Metabolomics-Based Profiling via a Chemometric Approach to Investigate the Antidiabetic Property of Different Parts and Origins of Pistacia lentiscus L.
Abstract
:1. Introduction
- (i)
- Evaluate α-glucosidase inhibition in P. lentiscus samples and identify compounds more positively correlated to α-glucosidase inhibition using a metabolomic approach;
- (ii)
- Study the variation of this inhibition according to the organ (i.e., leaf, stem bark and fruit) and the geographical origin (i.e., mountain and littoral) of the plant P. lentiscus. This metabolomic method was based on the link between phytochemical profiling by ultra-high-performance liquid chromatography coupled to high-resolution mass spectrometry (UHPLC-ESI-HRMS) and α-glucosidase dosage data.
2. Materials and Methods
2.1. Chemicals
2.2. Plant Collection
2.3. Drying and Extraction
2.4. UHPLC-ESI-HRMS Analysis of the Extracts
2.5. Glucosidase-Inhibitory Activity
2.6. Metabolomic and Chemometric Analysis
2.7. Intensity Variation of the Metabolites Identified
3. Results
3.1. α-Glucosidase-Inhibitory Activity
3.2. Metabolomic and Chemometric Analysis
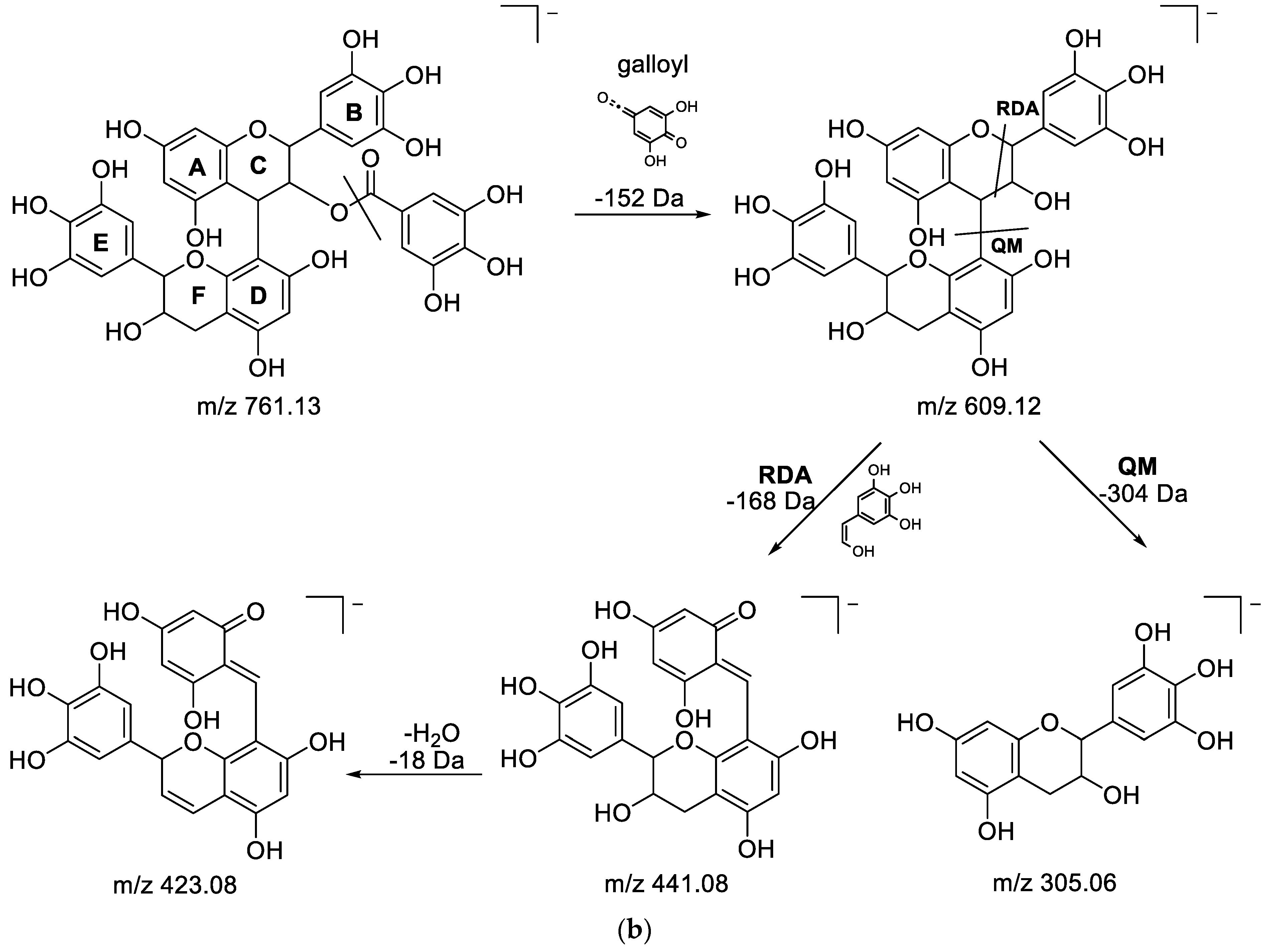
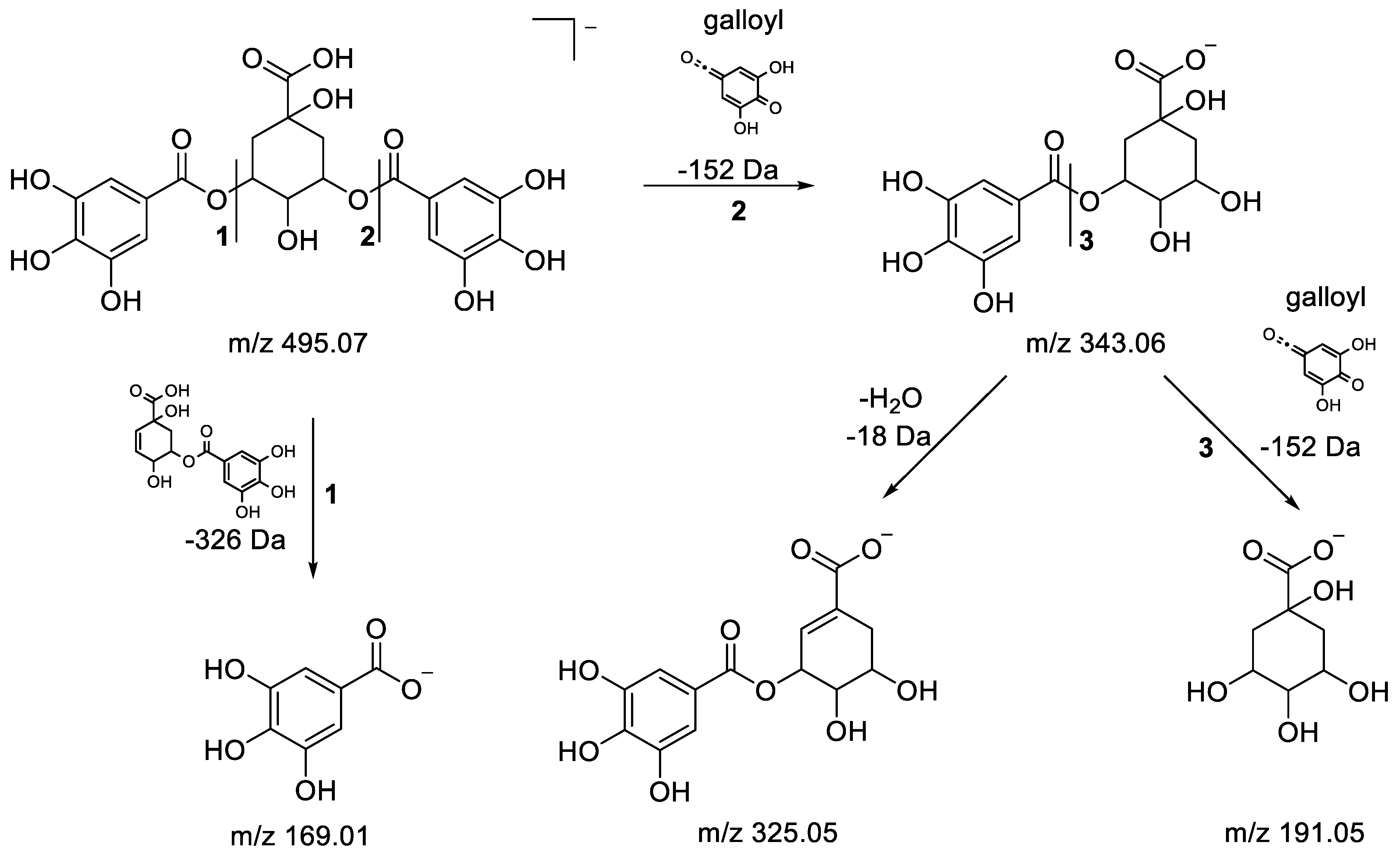
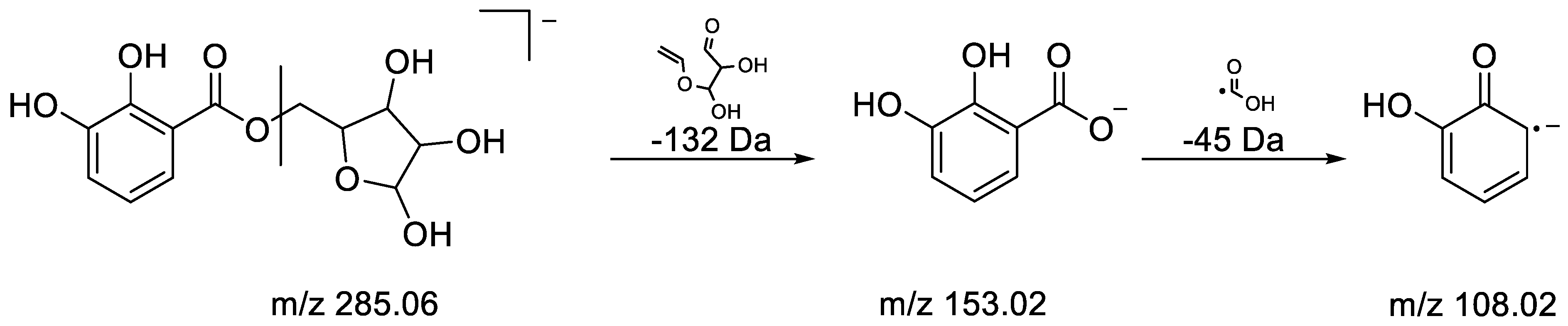
3.3. UHPLC-ESI-HRMS Identification of α-Glucosidase Inhibitory Metabolites
3.4. Intensity Variation of the Metabolites Identified
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benalla, W.; Bellahcen, S.; Bnouham, M. Antidiabetic medicinal plants as a source of alpha glucosidase inhibitors. Curr. Diabetes Rev. 2010, 6, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.A.; Alcolado, J.C. Animal models of diabetes mellitus. Diabet. Med. 2005, 22, 359–370. [Google Scholar] [CrossRef]
- Sudesna, C.; Khunti, K.; Davies, M.J. Type 2 diabetes. Lancet 2017, 389, 2239–2251. [Google Scholar]
- Vinholes, J.; Vizzotto, M. Synergisms in alpha-glucosidase inhibition and antioxidant activity of camellia sinensis l. kuntze and Eugeniauniflora L. ethanolic extracts. Pharmacogn. Res. 2017, 9, 101. [Google Scholar] [CrossRef]
- Hedrington, M.S.; Davis, S.N. Considerations when using alpha-glucosidase inhibitors in the treatment of type 2 diabetes. Expert Opin. Pharmacother. 2019, 20, 2229–2235. [Google Scholar] [CrossRef]
