both (GlcNAc)5 and β-chitin, showing 1.57-fold and 1.75-fold higher activity than the control groups (p < 0.01), respectively (Figure 6A). This indicates that, without the transmembrane helix structure, TwCDA-S had higher deglycosylation activity. We then detected its chitinolytic activity at different temperatures with (GlcNAc)5 as the substrate, and found that the optimal reaction temperature was 35 °C (Figure 6B). It was found that TwCDA-S had the highest chitinolytic activity when (GlcNAc)5 was used as the substrate. All of the source data of the enzyme activity under different substrates and different temperatures are shown in Tables S8–S11. The molecular weight of the reaction product was analyzed by LC-coTOF-MS, with the inactivated TwCDA-S and (GlcNAc)5 solution as a control. A clear characteristic peak appeared at approximately 11 min in the experimental group, with a predicted molecular weight of 222.1122, which is in line with the MW of chitin monomer N-acetylglucosamine (C8H15NO6) (Figure 7).
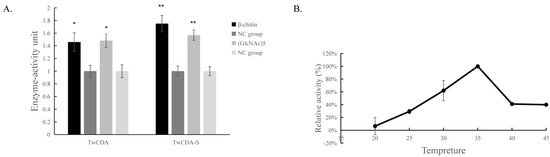
Figure 6.
Chitinolytic activity of TwCDAs. (A) Enzymatic activities of TwCDA and TwCDA-S with β-chitin as substrate (black bar); enzymatic activities of TwCDA and TwCDA-S with (GlcNAc)5 as substrate (gray bar); control group (dark graybar and light gray bar). *, p < 0.05; **, p < 0.01. (B) Influence of temperature (20–45 °C) on the activities of TwCDA-S with (GlcNAc)5 as substrate. The error bars are the mean ± SD of three technical replicates for each sample.
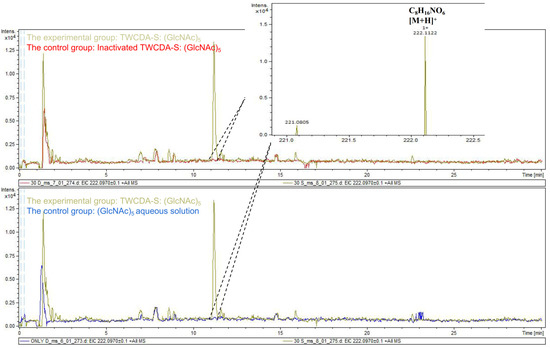
Figure 7.
LC-coTOF-MS chromatograms. TwCDA-S and (GlcNAc)5 were taken as the experimental group (green curves), and inactivated TwCDA-S (above panel) and (GlcNAc)5 aqueous solution (below panel) were taken as the control group, respectively. The characteristic peak appeared at around 11 min in the experimental group, with the predicted molecular weight of 222.1122, which is in line with the MW of chitin monomer N-acetylglucosamine. Note the molecular ion species formed: [M + H]+.
4. Discussion
Diatoms represent an important source of β-chitin, which has been reported to be a better material for application compared with α-chitin [50]. β-Chitin displays higher reactivity for deacetylation and chemical modification due to its weak intermolecular interactions [7]. Although the biosynthesis and degradation pathway of chitin has been extensively investigated, especially in fungi and insects, there are few reports on chitin-relevant enzymes from β-chitin producers [36]. Chitosan and chitooligosaccharide are two of the most important derivatives and have great potential in the biomedical field. In their in vitro synthesis, chitin deacetylase (CDA) plays a crucial role in the enzymatic deacetylation of chitin polymers. At present, the research on the CDA mainly focuses on enzyme function analysis and crystal structure establishment [33,51]. In fungi and insects, CDAs may play a role in morphogenesis, whereas bacterial CDAs may contribute to chitin degradation for nutritional purposes [28,32]. In this study, we isolated a CDA gene from T. weissflogii, a high producer of β-chitin, and explored its structural and enzymatic properties.
