Abstract
Western diets high in sugars and saturated fats have been reported to induce metabolic and inflammatory impairments that are associated with several age-related disorders, including Alzheimer’s disease (AD) and type 2 diabetes (T2D). The apolipoprotein E (APOE) genotype is associated with metabolic and inflammatory outcomes that contribute to risks for AD and T2D, with the APOE4 genotype increasing risks relative to the more common APOE3 allele. In this study, we investigated the impacts of the APOE genotype on systemic and neural effects of the Western diet. Female mice with knock-in of human APOE3 or APOE4 were exposed to control or Western diet for 13 weeks. In the control diet, we observed that APOE4 mice presented with impaired metabolic phenotypes, exhibiting greater adiposity, higher plasma leptin and insulin levels, and poorer glucose clearance than APOE3 mice. Behaviorally, APOE4 mice exhibited worse performance in a hippocampal-dependent learning task. In visceral adipose tissue, APOE4 mice exhibited generally higher expression levels of macrophage- and inflammation-related genes. The cerebral cortex showed a similar pattern, with higher expression of macrophage- and inflammation-related genes in APOE4 than APOE3 mice. Exposure to the Western diet yielded modest, statistically non-significant effects on most metabolic, behavioral, and gene expression measures in both APOE genotypes. Interestingly, the Western diet resulted in reduced gene expression of a few macrophage markers, specifically in APOE4 mice. The observed relative resistance to the Western diet suggests protective roles of both female sex and young adult age. Further, the data demonstrate that APOE4 is associated with deleterious systemic and neural phenotypes and an altered response to a metabolic stressor, findings relevant to the understanding of interactions between the APOE genotype and risks for metabolic disorders.
1. Introduction
Apolipoprotein E (ApoE) is a cholesterol transport protein that is coded for by three different alleles of the APOE gene, APOE2, APOE3, and APOE4. These APOE alleles generate ApoE proteins that vary at amino acids 112 and 158 and show significant functional differences [1]. Importantly, APOE genotype impacts well established associations among Alzheimer’s disease (AD), type 2 diabetes (T2D), and cardiovascular disease (CVD) [2]. APOE3 is the most common allele in human populations and is used as the reference for establishing relative risk. In comparison to APOE3, APOE4 increases risk as much as 15-fold for AD [3,4,5] and two- to three-fold for T2D and CVD [6]. Conversely, APOE2 is associated with decreased risks for AD [7,8] and CVD [9]. Interestingly, the APOE genotype shows parallel relationships with lifespan in both humans and mice. APOE2 is associated with increased longevity, and APOE4 is associated with decreased longevity [10,11]. The mechanism(s) by which APOE affects aging and age-related disease risks is unclear, but ApoE modulates aspects of metabolism [12], lipid dynamics, and inflammation, all of which are also implicated in pathways relevant to these age-related diseases.
Obesity is an important factor that links APOE, AD, CVD, and T2D. Risks for AD, CVD, and T2D are all strongly increased by obesity [13,14,15], interactions that can be impacted by the APOE genotype. For example, abundant evidence indicates that relationships between obesity and AD risk can be worsened by APOE4 [16,17]. Both APOE4 and obesity affect similar pathways associated with metabolic stress, including inflammation. Many prior studies have shown higher markers of inflammation in APOE4 carriers [18,19,20,21], although the relationship between the APOE genotype and inflammatory tone appears to be complex and may depend upon context [22,23,24]. Inflammatory pathways are implicated in pathological and normal age-related cognitive decline [25,26,27] and represent one mechanism by which obesity and APOE may interact to affect vulnerability to AD and other age-related metabolic disorders.
In the present study, we investigated the effects of the APOE genotype on systemic and neural responses to an obesogenic diet. As APOE4-associated cognitive [28,29] and metabolic [30] risks are more robust in females, we studied female mice. In prior work with AD transgenic mice with knock-in of human APOE3 or APOE4, we observed that young adult female APOE3 mice showed more metabolic dysfunction and greater AD-related pathology and behavioral impairment in response to an obesogenic diet than APOE4 females [31]. However, because even early AD-related pathology can result in systemic metabolic impairments [32], understanding the metabolic consequences of APOE genotype requires analyses in the absence of AD transgenes. This was accomplished by assessing metabolic and cognitive responses in APOE knock-in mice on a 13-week exposure to control versus Western diet that is high in both sugars and saturated fats [28,29,30]. Our findings provide new insights into the effects of APOE genotype on metabolic stressors associated with AD risk.
2. Methods
2.1. Animals
A colony of EFAD mice [33] were maintained at vivarium facilities at the University of Southern California from breeder mice generously provided by Dr. Mary Jo LaDu (University of Illinois at Chicago). EFAD mice are heterozygous for 5xFAD transgenes and homozygous for knock-in of human APOE3 or APOE4. Breeding strategies for EFAD mice yield litters that are ~50% 5xFAD+/− (EFAD) and ~50% 5xFAD−/− (EFAD-non-carriers, EFAD-NC). The latter are homozygous for APOE, but they lack AD-related transgenes. In this study, female EFAD-NC mice homozygous for APOE3 (APOE3 mice) or APOE4 (APOE4 mice) were maintained under controlled temperature, a 12:12 light/dark schedule (lights on at 06:00), and with ad libitum access to food and water. At 2.5 months of age, mice were randomly assigned (n = 7–8/group) to either ingredient-matched control (10% calories from fat and 7% from sugar; catalog #D12450J, Research Diets, Inc., New Brunswick, NJ, USA) or Western (WD; 45% calories from fat and 17% from sugar; catalog #D12451, Research Diets, Inc.) diets. Body weight was monitored weekly for the 13-week diet exposure period. All procedures were conducted under a protocol approved by the USC Institution for Animal Care and Use Committee and under the supervision of USC veterinarians.
2.2. Glucose Tolerance Test
A glucose tolerance test was performed 12 weeks after the start of the diets. Animals were fasted overnight (~16 h) and orally gavaged with 2 g/kg D-glucose. Blood glucose levels were measured at 0, 15-, 30-, 60-, and 120-min following glucose administration. Five microliters of blood were collected on a glucose test strip and assayed using a Precision Xtra glucose monitor (Abbott Laboratories, Abbott Park, IL, USA).
2.3. Tissue Collection
At the end of the 13-week treatment period, APOE3 and APOE4 mice were euthanized by carbon dioxide overdose, following overnight fasting, after which the brain was rapidly removed, and the cerebral cortex was dissected and stored as frozen. Blood was collected and kept on ice prior to centrifugation to collect plasma, which was stored in aliquots at −80° until assayed. The retroperitoneal and visceral (including gonadal and uterine fat) fat pads were dissected, weighed, and frozen.
2.4. Insulin, Leptin, and Cholesterol Assays
Fasting levels of insulin and leptin were determined using rat/mouse insulin (Millipore, Catalog #EZRMI-13K) and mouse leptin (Millipore, Catalog # EZML-82K) ELISA kits, according to the manufacturers’ instructions, using plasma collected at euthanization. Plasma cholesterol was measured using a colorimetric cholesterol quantitation assay kit (Abcam, Catalog #ab65359), according to the manufacturer’s directions. Two APOE3 control and two APOE3 WD animals exhibited negative insulin concentrations and were excluded from insulin analyses. One APOE3 control animal exhibited a negative leptin concentration and was excluded from leptin analysis.