- Nakhaee, A.; Sanjari, M. Evaluation of effect of acarbose consumption on weight losing in non-diabetic overweight or obese patients in Kerman. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2013, 18, 391. [Google Scholar]
- Kast, R. Acarbose related diarrhea: Increased butyrate upregulates prostaglandin. E. Inflamm. Res. 2002, 51, 117–118. [Google Scholar] [CrossRef]
- Apostolidis, E.; Kwon, Y.I.; Shetty, K. Inhibitory potential of herb, fruit, and fungal-enriched cheese against key enzymes linked to type 2 diabetes and hypertension. Innov. Food Sci. Emerg. Technol. 2007, 8, 46–54. [Google Scholar] [CrossRef]
- Andrade, R.J.; Lucena, M.; Vega, J.L.; Torres, M. Acarbose-associated hepatotoxicity. Diabetes Care 1998, 21, 2029. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Ohhira, M.; Miyokawa, N.; Kitamori, S.; Kohgo, Y. Acarbose- induced hepatic injury. Lancet 1998, 351, 340. [Google Scholar] [CrossRef] [PubMed]
- Gentile, S.; Turco, S.; Guarino, G.; Sasso, F.C.; Torella, R. Aminotransferase activity and acarbose treatment in patients with type 2 diabetes. Diabetes Care 1999, 22, 1217. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Remorini, D.; Pinelli, P.; Agati, G.; Saracini, E.; Traversi, M.L.; Massai, R. Morpho-anatomical, physiological and biochemical adjustments in response to root zone salinity stress and high solar radiation in two Mediterranean evergreen shrubs, Myrtuscommunis and Pistacia lentiscus. New Phytol. 2006, 170, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, H.; Renaud, J.; Herchi, W.; Boukhchina, S.; Mayer, P. Triacylglycerols and aliphatic alcohols from fruits of three Tunisian Pistacia lentiscus populations. J. Sci. Food Agric. 2015, 95, 2028–2032. [Google Scholar] [CrossRef] [PubMed]
- Nabila, B.; Fawzia, A.B.; Tatjana, K.P. Antioxidant and antimicrobial activities of the Pistacia lentiscus and Pistacia atlantica extracts. Afr. J. Pharm. Pharmacol. 2008, 2, 22–28. [Google Scholar]
- Boudieb, K.; Kaki, S.A.S.A.; Amellal-Chibane, H. Traditional uses, phytochemical study and morphological characterization of Pistacia lentiscus L. fruits from three areas of northern Algeria. J. Appl. Biosci. 2019, 135, 13788–13797. [Google Scholar] [CrossRef]