Most CDAs have a conserved (α/β)8 barrel fold with six or seven strands and encompass a mononuclear metalloenzyme that employs a conserved His–His–Asp zinc-binding triad to carry out acid/base catalysis [33,51,52]. In this study, TwCDA had a characteristic of the NodB homology domain, adopting a fold reminiscent of an (α/β)8 topology, which is consistent with CDA structure studies. The full-length sequence contains 658 amino acids, but there are 403 amino acid residues in its N-terminal region, whose tertiary structure is not predicted, denoting the unstructured nature of this region [51]. The active site of the His–His–Asp zinc-binding triad was not found in TwCDA. Although Strunk et al. [53] reported that the conserved Asp–His–His triad was indispensable for a functional polysaccharide deacetylase, CDAs from diatoms seem to be an exception. This triad also disappeared in TpCDA (JGI Reference Sequence: Thaps3_J24880), which has been verified to catalyze the deacetylation of (GlcNAc)5 and the partially acetylated chitosan polymer [36]. Together with the detection of the deacetylase activity of the TwCDA in this study, we presumed that acid/base catalytic sites might be diversified and HHD is not conservative and indispensable in diatoms.
The putative CDA sequences from the MMETSP, PhycoSM, and PLAZA databases show that the CDAs are dominated by sequences from marine centric diatoms and the Thalassiosirales contain the most abundant CDA homologues. Sequence homology alignment found that CDAs were retrieved from two independent divisions: centric diatoms and pennate diatoms. In pennate diatoms, we first reported the presence of CDA homologous sequences in Synedra acus, which expanded the taxonomic distribution of CDA in diatoms and provided new sources of novel CDA enzymes. A phylogenetic tree found that TwCDA was closely related to T. pseudonana CDA and C. cryptica CDA. The evolutionary position of C. cryptica is adjacent to T. pseudonana. This was consistent with the finding that T. pseudonana likely descended from a freshwater ancestor in Cyclotella [54].
TwCDA was predicted to have a structure of eight transmembrane helices. The prediction of subcellular localization showed that TwCDA was potentially located in the endoplasmic reticulum. This was in accordance with the results of Shao et al. [36], who reported that the subcellular localization of T. pseudonana CDA–GFP fusion proteins is in the ER. These transmembrane helix structures may cause difficulties for our protein expression in vitro. However, producing a prokaryotic membrane protein in the simple, flexible, and inexpensive E. coli system has already proven successful in many cases [55,56,57]. Unfortunately, the yield and quality of membrane proteins, especially eukaryotic ones, are often insufficient for structural and functional studies in E. coli [58]. To address this issue, Wagner et al. [59] found that membrane protein production can be increased in E. coli through the rational design engineering of strains or by modifying target proteins. In addition, Brosson et al. [60] studied a CDA with transmembrane helices in the Encephalitozoon cuniculi, produced a recombinant form of the EcCDA protein that was slightly truncated to exclude transmembrane helices, and obtained a high yield of purified protein. Therefore, we removed as many transmembrane helices as possible on the basis of preserving the complete functional domain and expressed it in E. coli. We simultaneously expressed TwCDA with the full length of ORF and TwCDA-S with the transmembrane structure removed, and carried out enzyme activity detection. It was found that TwCDA-S had a higher expression level and higher chitinolytic activity. Although TwCDA retains its transmembrane helical structure, its expression level is acceptable and it has higher deacetylase activity.
We found that TwCDA-S had optimum chitinolytic enzyme activity at 35 °C. At present, the optimal conditions for the enzyme activity of chitinase have been studied in animals, plants, bacteria, and fungi. According to the previous results, the optimal temperature is 25–65 °C [61,62,63]. The optimum temperature of 35 °C measured in our study is within this range. This is consistent with the optimum temperature of CDAs from Francisella tularensis (35 °C) and Coprinellus congregatus (35 °C) [64,65].