2.5. Behavior
Mice were behaviorally assessed at week 11 using object placement (NOP) and novel object recognition (NOR), tasks that assess cognition and learning and memory involving hippocampal and parahippocampal regions [34]. Mice were habituated to the testing arena without objects for 5 min per day for three days prior to testing. Before testing each day, animals were habituated to the room for at least 30 min. On the test day, two identical objects were placed into the box, and the mice were allowed to explore for five minutes (training). Twenty minutes later, the animals were placed again in the chamber with the same objects, but one moved 90° in the chamber (NOP) and was allowed to explore for five minutes. Twenty minutes after NOP, the mice were placed in the chamber with one familiar object and one novel object for five minutes (NOR). The time the mice spent exploring the objects was recorded. The discrimination index was calculated as (time with novel—time with familiar)/total time with objects. The recognition index was calculated as time with novel/total time with objects. Mice were excluded from analysis if they spent less than 6 s total exploring the objects. In total, two mice were excluded from NOP, and four mice were excluded from NOR.
2.6. Quantitative Real-Time PCR
RNA was extracted from visceral adipose tissue or the cerebral cortex by lysis with TRIzol reagent (Life Technologies, Carlsbad, CA, USA, Cat #15596018), according to the manufacturer’s protocol, except for in adipose tissue. For adipose tissue, tissue was lysed using a dounce homogenizer for 20 strokes. The solution was centrifuged for 10 min, and the top layer (fat) was removed prior to the addition of chloroform. After the completion of the TRI-zol protocol, potential genomic DNA contamination was removed from all samples by an RNase-free DNase treatment (Lucigen, Middlesex, UK, Cat #D9905K). Purified RNA (1 μg) was used for reverse transcription using the iScript synthesis system (Bio-Rad, Hercules, CA, USA, Cat #1708891). The resulting cDNA was used for quantitative PCR using a Bio-Rad CFX96 Touch Real-Time PCR Detection System. Standard PCR cycling protocols were used, consisting of 40 cycles, with denaturation at 95 °C and annealing at 60 °C. The amplification efficiency was estimated from the standard curve for each gene. All primers have an efficiency of 88–115%. Relative quantification of mRNA levels was determined by the ΔΔCt method. All groups were compared to the APOE3 control. All experimental primers were compared to the expression of β-actin in the brain and the average expression of succinate dehydrogenase complex, subunit A (SDHA), and hypoxanthine guanine phosphoribosyltransferase (HPRT) in adipose tissue. The expression of control primers showed no significant differences according to the APOE genotype or diet (2-way ANOVA). Primer sequences (Life Technologies) are listed in Table 1.

Table 1.
Primer sequences used in PCR analyses.
2.7. Statistics
All data are reported as means + standard errors of the mean. Raw data were analyzed using GraphPad Prism 8. Most data were analyzed using two-way ANOVAs (APOE × diet), with Tukey’s post hoc tests where applicable. Changes in body weight and glucose tolerance over time were analyzed using three-way repeated measure ANOVAs (APOE × diet × time). Initial body weights were compared by t-test. Comparisons with p < 0.05 were considered statistically significant. Because several comparisons yielded p values only slightly above the significance limit, outcomes with p ≤ 0.10 are also noted. All relevant statistics are listed in Table 2 and noted in the figures.

Table 2.
Statistical analyses with significant or nearly significant outcomes.
3. Results
3.1. Metabolic Consequences of APOE Genotype and Western Diet
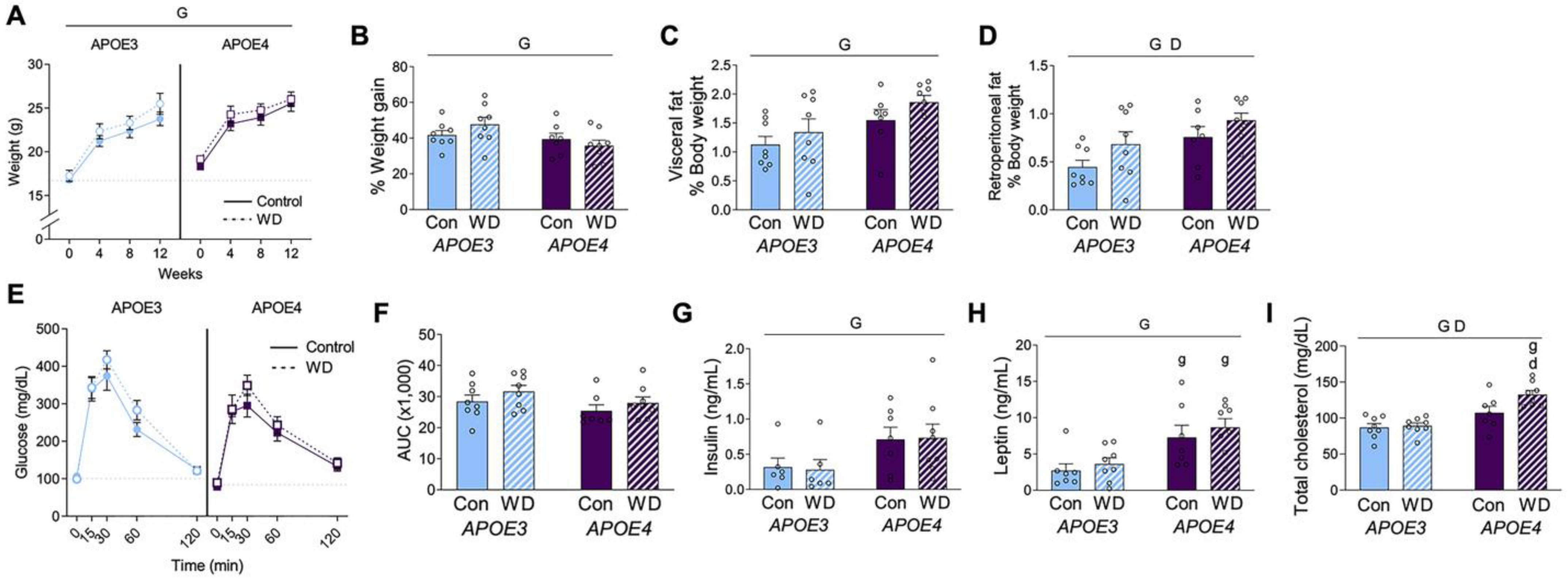
Female mice were maintained on control or Western diet (WD) for 13 weeks. At the start of the treatment period, APOE4 mice had significantly heavier body weight than APOE3 mice (Figure 1A; t-test, p < 0.001). There was a significant effect of APOE genotype on body weight over the course of the study (see Table 2 for statistical analyses). By the end of the 13-week treatment period, APOE3 and APOE4 mice were of similar weight. Body weight increased across all groups regardless of diet, although the percent weight change was significantly greater in APOE3 mice compared to APOE4 mice (Figure 1B). There was a significant main effect of genotype on adiposity, with APOE4 mice exhibiting significantly larger visceral and retroperitoneal fat pads (normalized to body weight; Figure 1C,D, respectively). Diet did not significantly affect body weight or visceral fat mass, but it was associated with increased retroperitoneal fat.
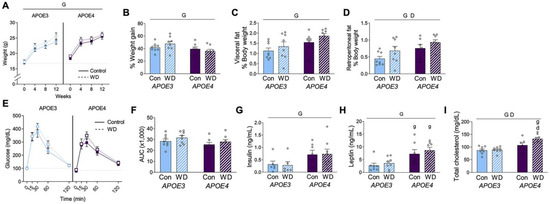
Figure 1.