- Saiah, H.; Allem, R.; Kebir, F.Z.R. Antioxidant and antibacterial activities of six Algerian medicinalplants. Int. J. Pharm. Pharm. Sci. 2016, 8, 367–374. [Google Scholar]
- Bouyahya, A.; El Omari, N.; Elmenyiy, N.; Guaouguaou, F.E.; Balahbib, A.; Belmehdi, O.; Salhi, N.; Imtara, H.; Mrabti, H.N.; El-Shazly, M.; et al. Moroccan antidiabetic medicinal plants: Ethnobotanical studies, phytochemical bioactive compounds, preclinical investigations, toxicological validations and clinical evidences; challenges, guidance and perspectives for future management of diabetes worldwide. Trends Food Sci. Technol. 2021, 115, 147–254. [Google Scholar]
- El Bishbishy, M.H.; Gad, H.A.; Aborehab, N.M. Chemiometric discrimination of three Pistacia species via their metabolic profiling and their possible in vitro effects on memory functions. J. Pharm. Biomed. Anal. 2020, 177, 112840. [Google Scholar] [CrossRef]
- Umadevi, I.; Daniel, M.; Sabnis, S.D. Chemotaxonomic studies on some members of Anacardiaceae. Proc. Plant Sci. 1988, 98, 205–208. [Google Scholar] [CrossRef]
- Romani, A.; Pinelli, P.; Galardi, C.; Mulinacci, N.; Tattini, M. Identification and quantification of galloyl derivatives, flavonoid glycosides and anthocyanins in leaves of Pistacia lentiscus L. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2002, 13, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Yemmen, M.; Landolsi, A.; Hamida, J.B.; Mégraud, F.; Ayadi, M.T. Antioxidant activities, anticancer activity and polyphenolics profile, of leaf, fruit and stem extracts of Pistacia lentiscus from Tunisia. Cell. Mol. Biol. 2017, 63, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Mehenni, C.; Atmani-Kilani, D.; Dumarçay, S.; Perrin, D.; Gérardin, P.; Atmani, D. Hepatoprotective and antidiabetic effects of Pistacia lentiscus leaf and fruit extracts. J. Food Drug Anal. 2016, 24, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Trifan, A.; Zengin, G.; Sinan, K.I.; Wolfram, E.; Skalicka-Woźniak, K.; Luca, S.V. LC-HRMS/MS phytochemical profiling of Symphytumofficinale L. and Anchusaochroleuca M. Bieb.(Boraginaceae): Unveiling their multi-biological potential via an integrated approach. J. Pharm. Biomed. Anal. 2021, 204, 114283. [Google Scholar] [CrossRef] [PubMed]