Previous results have shown that most CDAs had high activity on chitooligosaccharides, water-soluble chitosan, and colloidal chitin, but low or no activity toward chitin polymer [30,66]. Research on chitin deacetylase has mostly used chitin oligosaccharide or chitosan as a substrate, but rarely chitin. Even in functional CDA studies in diatoms, diatom CDAs only had enzyme activities on partially deacetylated chitosan of DA20 and DA60, but there is no report of chitin polymers [36]. Chitin polymers are difficult to catalyze by CDA, due to the stable structure and insolubility. For example, Cord-Landwehr et al. [67] explored the activity of PesCDA from Pestalotiopsis sp., and revealed that this enzyme is inactive against insoluble α-chitin and β-chitin, slightly active against colloidal chitin, and active against soluble chitosan polymers. In our study, HPLC results revealed that TwCDA could catalyze both (GlcNAc)5 and β-chitin, but TwCDA-S can only catalyze (GlcNAc)5. We speculate that the N-terminal TMH or signal-anchoring sequence is essential for the catalysis of the polysaccharide. This speculation was supported by the previous report that a signal anchor is important for fungal CDAs to function efficiently at the cell surface [68]. The effective deacetylation of TwCDA on crystalline chitin substrates provides a new enzyme resource for producing chitin derivatives in vitro.
Most of the reports on CDAs are about its deacetylation activity; there have been few reports on its chitinolytic activity [46]. Chitinase activity can be divided into two types: exochitinase activity and endochitinase activity. Endochitinase activity randomly cleaves chitin at internal sites, whereas exochitinase acts at the end point of chitin oligosaccharides to liberate (GlcNAc)2 or GlcNAc [69]. Thus, the finding of a chitinolytic enzyme with exochitinase activity could be promising in regard to its potential for the production of GlcNAc by the enzymatic method [70]. Since we detected the presence of GlcNAc monosaccharide in the reaction mixture using LC-coTOF-MS, we speculated that TwCDA might display exochitinase activity. This makes TwCDA a more important candidate enzyme to be co-utilized to produce chitooligosaccharide, the most promising chitin derivative. However, glucosamine was not detected, presumably because TwCDA has no deacetylation activity on monosaccharide, which is consistent with previous studies showing that most CDAs have no deacetylation activity against GlcNAc monosaccharide [33,67,71]. However, our enzyme activity reaction was carried out for the crude enzyme solution, not the purified proteins. We need to further analyze the enzyme kinetic parameters on the basis of pure TwCDA for a deeper understanding of TwCDA.
In summary, a putative TwCDA gene was identified and characterized, and the catalytic products of TwCDA recombinant protein were explored. To our knowledge, this is the first analysis of chitin deacetylase with both chitin deacetylase and chitinase activities from diatoms, and the first report of a CDA that could catalyze β-chitin polymer. Although we used a crude TwCDA preparation instead of a pure enzyme in this study, it is worth mentioning that crude enzymes are often used in industry [72]. Therefore, TwCDA could be used to prepare chitosan in combination with partial chemical treatment or pretreated chitin, which needs to be carried out industrially in the near future.