Metabolic profiles in APOE mice after Western diet. (A) Body weight was measured at the start and at four-week intervals in female APOE mice on control (Con; open circles) and Western diet (WD; filled squares). The dotted line indicates the initial weight of the APOE3 mice. (B) The percent weight change was calculated from the beginning to the end of the experimental period. (C,D) Fat pads from (C) visceral and (D) retroperitoneal depots were removed and weighed at the conclusion of the experiment. Values are expressed as a percentage of body weight. (E) Results from an oral glucose tolerance test that was administered after 12 weeks of diet. (F) The area under the curve was calculated for the GTT. (G–I) Plasma was collected under fasting conditions at the end of the experiment and measured for (G) insulin, (H) leptin, and (I) cholesterol. Data are shown as individual values (open circles) and mean values (+SEM) from n = 7–8 mice per group. “G” indicates a significant main effect of the APOE genotype, “D” indicates a significant main effect of diet, “g” indicates a significant difference from the APOE3 mice on the same diet, “d” indicates a significant difference from animals with the same APOE genotype on the control diet. Significance is p < 0.05.
An oral glucose tolerance test was administered 12 weeks after the start of diet (Figure 1E). The area under the curve appeared to be greater in APOE3 mice, but this was not statistically significant (Figure 1F). Terminal levels of plasma insulin and leptin were significantly higher in APOE4 females (Figure 1G,H), with no significant effect of diet in either genotype. Plasma cholesterol was significantly increased in APOE4 females and by WD, though this diet increase was driven by a change in the APOE4 mice (Figure 1I).
3.2. Cognitive Effects of APOE Genotype and Western Diet in APOE Mice on the Western Diet
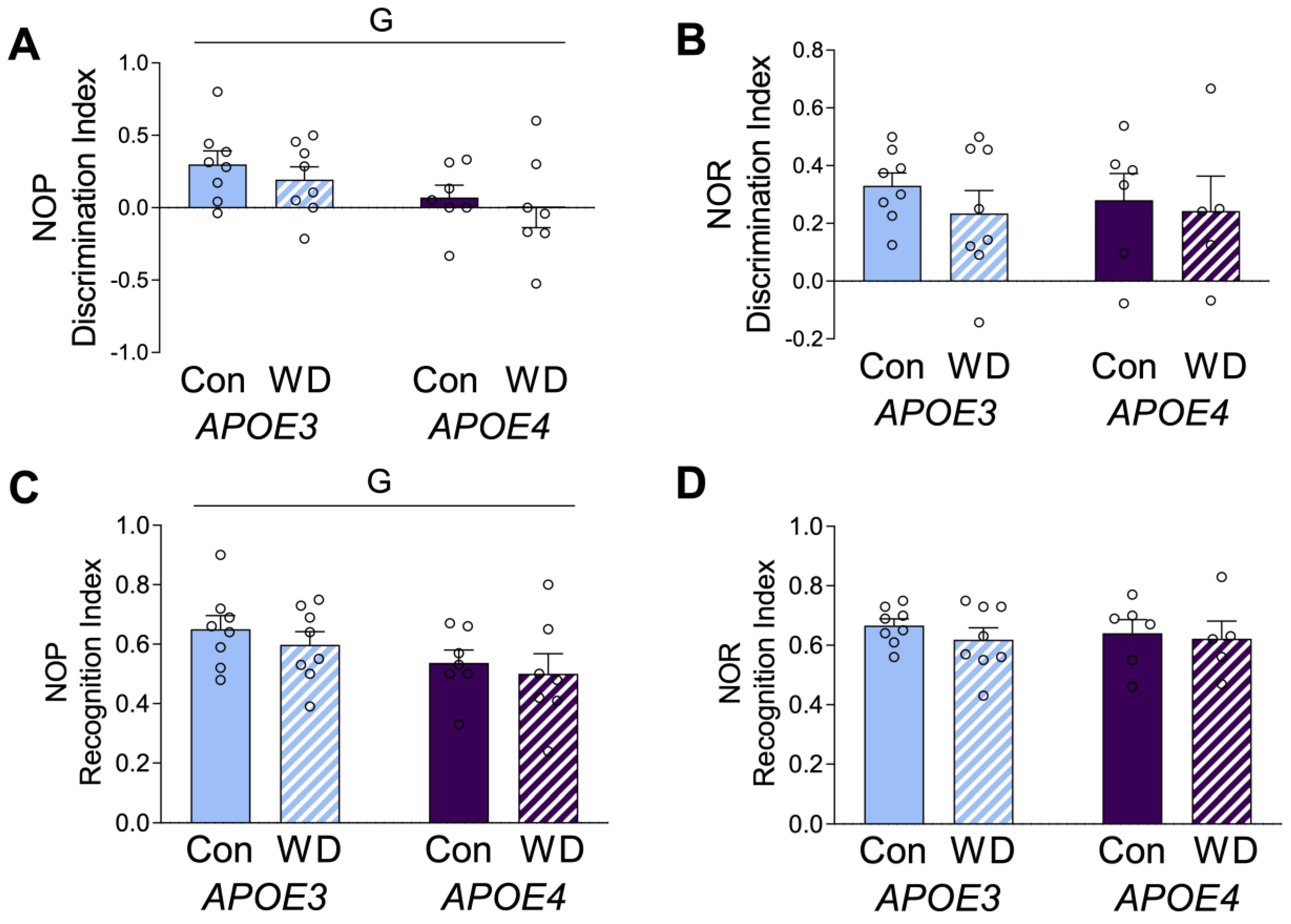
APOE mice were tested on the novel object placement and novel object recognition tests. Novel object placement primarily evaluates spatial learning and relies mostly on hippocampal learning. Novel object recognition relies on non-spatial learning and engages multiple brain regions [35]. APOE4 animals showed significantly poorer performance in novel object placement (Figure 2A,C), but there was no genotype difference in performance in novel object recognition (Figure 2B,D). There were no significant effects of diet on either behavioral measure, which are presented as both discrimination index (Figure 2A,B) and recognition index (Figure 2C,D).
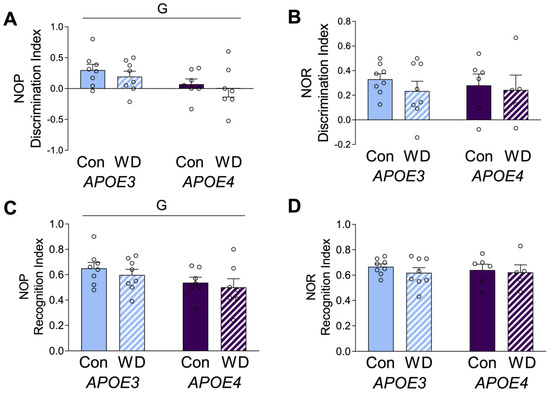
Figure 2.
Behavioral effects in APOE mice after the Western diet. Animals were tested with (A,C) novel object placement (NOP) and (B,D) novel object recognition (NOR) behavioral tasks. The discrimination index is represented in (A,B), with values > 0 indicating positive performance. The recognition index is represented in (C,D). Data are shown as individual values (open circles) and mean values (+SEM), from n = 5–8 mice per group. “G” indicates a significant main effect of the APOE genotype. Significance is p < 0.05.