- Rocchetti, G.; Miras-Moreno, M.B.; Zengin, G.; Senkardes, I.; Sadeer, N.B.; Mahomoodally, M.F.; Lucini, L. UHPLC-QTOF-MS phytochemical profiling and in vitro biological properties of Rhamnuspetiolaris (Rhamnaceae). Ind. Crops Prod. 2019, 142, 111856. [Google Scholar] [CrossRef]
- Farag, M.A.; Sakna, S.T.; El-Fiky, N.M.; Shabana, M.M.; Wessjohann, L.A. Phytochemical, antioxidant and antidiabetic evaluation of eight Bauhinia L. species from Egypt using UHPLC–PDA–qTOF-MS and chemiometrics. Phytochemistry 2015, 119, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Syabana, M.A.; Yuliana, N.D.; Batubara, I.; Fardiaz, D. α-glucosidase inhibitors from Syzygiumpolyanthum (Wight) Walp leaves as revealed by metabolomics and in silico approaches. J. Ethnopharmacol. 2022, 282, 114618. [Google Scholar] [CrossRef] [PubMed]
- Assefa, S.T.; Yang, E.Y.; Asamenew, G.; Kim, H.W.; Cho, M.C.; Lee, J. Identification of α-Glucosidase Inhibitors from Leaf Extract of Pepper (Capsicum spp.) through Metabolomic Analysis. Metabolites 2021, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Sehaki, C.; Jullian, N.; Ayati, F.; Fernane, F.; Gontier, E. A Review of Pistacia lentiscus Polyphenols: Chemical Diversity and Pharmacological Activities. Plants 2023, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Foddai, M.; Kasabri, V.; Afifi, F.U.; Azara, E.; Petretto, G.L.; Pintore, G. In vitro inhibitory effects of Sardinian Pistacia lentiscus L. and Pistacia terebinthus L. on metabolic enzymes: Pancreatic lipase, α-amylase, and α-glucosidase. Starch-Stärke 2015, 67, 204–212. [Google Scholar] [CrossRef]
- Benhammou, N.; Bekkara, F.A.; Panovska, T.K. Antiradical capacity of the phenolic compounds of Pistacia lentiscus L. and Pistacia atlanticadesf. Adv. Food Sci. 2007, 29, 155–161. [Google Scholar]
- Ghenima, A.I.; Idir, M.; Nadjet, M.G.; Samia, M.A.; Mihoub, Z.M.; Karim, H. In vitro evaluation of biological activities of Pistacia lentiscus aqueous extract. Int. J. Pharm. Pharm. Sci. 2015, 7, 133–139. [Google Scholar]