5. Conclusions
In this study, a chitin deacetylase (TwCDA) from Thalassiosira weissflogii was heterologously expressed in vitro for functional analysis. The results showed that both the full-length sequence (TwCDA) and the N-terminal truncated sequence (TwCDA-S) had chitin deacetylase and chitinolytic activities after expression in Escherichia coli. TwCDA and TwCDA-S could catalyze the deacetylation of oligosaccharide (GlcNAc)5. TwCDA had higher deacetylase activity, and also catalyzed the deacetylation of the β-chitin polymer. TwCDA-S had high chitinolytic activity for (GlcNAc)5, and the optimal reaction temperature was 35 °C. Altogether, we isolated a chitin deacetylase from a marine diatom, which can catalyze the deacetylation and degradation of chitin and chitin oligosaccharides. The relevant results lay a foundation for the internal regulation mechanism of chitin metabolism in diatoms, and provide a candidate enzyme for the green industrial preparation of chitosan and chitin oligosaccharides. However, our enzyme activity reaction was carried out on the basis of a crude enzyme solution, and a sufficient quantity of active recombinant enzyme was not obtained after purification. We need to find solutions regarding how to retain the enzymatic activity during the purification process and further analyze the enzyme kinetic parameters for a deeper understanding of TwCDA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo13030429/s1, Table S1. Nutrient and metal concentration in optimized f/2 liquid medium. Table S2. Diatom sequences with high homology to the full-length amino acid sequences of TwCDA retrieved from PhycoCosm. Table S3. Diatom sequences with high homology to the full-length amino acid sequences of TwCDA retrieved from MMETSP. Table S4. Diatom sequences with high homology to the full-length amino acid sequences of TwCDA retrieved from PLAZA. Table S5. Diatom TwCDA homologous sequences retrieved in PhycoCosm Bacillariophyta database. Table S6. Diatom TwCDA homologous sequences retrieved in MMETSP database. Table S7. Diatom TwCDA homologous sequences retrieved in PLAZA diatoms 1.0 database. Table S8. Standard curves of different concentrations of N-acetylglucosamine and absorption value. Table S9. The source data of enzyme activity under different substrates. Table S10. The source data of enzyme activity under different temperatures. Table S11. The source data of SPSS of enzyme activity under different substrates. Figure S1. Multiple sequence alignment of TwCDA with T. pseudonana CDA (A) and C. cryptica CDA (B), respectively. Figure S2. Original Western blots of recombinant proteins. Figure S3. HPLC chromatograms and acetic acid standard curve. Figure S4. GC-MS of experimental group (above figure) and control group (below figure). Figure S5. Chitinolytic specific activity of TwCDAs. Enzymatic specific activities of TwCDA and TwCDA-S with β-chitin as substrate (black bar); enzymatic activities of TwCDA and TwCDA-S with (GlcNAc)5 as substrate (gray bar). The enzyme activities in the four experimental groups were significantly higher than those in the control group (dark gray bar and light gray bar). *, p < 0.05; **, p < 0.01.
Author Contributions
Conceptualization, M.C., Z.S. and D.D.; methodology, M.C. and Z.S.; software, M.C., X.W., S.L. and C.L.; formal analysis, M.C.; writing—original draft preparation, M.C.; writing—review and editing, Z.S.; funding acquisition, Z.S.; supervision, all. All authors have read and agreed to the published version of the manuscript.
Funding
This work received financial support from the National Natural Science Foundation of China (no. 41806175) and the Youth Innovation Promotion Association, Chinese Academy of Sciences (no. 2022205).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the findings of this study are available in public databases, within the paper, and within its Supplementary Materials published online.
Acknowledgments
We thank Xiaohui Li from Ningbo University, China, for providing the T. weissflogii 9021 strain.
Conflicts of Interest
No conflict of interest declared.