3.3. Adipose Gene Expression with the APOE Genotype and the Western Diet
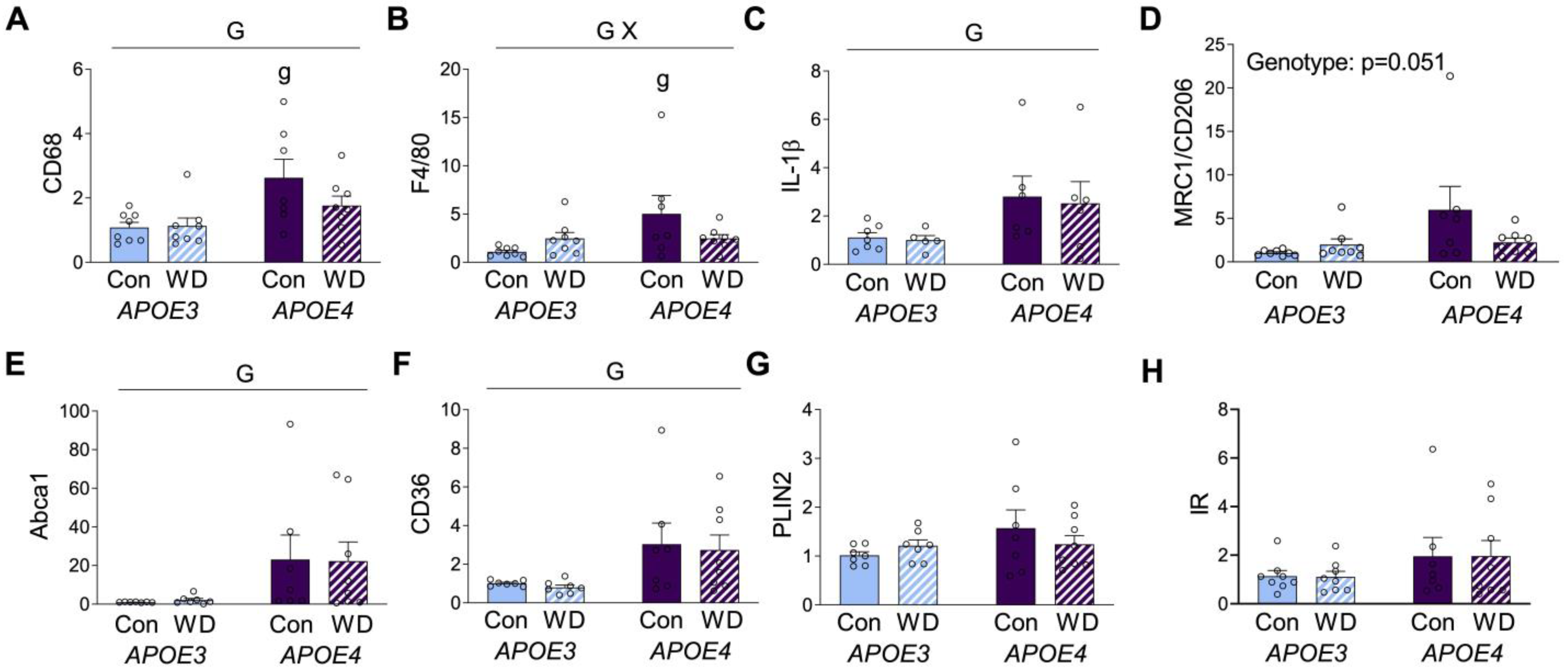
As both obesogenic diets and the APOE genotype are known to affect adiposity, we examined mRNA expression levels of several genes in visceral adipose tissue. We first assessed two well-established macrophage markers, CD68 and F4/80. There was a main effect of genotype, such that expression of both was higher in APOE4 mice (Figure 3A,B). Interestingly, post hoc analyses showed significant APOE3 vs. APOE4 differences in CD68 and F4/80 expression only under the control diet. Although there was no significant main effect of diet on CD68 and F4/80 expression, there was a significant interaction for F4/80 expression, wherein WD was associated with increased levels in APOE3, but lower levels in APOE4 relative to the control diet (Figure 3B). CD68 showed a similar trend of lower expression, with WD only in APOE4 mice (Figure 3A).
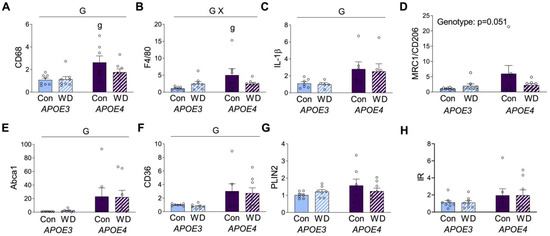
Figure 3.
Gene expression levels in visceral adipose tissue. Results of mRNA quantification studies in APOE3 (blue) and APOE4 (purple) mice maintained on control (Con; solid bars) or Western (WD; hatched bars) diets. Gene targets included the macrophage markers (A) CD68 and (B) F4/80, the inflammation-related markers (C) IL-1β and (D) CD206, and the metabolic macrophage markers (E) Abca1, (F) CD36, (G) PLIN2, and (H) insulin receptor (IR). Data show fold difference as individual values (open circles) and means (+SEM) relative to the Con APOE3 group from n = 7–8 mice per group. “G” indicates a significant main effect of APOE genotype, “X” indicates a significant interaction effect between APOE and diet, and “g” indicates a significant difference from APOE3 mice on the same diet. Significance is p < 0.05.
Macrophages have traditionally been separated into M1 and M2 phenotypes, which are characterized by predominantly pro- or anti-inflammatory properties, respectively. Although an oversimplification, these categories can provide insights into macrophage biology. The M1 marker interleukin-1β (IL-1β) was significantly increased in APOE4 mice, regardless of diet (Figure 3C). Expression of the M2 marker mannose receptor C type 1 or cluster of differentiation 206 (MRC or CD206) showed a pattern similar to CD68 and F4/80, with a nonsignificant trend (p = 0.056) of higher levels in APOE4 mice under control diet, but not for those of WD (Figure 3D). Macrophages can also exhibit metabolic phenotypes that are associated with obesity. The main markers of these metabolic macrophages are ATP binding cassette subfamily A type 1 (ABCA1), cluster of differentiation 36 (CD36), and perilipin 2 (PLIN2) [36], although these markers are not exclusive to macrophages and can also be expressed by adipocytes. APOE4 mice showed higher expression of both ABCA1 and CD36, with no significant modulatory effect of diet (Figure 3E,F). PLIN2 expression did not significantly differ across APOE or diet (Figure 3G). Further, given the observed differences in insulin and glucose levels, we probed for insulin receptor (IR) expression in the adipose tissue, but we saw no significant differences (Figure 3H).