- Amessis-Ouchemoukh, N.; Madani, K.; Falé, P.L.; Serralheiro, M.L.; Araújo, M.E.M. Antioxidant capacity and phenolic contents of some Mediterranean medicinal plants and their potential role in the inhibition of cyclooxygenase-1 and acetylcholinesterase activities. Ind. Crops Prod. 2014, 53, 6–15. [Google Scholar] [CrossRef]
- Belhachat, D.; Aid, F.; Mekimene, L.; Belhachat, M. Phytochemical screening and in vitro antioxidant activity of Pistacia lentiscus berries ethanolic extract growing in Algeria. Mediterr. J. Nutr. Metab. 2017, 10, 273–285. [Google Scholar] [CrossRef]
- Salhi, A.; Bellaouchi, R.; El Barkany, S.; Rokni, Y.; Bouyanzer, A.; Asehraou, A.; Amhamdi, H.; Hammouti, B. Total phenolic content, antioxidant and antimicrobial activities of extracts from Pistacia lentiscus leaves. Casp. J. Environ. Sci. 2019, 17, 189–198. [Google Scholar]
- Remila, S.; Atmani-Kilani, D.; Delemasure, S.; Connat, J.L.; Azib, L.; Richard, T.; Atmani, D. Antioxidant, cytoprotective, anti-inflammatory and anticancer activities of Pistacia lentiscus (Anacardiaceae) leaf and fruit extracts. Eur. J. Integr. Med. 2015, 7, 274–286. [Google Scholar] [CrossRef]
- Azib, L.; Debbache-Benaida, N.; Da Costa, G.; Atmani-Kilani, D.; Saidene, N.; Ayouni, K.; Richard, T.; Atmani, D. Pistacia lentiscus L. leaves extract and its major phenolic compounds reverse aluminium-induced neurotoxicity in mice. Ind. Crops Prod. 2019, 137, 576–584. [Google Scholar] [CrossRef]
- Bakli, S.; Daoud, H.; Amina, Z.; Nouari, S.; Asma, B.; Soufiane, G.; Oumaima, N. Antimicrobial and AntioxidantActivities of FlavonoidsExtractedfrom Pistacia lentiscus L., Leaves. J. Drug Deliv. Ther. 2020, 10, 83–89. [Google Scholar] [CrossRef]
- Iauk, L.; Ragusa, S.; Rapisarda, A.; Franco, S.; Nicolosi, V.M. In vitro antimicrobial activity of Pistacia lentiscus L. extracts: Preliminary report. J. Chemother. 1996, 8, 207–209. [Google Scholar] [CrossRef]
- Alhadad, A.O.; Salem, G.S.; Elmhdwi, M.F.; Hussein, S.M.; Elshareef, S.M. Assessments of Antibacterial and Antioxidant Properties in the Methanolic and Aqueous Leaf Extracts of Pistacia lentiscus against Different Antibiotic Resistance Pathogenic Bacteria. Adv. Biosci. Biotechnol. 2022, 13, 113–133. [Google Scholar] [CrossRef]