References
- Minke, R.; Blackwell, J. The structure of α-chitin. J. Mol. Biol. 1978, 120, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Aam, B.B.; Heggset, E.B.; Norberg, A.L.; Sørlie, M.; Vårum, K.M.; Eijsink, V.G.H. Production of chitooligosaccharides and their potential applications in medicine. Mar. Drugs 2010, 8, 1482–1517. [Google Scholar] [CrossRef]
- Souza, C.P.; Almeida, B.C.; Colwell, R.R.; Rivera, I.N.G. The importance of chitin in the marine environment. Mar. Biotechnol. 2011, 13, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, R.A.G.; Tuveng, T.R.; Antonsen, S.G.; Eijsink, V.G.H.; Sorlie, M. Can we make chitosan by enzymatic deacetylation of chitin? Molecules 2019, 24, 3862. [Google Scholar] [CrossRef]
- Svitil, A.L.; Chadhain, S.M.N.; Moore, J.A.; Kirchman, D.L. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 1997, 63, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Lamarque, G.; Cretenet, M.; Viton, C.; Domard, A. New route of deacetylation of alpha- and beta-chitins by means of freeze-pump out-thaw cycles. Biomacromolecules 2005, 6, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Pino, Z.; Shirai, K.; Arias, L.; Vazquez-Torres, H. Effect of water quality and particle size on the production of chitosan from beta-chitin isolated from jumbo squid processing wastes (Dosidicus gigas). Rev. Mex. Ing. Quim. 2008, 7, 299–307. [Google Scholar]
- Imai, T.; Watanabe, T.; Yui, T.; Sugiyama, J. The directionality of chitin biosynthesis: A revist. Biochem. J. 2003, 374, 755–760. [Google Scholar] [CrossRef]
- Cuong, H.N.; Minh, N.C.; Hoa, N.V.; Trang, T.S. Preparation and characterization of high purity β-chitin from squid pens (Loligo chenisis). Int. J. Biol. Macromol. 2016, 93, 442–447. [Google Scholar] [CrossRef]
- Bowler, C.; Vardi, A.; Allen, A.E. Oceanographic and biogeochemical insights from diatom genomes. Ann. Rev. Mar. Sci 2010, 2, 333–365. [Google Scholar] [CrossRef]
- Clement, R.; Dimnet, L.; Maberly, S.C.; Gontero, B. The nature of the CO2-concentrating mechanisms in a marine diatom, Thalassiosira pseudonana. New Phytol. 2016, 209, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Li, F.T.; Beardall, J.; Collins, S.; Gao, K.S. Decreased photosynthesis and growth with reduced respiration in the model diatom Phaeodactylum tricornutum grown under elevated CO2 over 1800 generations. Glob. Chang. Biol. 2017, 23, 127–137. [Google Scholar] [CrossRef]
- McLachlan, J.; McInnes, A.G.; Falk, M. Studies on the chitan (chitin: Poly-N-acetylglucosamine) fibers of the diatom Thalassiosira fluviatilis hustedt: I production and isolation of chitan fibers. Can. J. Bot. 1965, 43, 707–713. [Google Scholar] [CrossRef]
- Brunner, E.; Richthammer, P.; Ehrlich, H.; Paasch, S.; Simon, P.; Ueberlein, S.; van Pee, K.H. Chitin-based organic networks: An integral part of cell wall biosilica in the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 2009, 48, 9724–9727. [Google Scholar] [CrossRef]
- Longnecker, K.; Soule, M.C.K.; Kujawinski, E.B. Dissolved organic matter produced by Thalassiosira pseudonana. Mar. Chem. 2015, 168, 114–123. [Google Scholar] [CrossRef]
- Cheng, H.; Shao, Z.; Lu, C.; Duan, D. Genome-wide identifification of chitinase genes in Thalassiosira pseudonana and analysis of their expression under abiotic stresses. BMC Plant Biol. 2021, 21, 87. [Google Scholar] [CrossRef]
- Yuan, Y.; Rong, Y.; Yang, F.; Chen, L.; Shi, W.; Wang, Z. The separation and extraction methods of chitin/chitosan and its application in food industry. Food Ferm. Ind. 2020, 46, 252–258. [Google Scholar]