3.4. Cortex Gene Expression with APOE Genotype and Western Diet
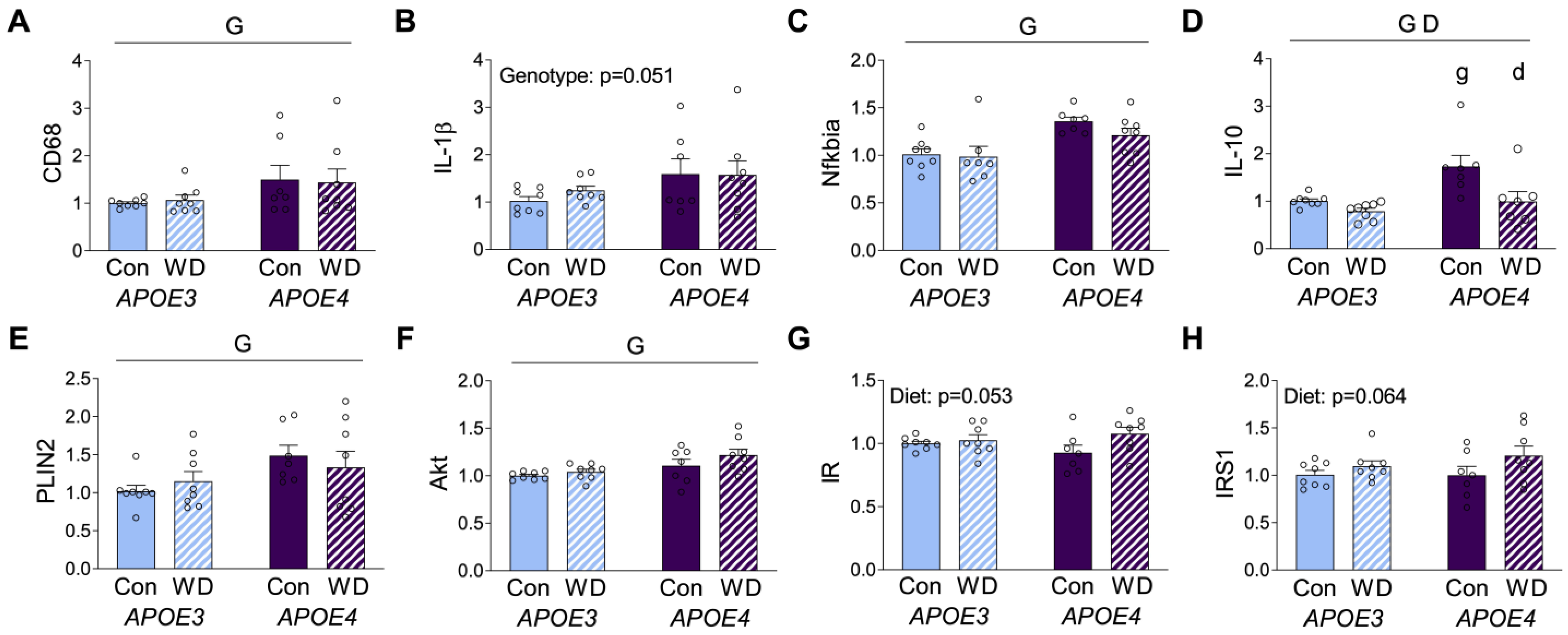
In order to investigate diet and APOE genotype interactions in the brain, mRNA expression of selected inflammatory and metabolic genes was probed in the cerebral cortex. The macrophage marker CD68 showed higher levels in APOE4 mice, with no significant effect of diet (Figure 4A). The pro-inflammatory cytokine IL-1β exhibited a similar pattern of increased expression in APOE4 mice, although it did not meet the significance criterion (p = 0.051, Figure 4B). Likewise, the inflammatory regulator NFκB inhibitor alpha (NFκBIA) that modulates NFκB activity was also significantly higher in APOE4 mice, but it was not significantly affected by WD in either genotype (Figure 4C). The anti-inflammatory cytokine interleukin-10 (IL-10) showed significant effects regarding both APOE genotype and diet, with APOE4 increasing and WD decreasing expression levels (Figure 4D).
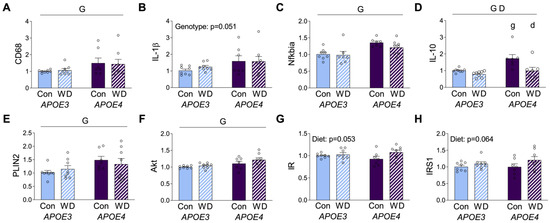
Figure 4.
RNA expression levels in cerebrocortical brain tissue. Results of mRNA quantification studies in APOE3 (blue) and APOE4 (purple) mice maintained on control (Con; solid bars) or Western (WD; hatched bars) diets. Gene targets included the macrophage marker, (A) CD68, the inflammatory markers (B) IL-1β, (C) Nfkbia, and (D) IL-10, and the metabolic markers (E) PLIN2, (F) Akt, (G) IR, and (H) IRS1. Data show fold difference as individual values (open circles) and mean values (+SEM) relative to the Con APOE3 group from n = 7–8 mice per group. “G” indicates a significant main effect of APOE genotype, “D” indicates a significant main effect of diet, “g” indicates a significant difference from APOE3 mice on the same diet, and “d” indicates a significant difference from animals with the same APOE genotype on the control diet. Significance is p < 0.05.
Upon assessing metabolic markers, we first examined the metabolic macrophage markers, ABCA1 and CD36, and we found that they were not affected by APOE or diet (data not shown). However, PLIN2 was increased in the cortex of APOE4 relative to APOE3, but it was not affected by WD (Figure 4E), confirming recent work showing greater expression of this protein in APOE4 astrocytes [37]. We next probed expression of the insulin related genes AKT, insulin receptor (IR), and insulin receptor substrate-1 (IRS-1). AKT expression showed modest, but significantly higher levels, in APOE4 mice with no effect of diet (Figure 4F). In contrast, both IR and IRS-1 showed no APOE genotype differences and nonsignificant trends (p = 0.053 and p = 0.064, respectively) towards increased expression with WD (Figure 4G,H).
4. Discussion
Peripheral and neural effects of the APOE genotype and the Western diet were explored in young adult female mice. Interestingly, there were few effects of diet in either the APOE3 or APOE4 mice, although APOE4 mice were relatively impaired regarding most measures. APOE4 mice had larger fat pads and worse metabolic and cognitive phenotypes. In addition, APOE4 mice generally exhibited higher expression of macrophage and inflammation-related markers in adipose and brain tissue. For a subset of these markers, the Western diet was associated with reduced expression, specifically within APOE4 mice. Note that these findings are limited to gene expression and may not fully reflect changes detectable by histological analyses.
The generally modest effects of Western diet in female mice observed in this study are not altogether surprising. Young females have been shown previously to be resistant to many of the effects of high-fat diets [38,39,40]. Indeed, in both wild type [41] and AD transgenic [42] mice, females have been reported to exhibit little metabolic response to obesogenic diets. This effect appears to be age-dependent, such that, by middle age, the sex difference partially reverses, yielding greater diet-induced effects in females than in males [43]. These sex differences may be due, in part, to the effects of estrogens, which are much higher in young adult females, but they decline in abundance and or efficacy with increasing age. Estrogens have effects on both the brain and peripheral tissues implicated in protecting females from many of the deleterious effects of obesogenic diets [44,45,46].