- Mansour-Djaalab, H.; Kahlouche-Riachi, F.; Djerrou, Z.; Serakta-Delmi, M.; Hamimed, S.; Trifa, W.; Djaalab, I.; HamdiPacha, Y. In vitro evaluation of antifungal effects of Lawsoniainermis, Pistacia lentiscus and Juglans regia. Int. J. Med. Aromat. Plants 2012, 2, 263–268. [Google Scholar]
- Kirollos, F.N.; Elhawary, S.S.; Salama, O.M.; Elkhawas, Y.A. LC-ESI-MS/MS and cytotoxic activity of three Pistacia species. Nat. Prod. Res. 2019, 33, 1747–1750. [Google Scholar] [CrossRef]
- Pacifico, S.; Piccolella, S.; Marciano, S.; Galasso, S.; Nocera, P.; Piscopo, V.; Fiorentino, A.; Monaco, P. LC-MS/MS profiling of a mastic leaf phenol enriched extract and its effects on H2O2 and Aβ (25–35) oxidative injury in SK-B-NE (C)-2 cells. J. Agric. Food Chem. 2014, 62, 11957–11966. [Google Scholar] [CrossRef] [PubMed]
- Siano, F.; Cutignano, A.; Moccia, S.; Russo, G.L.; Volpe, M.G.; Picariello, G. (Phytochemical characterization and effects on cell proliferation of lentisk (Pistacia lentiscus) berry oil: A revalued source of phenolic. Plant Foods Hum. Nutr. 2020, 75, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Bouriche, H.; Saidi, A.; Ferradji, A.; Belambri, S.A.; Senator, A. Anti-inflammatory and immunomodulatory properties of Pistacia lentiscus extracts. J. Appl. Pharm. Sci. 2016, 6, 140–146. [Google Scholar] [CrossRef]
- Bouriche, H.; Khalfaoui, S.; Meziti, H.; Senotor, A. Anti-inflammatory activity of acetonic extract of Pistacia lentiscus fruits. TOJSAT 2013, 3, 40–48. [Google Scholar]
- Dellai, A.; Souissi, H.; Borgi, W.; Bouraoui, A.; Chouchane, N. Antiinflammatory and antiulcerogenic activities of Pistacia lentiscus L. leaves extracts. Ind. Crops Prod. 2013, 49, 879–882. [Google Scholar] [CrossRef]
- Parizad, S.; Dizadji, A.; Habibi, M.K.; Winter, S.; Kalantari, S.; Movi, S.; Tendero, K.L.; Alonso, G.; Moratalla-Lopez, N. The effects of geographical origin and virus infection on the saffron (Crocus sativus L.) quality. Food Chem. 2019, 295, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.K.; Jamali, A.; Grand, E.; Morreel, K.; Marcelo, P.; Gontier, E.; Dauwe, R. Phenylpropanoid profiling reveals a class of hydroxycinnamoylglucaric acid conjugates in Isatistinctoria leaves. Phytochemistry 2017, 144, 127–140. [Google Scholar] [CrossRef]