- Guan, Z.W.; Feng, Q. Chitosan and chitooligosaccharide: The promising non-plant-derived prebiotics with multiple biological activities. Int. J. Mol. Sci. 2022, 23, 6761. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Zhao, L. The history of chito/chitin oligosaccharides and Its monomer. In The Oligosaccharides of Chitin and Chitosan: Bio-Manufacture and Applications; Zhao, L., Ed.; Springer: Singapore, 2019; pp. 3–14. [Google Scholar]
- Satitsri, S.; Muanprasat, C. Chitin and chitosan derivatives as biomaterial resources for biological and biomedical applications. Molecules 2020, 25, 5961. [Google Scholar] [CrossRef]
- Muanprasat, C.; Chatsudthipong, V. Chitosan oligosaccharide: Biological activities and potential therapeutic applications. Pharmacol. Ther. 2017, 170, 80–97. [Google Scholar] [CrossRef]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Eijsink, V.G. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, I.; Bouriotis, V. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef]
- Araki, Y.; Ito, E. A pathway of chitosan formation in Mucor rouxii: Enzymatic deacetylation of chitin. Biochem. Biophys. Res. Commun. 1974, 56, 669–675. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ohnishi-Kameyama, M.; Hayashi, K.; Mori, Y. Cloning and expression of chitin deacetylase gene from a Deuteromycete, Colletotrichum lindemuthianum. J. Biosci. Bioeng. 1999, 87, 418–423. [Google Scholar] [CrossRef]
- Grifoll-Romero, L.; Pascual, S.; Aragunde, H.; Biarnés, X.; Planas, A. Chitin deacetylases: Structures, specificities, and biotech applications. Polymers 2018, 10, 352. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.R.; Liu, W.M.; Zhao, X.M.; Zhang, M.; Li, D.Q.; Zuber, R.; Ma, E.B.; Zhu, K.Y.; Moussian, B.; Zhang, J.Z. LmCDA1 organizes the cuticle by chitin deacetylation in Locusta migratoria. Insect. Mol. Biol. 2019, 28, 301–312. [Google Scholar] [CrossRef]
- Yu, H.Z.; Li, N.Y.; Li, B.; Toufeeq, S.; Xie, Y.X.; Huang, Y.L.; Du, Y.M.; Zeng, X.D.; Zhu, B.; Lu, Z.J. Immune Functional Analysis of Chitin Deacetylase 3 from the Asian Citrus Psyllid Diaphorina citri. Int. J. Mol. Sci. 2020, 21, 64. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, Z.; Wu, H.; Fu, C.R.; Li, Y.Z.; Lu, X.J.; Guo, W. RNA interference-mediated functional characterization of Group I chitin deacetylases in Holotrichia parallela Motschulsky. Pestic. Biochem. Physiol. 2021, 173, 104770. [Google Scholar] [CrossRef]
- Ma, Q.; Gao, X.; Bi, X.; Tu, L.; Xia, M.; Shen, Y.B.; Wang, M. Isolation, characterisation, and genome sequencing of Rhodococcus equi: A novel strain producing chitin deacetylase. Sci. Rep. 2020, 10, 4329. [Google Scholar] [CrossRef] [PubMed]
- Linhorst, M.; Wattjes, J.; Moerschbacher, B.M. Chitin Deacetylase as a Biocatalyst for the Selective N-Acylation of Chitosan Oligo- and Polymers. ACS Catal. 2021, 11, 14456–14466. [Google Scholar] [CrossRef]
- Yang, G.; Wang, Y.H.; Fang, Y.W.; An, J.; Hou, X.Y.; Lu, J.; Zhu, R.J.; Liu, S. A novel potent crystalline chitin decomposer: Chitin deacetylase from Acinetobacter schindleri MCDA01. Molecules 2022, 27, 5354. [Google Scholar] [CrossRef] [PubMed]
- Hembach, L.; Bonin, M.; Gorzelanny, C.; Moerschbacher, B.M. Unique subsite specificity and potential natural function of a chitosan deacetylase from the human pathogen Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 2020, 117, 3551–3559. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.B.; Zhou, C.; Gong, M.F.; Yang, H.; Long, G.Y.; Jin, D.C. Identification and RNAi-Based Functional Analysis of Four Chitin Deacetylase Gaenes in Sogatella furcifera (Hemiptera: Delphacidae). J. Insect. Sci. 2021, 21, 9. [Google Scholar] [CrossRef]