Prior studies of obesogenic diets in APOE knock-in mice suggest both APOE genotype and sex differences are relevant predictors of vulnerability. Perhaps the most consistent finding has been greater weight gain in APOE3 mice [47,48], which aligns with our observation that APOE3 females on both diets showed proportionally larger increases in body weight. Note, however, that APOE4 mice had a significantly higher body weight at the start of the study, indicating complexity in the effects of the APOE genotype and body weight. The literature on APOE-dependent metabolic differences is somewhat mixed, with evidence of both similar outcomes in APOE3 vs. APOE4 mice [49,50] and greater metabolic impairments in APOE4 mice [51,52]. Although we observed only modest metabolic effects of the Western diet, our results support prior observations of metabolic impairments in APOE4 mice. In one study that compared both sexes in adult, but not aged APOE mice, male mice showed stronger diet-induced metabolic responses, with greater susceptibility in APOE4 mice, whereas females showed similar metabolic responses across APOE genotypes [49]; our results align with the latter finding. Interestingly, estrogen status may be important for not only the metabolic resistance of females, but perhaps also for masking APOE4 vulnerability. In a recent study, an experimental model of menopause/ovarian failure, combined with high-fat diet, worsened cognitive outcomes more strongly in APOE4 females [53]. There have also been reports of obesogenic diets in APOE knock-in mice crossed into AD transgenic models. In the EFAD mouse model, the Western diet yielded similar or greater metabolic impairments in APOE3 mice of both sexes, but it increased AD-related pathology only in APOE3 for females [31] and APOE4 for males [54]. In APP/PS1ΔE9 mice with human APOE, the obesogenic diet increased pathology only in the APOE4 genotype, and this had a stronger effect in males [55]. Collectively, our results complement an increase in the literature that is defining the modulatory roles of sex and APOE genotype on their effects on metabolic stress that is linked to disease risk.
Particularly interesting are our findings on the mRNA expression of macrophage- and inflammation-related genes. We observed that levels of these markers were generally higher in adipose tissue and cerebral cortex of APOE4 mice, a finding consistent with prior findings, indicating an exaggerated inflammatory tone with the APOE4 genotype [56,57]. Perhaps unexpected was the observed general trend of lower adipose levels of macrophage and inflammation markers with Western diet, specifically in APOE4 mice. The cerebral cortex showed limited diet effects; the hippocampus was not examined, but prior work indicates similar effects of obesogenic diet on the cortex and the hippocampus [58,59]. Consistent with our results, lower levels of inflammation-related factors in the liver and the plasma in APOE4 relative to APOE3 mice following the Western diet have been reported [23]. In a recent study, cultured microglia from APOE4 mice have been shown to have increased cytokine production and lipid accumulation compared to APOE3 microglia in the absence of other stressors [60]. However, the obesogenic diet was reported to result in both microglial and astrocytic gliosis in APOE3, but not APOE4, mice [61]. Further, the obesogenic diet in female APOE4 mice reduced the microglia/macrophage marker CD68 [62]. Together, these findings may suggest that APOE4 is associated with an altered inflammation-related response to metabolic stressors.
In summary, these results extend and confirm previous evidence that APOE4 yields systemic and neural phenotypes characterized by metabolic and cognitive impairments even in the absence of significant aging or AD-related pathology. Although obesogenic diets are known to induce metabolic and cognitive dysfunctions, the Western diet caused only very modest changes in this study. We suggest that the observed resistance to the obesogenic diet is related to both female sex and young adult age of the APOE mice. It is important to interpret the cognitive data with caution, as a relatively small number of mice were tested. Our finding that the Western diet reduces expression of inflammatory markers, specifically in APOE4 mice, suggests an APOE4-dependent alteration in inflammatory responses to metabolic stress, which in turn implicates inflammatory more than metabolic pathways in contributing to the impacts of the APOE genotype on the consequences of the obesogenic diet.
Author Contributions
Conceptualization, A.C. and C.J.P.; methodology, A.C. and C.J.P.; validation, A.C. and C.J.P.; formal analysis, A.C.; investigation, A.C.; resources, C.J.P.; data curation, A.C.; writing—original draft preparation, A.C.; writing—review and editing, A.C. and C.J.P.; visualization, A.C.; supervision, C.J.P.; project administration, C.J.P.; funding acquisition, C.J.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the NIH/NIA (P01AG026572, RF1AG058068, T32AG05237) and Cure Alzheimer’s Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
All procedures were conducted under a protocol approved by the USC Institution for Animal Care and Use Committee (Approval Code: 20648) and under the supervision of USC veterinarians.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
APOE (EFAD) mice were provided through a Material Transfer Agreement with Mary Jo LaDu (University of Illinois Chicago).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Husain, M.A.; Laurent, B.; Plourde, M. APOE and Alzheimer’s Disease: From Lipid Transport to Physiopathology and Therapeutics. Front. Neurosci. 2021, 15, 630502. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.Y.; Snyder, P.J.; Wu, W.; Zhang, M.; Echeverria, A.; Alber, J. Pathophysiologic relationship between Alzheimer’s disease, cerebrovascular disease, and cardiovascular risk: A review and synthesis. Alzheimers Dement. 2017, 7, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Payami, H.; Montee, K.R.; Kaye, J.A.; Bird, T.D.; Yu, C.E.; Wijsman, E.M.; Schellenberg, G.D. Alzheimer’s disease, apolipoprotein E4, and gender. JAMA 1994, 271, 1316–1317. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Tian, L.; Henderson, V.W.; Greicius, M.D. Alzheimer’s Disease Neuroimaging Initiative Investigators Sex modifies the APOE-related risk of developing Alzheimer disease. Ann. Neurol. 2014, 75, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, J.; Weng, R.; Gu, X.; Zhong, Z. Apolipoprotein E gene polymorphism and the risk of cardiovascular disease and type 2 diabetes. BMC Cardiovasc. Disord. 2019, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Qian, J.; Monsell, S.E.; Betensky, R.A.; Hyman, B.T. APOEepsilon2 is associated with milder clinical and pathological Alzheimer disease. Ann. Neurol. 2015, 77, 917–929. [Google Scholar] [CrossRef]
- Reiman, E.M.; Arboleda-Velasquez, J.F.; Quiroz, Y.T.; Huentelman, M.J.; Beach, T.G.; Caselli, R.J.; Chen, Y.; Su, Y.; Myers, A.J.; Hardy, J.; et al. Alzheimer’s Disease Genetics Consortium Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 2020, 11, 667–668. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.V.; Kwok, M.K.; Schooling, C.M. Age and sex specific effects of APOE genotypes on ischemic heart disease and its risk factors in the UK Biobank. Sci. Rep. 2021, 11, 9229. [Google Scholar] [CrossRef]
- Li, Z.; Shue, F.; Zhao, N.; Shinohara, M.; Bu, G. APOE2: Protective mechanism and therapeutic implications for Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 63–64. [Google Scholar] [CrossRef]
- Shinohara, M.; Kanekiyo, T.; Tachibana, M.; Kurti, A.; Shinohara, M.; Fu, Y.; Zhao, J.; Han, X.; Sullivan, P.M.; Rebeck, G.W.; et al. APOE2 is associated with longevity independent of Alzheimer’s disease. Elife 2020, 9, e62199. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, M.; Sato, N. The Roles of Apolipoprotein E, Lipids, and Glucose in the Pathogenesis of Alzheimer’s Disease. Adv. Exp. Med. Biol. 2019, 1128, 85–101. [Google Scholar] [PubMed]
- Whitmer, R.A.; Gunderson, E.P.; Barrett-Connor, E.; Quesenberry, C.P.J.; Yaffe, K. Obesity in middle age and future risk of dementia: A 27 year longitudinal population based study. BMJ 2005, 330, 1360. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.W.F.; Kannel, W.B. Obesity, diabetes, and risk of cardiovascular disease in the elderly. Am. J. Geriatr. Cardiol. 2002, 11, 119–124. [Google Scholar] [CrossRef]
- Golay, A.; Ybarra, J. Link between obesity and type 2 diabetes. Best Pract. Res. Clin. Endocrinol. Metab. 2005, 19, 649–663. [Google Scholar] [CrossRef]
- Moser, V.A.; Pike, C.J. Obesity and sex interact in the regulation of Alzheimer’s disease. Neurosci. Biobehav. Rev. 2016, 67, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.S.; Rebeck, G.W. The Synergistic Effects of APOE Genotype and Obesity on Alzheimer’s Disease Risk. Int. J. Mol. Sci. 2018, 20, 63. [Google Scholar] [CrossRef]
- Tao, Q.; Ang, T.F.A.; DeCarli, C.; Auerbach, S.H.; Devine, S.; Stein, T.D.; Zhang, X.; Massaro, J.; Au, R.; Qiu, W.Q. Association of Chronic Low-grade Inflammation with Risk of Alzheimer Disease in ApoE4 Carriers. JAMA Netw. Open 2018, 1, e183597. [Google Scholar] [CrossRef]
- Ophir, G.; Amariglio, N.; Jacob-Hirsch, J.; Elkon, R.; Rechavi, G.; Michaelson, D.M. Apolipoprotein E4 enhances brain inflammation by modulation of the NF-kappaB signaling cascade. Neurobiol. Dis. 2005, 20, 709–718. [Google Scholar] [CrossRef]
- Chen, S.; Averett, N.T.; Manelli, A.; Ladu, M.J.; May, W.; Ard, M.D. Isoform-specific effects of apolipoprotein E on secretion of inflammatory mediators in adult rat microglia. J. Alzheimers Dis. 2005, 7, 25–35. [Google Scholar] [CrossRef]
- Iannucci, J.; Sen, A.; Grammas, P. Isoform-Specific Effects of Apolipoprotein E on Markers of Inflammation and Toxicity in Brain Glia and Neuronal Cells In Vitro. Curr. Issues Mol. Biol. 2021, 43, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Marottoli, F.M.; Katsumata, Y.; Koster, K.P.; Thomas, R.; Fardo, D.W.; Tai, L.M. Peripheral Inflammation, Apolipoprotein E4, and Amyloid-beta Interact to Induce Cognitive and Cerebrovascular Dysfunction. ASN Neuro 2017, 9, 1759091417719201. [Google Scholar] [CrossRef] [PubMed]
- Dose, J.; Schloesser, A.; Torres, G.G.; Venkatesh, G.; Hasler, R.; Flachsbart, F.; Lieb, W.; Nebel, A.; Rimbach, G.; Huebbe, P. On a Western diet, APOEvarepsilon4 is associated with low innate immune sensing, but not APOEvarepsilon3. J. Allergy Clin. Immunol. 2018, 142, 1346–1349.e9. [Google Scholar] [CrossRef]
- Mueller, T.; Fischer, J.; Gessner, R.; Rosendahl, J.; Bohm, S.; van Bommel, F.; Knop, V.; Sarrazin, C.; Witt, H.; Marques, A.M.; et al. Apolipoprotein E allele frequencies in chronic and self-limited hepatitis C suggest a protective effect of APOE4 in the course of hepatitis C virus infection. Liver Int. 2016, 36, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Teunissen, C.E.; van Boxtel, M.P.J.; Bosma, H.; Bosmans, E.; Delanghe, J.; De Bruijn, C.; Wauters, A.; Maes, M.; Jolles, J.; Steinbusch, H.W.M.; et al. Inflammation markers in relation to cognition in a healthy aging population. J. Neuroimmunol. 2003, 134, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Fard, M.T.; Cribb, L.; Nolidin, K.; Savage, K.; Wesnes, K.; Stough, C. Is there a relationship between low-grade systemic inflammation and cognition in healthy people aged 60–75 years? Behav. Brain Res. 2020, 383, 112502. [Google Scholar] [CrossRef]
- Holland, D.; Desikan, R.S.; Dale, A.M.; McEvoy, L.K. Alzheimer’s Disease Neuroimaging Initiative Higher rates of decline for women and apolipoprotein E epsilon4 carriers. AJNR Am. J. Neuroradiol. 2013, 34, 2287–2293. [Google Scholar] [CrossRef]
- Ungar, L.; Altmann, A.; Greicius, M.D. Apolipoprotein E, gender, and Alzheimer’s disease: An overlooked, but potent and promising interaction. Brain Imaging Behav. 2014, 8, 262–273. [Google Scholar] [CrossRef]
- Arnold, M.; Nho, K.; Kueider-Paisley, A.; Massaro, T.; Huynh, K.; Brauner, B.; MahmoudianDehkordi, S.; Louie, G.; Moseley, M.A.; Thompson, J.W.; et al. Sex and APOE epsilon4 genotype modify the Alzheimer’s disease serum metabolome. Nat. Commun. 2020, 11, 1148. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.; Pike, C.J. APOE genotype affects metabolic and Alzheimer-related outcomes induced by Western diet in female EFAD mice. FASEB J. 2019, 33, 4054–4066. [Google Scholar] [CrossRef] [PubMed]
- Macklin, L.; Griffith, C.M.; Cai, Y.; Rose, G.M.; Yan, X.; Patrylo, P.R. Glucose tolerance and insulin sensitivityare impaired in APP/PS1 transgenic mice prior to amyloid plaque pathogenesis and cognitive decline. Exp. Gerontol. 2017, 88, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Youmans, K.L.; Tai, L.M.; Nwabuisi-Heath, E.; Jungbauer, L.; Kanekiyo, T.; Gan, M.; Kim, J.; Eimer, W.A.; Estus, S.; Rebeck, G.W.; et al. APOE4-specific changes in Abeta accumulation in a new transgenic mouse model of Alzheimer disease. J. Biol. Chem. 2012, 287, 41774–41786. [Google Scholar] [CrossRef] [PubMed]
- Antunes, M.; Biala, G. The novel object recognition memory: Neurobiology, test procedure, and its modifications. Cogn. Process. 2012, 13, 93–110. [Google Scholar] [CrossRef]
- Denninger, J.K.; Smith, B.M.; Kirby, E.D. Novel Object Recognition and Object Location Behavioral Testing in Mice on a Budget. J. Vis. Exp. 2018, 141. [Google Scholar] [CrossRef]