- Li, X.G.; Diao, P.Y.; Chu, J.D.; Zhou, G.; Zhou, J.; Lin, H.; Chen, J.H.; Zeng, Q.F. Molecular characterization and function of chitin deacetylase-like from the Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. 2021, 256, 110612. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Thomas, Y.; Hembach, L.; Xing, X.; Duan, D.; Moerschbacher, B.M.; Bulone, V.; Tirichine, L.; Bowler, C. Comparative characterization of putative chitin deacetylases from Phaeodactylum tricornutum and Thalassiosira pseudonana highlights the potential for distinct chitin-based metabolic processes in diatoms. New Phytol. 2019, 221, 1890–1905. [Google Scholar] [CrossRef] [PubMed]
- Noishiki, Y.; Nishiyama, Y.; Wada, M.; Okada, S.; Kuga, S. Inclusion complex of β-chitin and aliphatic amines. Biomacromolecules 2003, 4, 944–949. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Worden, A.Z. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Sahoo, P.R.; Mishra, S.R.; Mohapatra, S.; Sahu, S.; Sahoo, G.; Behera, P.C. In silico structural and phylogenetic analysis of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in domestic animals. Indian J. Anim. Res. 2019, 53, 1607–1612. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Euk-mPLoc: A fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J. Proteome Res. 2007, 6, 1728–1734. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Schwede, T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.L.; Cai, A.N.; Chen, P.Q. Screening and properties of chitin deacetylase from Bacillus subtilis. Amino Acids Biot. Resour. 2007, 29, 30–32. [Google Scholar]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Walden, H.; Bell, G.S.; Russell, R.J.M.; Siebers, B.; Hensel, R.; Taylor, G.L. Tiny TIM: A small, tetrameric, hyperthermostable triosephosphate isomerase. J. Mol. Biol. 2001, 306, 745–757. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Hayes, R.D.; Calhoun, S.; Kamel, B.; Wang, A.; Ahrendt, S.; Dusheyko, S.; Nikitin, R.; Mondo, S.J.; Salamov, A.; et al. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res. 2021, 49, D1004–D1011. [Google Scholar] [CrossRef]
- Moon, H.; Choy, S.; Park, Y.; Jung, Y.M.; Koo, J.M.; Hwang, D.S. Different molecular interaction between collagen and α- or β-chitin in mechanically improved electrospun composite. Mar. Drugs 2019, 17, 318. [Google Scholar] [CrossRef]
- Giastas, P.; Andreou, A.; Papakyriakou, A.; Koutsioulis, D.; Balomenou, S.; Tzartos, S.J.; Bouriotis, V.; Eliopoulos, E.E. Structures of the peptidogly can N-acetylglucosamine deacetylase Bc1974 and its complexes with zinc metalloenzyme inhibitors. Biochemistry 2018, 57, 753–763. [Google Scholar] [CrossRef]
- Blair, D.E.; Hekmat, O.; Schuttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M.F. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef] [PubMed]
- Strunk, R.J.; Piemonte, K.M.; Petersen, N.M.; Koutsioulis, D.; Bouriotis, V.; Perry, K.; Cole, K.E. Structure determination of BA0150, a putative polysaccharide deacetylase from Bacillus anthracis. J. Struct. Biol. 2014, 70, 156–159. [Google Scholar]
- Alverson, A.J.; Beszteri, B.; Julius, M.L.; Theriot, E.C. The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol. Biol. 2011, 11, 125. [Google Scholar] [CrossRef]
- Klepsch, M.M.; Persson, J.O.; de Gier, J.W.L. Consequences of the overexpression of a eukaryotic membrane protein, the human KDEL receptor, in Escherichia coli. J. Mol. Biol. 2011, 407, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.F.; Wang, C.Z.; Wu, Y.H.; Cong, J.B.; Cheng, L.; Wang, M.Q.; Zhao, P.K.; Tang, L.; Zhang, C.G.; Wu, K. Tat peptide-mediated soluble expression of the membrane protein LSECtin-CRD in Escherichia coli. PLoS ONE 2013, 8, e83579. [Google Scholar] [CrossRef]