- Kratz, M.; Coats, B.R.; Hisert, K.B.; Hagman, D.; Mutskov, V.; Peris, E.; Schoenfelt, K.Q.; Kuzma, J.N.; Larson, I.; Billing, P.S.; et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014, 20, 614–625. [Google Scholar] [CrossRef]
- Farmer, B.C.; Kluemper, J.; Johnson, L.A. Apolipoprotein E4 Alters Astrocyte Fatty Acid Metabolism and Lipid Droplet Formation. Cells 2019, 8, 182. [Google Scholar] [CrossRef]
- Casimiro, I.; Stull, N.D.; Tersey, S.A.; Mirmira, R.G. Phenotypic sexual dimorphism in response to dietary fat manipulation in C57BL/6J mice. J. Diabetes Complicat. 2021, 35, 107795. [Google Scholar] [CrossRef]
- Gelineau, R.R.; Arruda, N.L.; Hicks, J.A.; Monteiro De Pina, I.; Hatzidis, A.; Seggio, J.A. The behavioral and physiological effects of high-fat diet and alcohol consumption: Sex differences in C57BL6/J mice. Brain Behav. 2017, 7, e00708. [Google Scholar] [CrossRef]
- Oraha, J.; Enriquez, R.F.; Herzog, H.; Lee, N.J. Sex-specific changes in metabolism during the transition from chow to high-fat diet feeding are abolished in response to dieting in C57BL/6J mice. Int. J. Obes. 2022, 46, 1749–1758. [Google Scholar] [CrossRef]
- Habib, S.M.; Zwicker, B.L.; Wykes, L.; Agellon, L.B. Sexually dimorphic response of mice to the Western-style diet caused by deficiency of fatty acid binding protein 6 (Fabp6). Physiol. Rep. 2021, 9, e14733. [Google Scholar] [CrossRef] [PubMed]
- Barron, A.M.; Rosario, E.R.; Elteriefi, R.; Pike, C.J. Sex-specific effects of high fat diet on indices of metabolic syndrome in 3xTg-AD mice: Implications for Alzheimer’s disease. PLoS ONE 2013, 8, e78554. [Google Scholar] [CrossRef]
- Salinero, A.E.; Anderson, B.M.; Zuloaga, K.L. Sex differences in the metabolic effects of diet-induced obesity vary by age of onset. Int. J. Obes. 2018, 42, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Riant, E.; Waget, A.; Cogo, H.; Arnal, J.; Burcelin, R.; Gourdy, P. Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology 2009, 150, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Yokota-Nakagi, N.; Takahashi, H.; Kawakami, M.; Takamata, A.; Uchida, Y.; Morimoto, K. Estradiol Replacement Improves High-Fat Diet-Induced Obesity by Suppressing the Action of Ghrelin in Ovariectomized Rats. Nutrients 2020, 12, 907. [Google Scholar] [CrossRef] [PubMed]
- Stubbins, R.E.; Holcomb, V.B.; Hong, J.; Nunez, N.P. Estrogen modulates abdominal adiposity and protects female mice from obesity and impaired glucose tolerance. Eur. J. Nutr. 2012, 51, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Huebbe, P.; Dose, J.; Schloesser, A.; Campbell, G.; Gluer, C.; Gupta, Y.; Ibrahim, S.; Minihane, A.; Baines, J.F.; Nebel, A.; et al. Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization-Studies in gene-targeted replacement mice. Mol. Nutr. Food Res. 2015, 59, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Arbones-Mainar, J.M.; Johnson, L.A.; Torres-Perez, E.; Garcia, A.E.; Perez-Diaz, S.; Raber, J.; Maeda, N. Metabolic shifts toward fatty-acid usage and increased thermogenesis are associated with impaired adipogenesis in mice expressing human APOE4. Int. J. Obes. 2016, 40, 1574–1581. [Google Scholar] [CrossRef]
- Jones, N.S.; Watson, K.Q.; Rebeck, G.W. Metabolic Disturbances of a High-Fat Diet Are Dependent on APOE Genotype and Sex. eNeuro 2019, 6, ENEURO.0267-19.2019. [Google Scholar] [CrossRef]
- To, A.W.M.; Ribe, E.M.; Chuang, T.T.; Schroeder, J.E.; Lovestone, S. The epsilon3 and epsilon4 alleles of human APOE differentially affect tau phosphorylation in hyperinsulinemic and pioglitazone treated mice. PLoS ONE 2011, 6, e16991. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Zhao, L. Human ApoE Isoforms Differentially Modulate Brain Glucose and Ketone Body Metabolism: Implications for Alzheimer’s Disease Risk Reduction and Early Intervention. J. Neurosci. 2018, 38, 6665–6681. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.K.; Uy, R.A.Z.; Vidoni, E.D.; Wilkins, H.M.; Archer, A.E.; Thyfault, J.P.; Miles, J.M.; Burns, J.M. Effect of APOE epsilon4 Genotype on Metabolic Biomarkers in Aging and Alzheimer’s Disease. J. Alzheimers Dis. 2017, 58, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Pontifex, M.G.; Martinsen, A.; Saleh, R.N.M.; Harden, G.; Tejera, N.; Muller, M.; Fox, C.; Vauzour, D.; Minihane, A. APOE4 genotype exacerbates the impact of menopause on cognition and synaptic plasticity in APOE-TR mice. FASEB J. 2021, 35, e21583. [Google Scholar] [CrossRef] [PubMed]
- Moser, V.A.; Pike, C.J. Obesity Accelerates Alzheimer-Related Pathology in APOE4 but not APOE3 Mice. eNeuro 2017, 4, ENEURO.0077-17.2017. [Google Scholar] [CrossRef]
- Nam, K.N.; Wolfe, C.M.; Fitz, N.F.; Letronne, F.; Castranio, E.L.; Mounier, A.; Schug, J.; Lefterov, I.; Koldamova, R. Integrated approach reveals diet, APOE genotype and sex affect immune response in APP mice. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 152–161. [Google Scholar] [CrossRef]
- Tzioras, M.; Davies, C.; Newman, A.; Jackson, R.; Spires-Jones, T. Invited Review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2019, 45, 327–346. [Google Scholar] [CrossRef]
- Tai, L.M.; Ghura, S.; Koster, K.P.; Liakaite, V.; Maienschein-Cline, M.; Kanabar, P.; Collins, N.; Ben-Aissa, M.; Lei, A.Z.; Bahroos, N.; et al. APOE-modulated Abeta-induced neuroinflammation in Alzheimer’s disease: Current landscape, novel data, and future perspective. J. Neurochem. 2015, 133, 465–488. [Google Scholar] [CrossRef]
- Tomiga, Y.; Yoshimura, S.; Ito, A.; Nakashima, S.; Kawanaka, K.; Uehara, Y.; Tanaka, H.; Higaki, Y. Exercise training rescues high fat diet-induced neuronal nitric oxide synthase expression in the hippocampus and cerebral cortex of mice. Nitric Oxide 2017, 66, 71–77. [Google Scholar] [CrossRef]
- Almeida-Suhett, C.P.; Graham, A.; Chen, Y.; Deuster, P. Behavioral changes in male mice fed a high-fat diet are associated with IL-1beta expression in specific brain regions. Physiol. Behav. 2017, 169, 130–140. [Google Scholar] [CrossRef]
- Machlovi, S.I.; Neuner, S.M.; Hemmer, B.M.; Khan, R.; Liu, Y.; Huang, M.; Zhu, J.D.; Castellano, J.M.; Cai, D.; Marcora, E.; et al. APOE confers transcriptomic and functional alterations to primary mouse microglia. Neurobiol. Dis. 2022, 164, e105615. [Google Scholar] [CrossRef]
- Jones, N.S.; Watson, K.Q.; Rebeck, G.W. High-fat diet increases gliosis and immediate early gene expression in APOE3 mice, but not APOE4 mice. J. Neuroinflamm. 2021, 18, 214. [Google Scholar] [CrossRef] [PubMed]
- Janssen, C.I.F.; Jansen, D.; Mutsaers, M.P.C.; Dederen, P.J.W.C.; Geenen, B.; Mulder, M.T.; Kiliaan, A.J. The Effect of a High-Fat Diet on Brain Plasticity, Inflammation and Cognition in Female ApoE4-Knockin and ApoE-Knockout Mice. PLoS ONE 2016, 11, e0155307. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).