- Tomatis, P.E.; Schutz, M.; Umudumov, E.; Pluckthun, A. Mutations in sigma 70 transcription factor improves expression of functional eukaryotic membrane proteins in Escherichia coli. Sci. Rep. 2019, 9, 2483. [Google Scholar] [CrossRef]
- Frelet-Barrand, A. Lactococcus lactis, an attractive cell factory for the expression of functional membrane proteins. Biomolecules 2022, 12, 180. [Google Scholar] [CrossRef]
- Wagner, S.; Baars, L.; Ytterberg, A.J.; Klussmeier, A.; Wagner, C.S.; Nord, O.; Nygren, P.A.; van Wijk, K.J.; de Gier, J.W. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteom. 2007, 6, 1527–1550. [Google Scholar] [CrossRef]
- Brosson, D.; Kuhn, L.; Prensier, G.; Vivare`s, C.P.; Texier, C. The putative chitin deacetylase of Encephalitozoon cuniculi: A surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 2005, 247, 81–90. [Google Scholar] [CrossRef]
- Leisner, J.J.; Vogensen, F.K.; Kollmann, J.; Aideh, B.; Vandamme, P.; Vancanneyt, M.; Ingmer, H. α-Chitinase activity among lactic acid bacteria. Syst. Appl. Microbiol. 2008, 31, 151–156. [Google Scholar] [CrossRef]
- Vincenzi, S.; Bierma, J.; Wickramasekara, S.I.; Curioni, A.; Gazzola, D.; Bakalinsky, A.T. Characterization of a grape class IV chitinase. J. Agric. Food Chem. 2014, 62, 5660–5668. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Y.; Fong, S.S. Cloning and characterization of a chitinase from Thermobifida fusca reveals Tfu_0580 as a thermostable and acidic endochitinase. Biotechnol. Rep. 2018, 19, e00274. [Google Scholar]
- Chandler, J.C.; Molins, C.R.; Petersen, J.M.; Belisle, J.T. Differential chitinase activity and production within Francisella species, subspecies, and subpopulations. J. Bacteriol. 2011, 193, 3265–3275. [Google Scholar] [CrossRef]
- Kang, Y.; Kim, H.; Choi, H.T. Biochemical characterization of chitinase 2 expressed during the autolytic phase of the inky cap, Coprinellus congregatus. Microb. Ecol. 2013, 51, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Arnold, N.D.; Bruck, W.M.; Garbe, D.; Brueck, T.B. Enzymatic modification of native chitin and conversion to specialty chemical Products. Mar. Drugs 2020, 18, 93. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Melcher, R.L.J.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef]
- Lee, S.; Kang, H.A.; Eyun, S.I. Evolutionary analysis and protein family classification of chitin deacetylases in Cryptococcus neoformans. J. Microbiol. 2020, 58, 805–811. [Google Scholar] [CrossRef]
- Zhang, A.; He, Y.M.; Wei, G.G.; Zhou, J.; Dong, W.L.; Chen, K.Q.; Ouyang, P.K. Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-d-glucosamine production. Biotechnol. Biofuels 2018, 11, 179. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, M.T.; Nguyen, V.B.; Vo, T.P.K.; Nguyen, A.D.; Wang, S.L. An exochitinase with N-acetyl-beta-glucosaminidase-like activity from shrimp head conversion by Streptomyces speibonae and its application in hydrolyzing beta-chitin powder to produce N-acetyl-d-glucosamine. Polymers 2019, 11, 1600. [Google Scholar] [CrossRef]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015, 5, 8716. [Google Scholar] [CrossRef]
- Sashiwa, H.; Fujishima, S.; Yamano, N.; Kawasaki, N.; Nakayama, A.; Muraki, E.; Hiraga, K.; Oda, K.; Aiba, S. Production of N-acetyl-d-glucosamine from α-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr. Res. 2002, 337, 761–763. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).