- Tchoumtchoua, J.; Mathiron, D.; Pontarin, N.; Gagneul, D.; Van Bohemen, A.I.; OtogoN’Nang, E.; Mesnard, F.; Petit, E.; Fontaine, J.X.; Roland, M.; et al. Phenolic profiling of flax highlights contrasting patterns in winter and spring varieties. Molecules 2019, 24, 4303. [Google Scholar] [CrossRef]
- Bachhawat, J.A.; Shihabudeen, M.S.; Thirumurugan, K. Screening of fifteen Indian ayurvedic plants for alpha-glucosidase inhibitory activity and enzyme kinetics. Int. J. Pharm. Pharm. Sci. 2011, 3, 267–274. [Google Scholar]
- Boccard, J.; Rutledge, D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Acta 2013, 769, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Thorsten, P. Calculate Pairwise Multiple Comparisons of Mean Rank Sums. R Package Version PMCMRplus 1.9.4. 2021. Available online: https://cran.r-project.org/web/packages/PMCMRplus/index.html (accessed on 1 February 2022).
- Graves, S.; Piepho, H.P.; Selzer, M.L. Package ‘multcompView’. Vis. Paired Comp. 2015. Available online: https://cran.stat.unipd.it/web/packages/multcompView/multcompView.pdf (accessed on 1 February 2022).
- Singh, A.; Kumar, S.; Kumar, B. LC-MS Identification of Proanthocyanidins in Bark and Fruit of six Terminalia species. Nat. Prod. Commun. 2018, 13, 555–560. [Google Scholar] [CrossRef]
- Escobar-Avello, D.; Lozano-Castellón, J.; Mardones, C.; Pérez, A.J.; Saéz, V.; Riquelme, S.; Von Bear, D.; Vallverdú-Queralt, A. Phenolic profile of grape canes: Novel compounds identified by LC-ESI-LTQ-Orbitrap-MS. Molecules 2019, 24, 3763. [Google Scholar]
- Abu-Reidah, I.M.; del Mar Contreras, M.; Arráez-Román, D.; Fernández-Gutiérrez, A.; Segura-Carretero, A. UHPLC-ESI-QTOF-MS-based metabolic profiling of Viciafaba L. (Fabaceae) seeds as a key strategy for characterization in foodomics. Electrophoresis 2014, 35, 1571–1581. [Google Scholar] [CrossRef]
- Jaiswal, R.; Jayasinghe, L.; Kuhnert, N. Identification and characterization of proanthocyanidins of 16 members of the Rhododendron genus (Ericaceae) by tandem LC–MS. J. Mass Spectrom. 2012, 47, 502–515. [Google Scholar] [CrossRef]
- D’Urso, G.; Montoro, P.; Piacente, S. Detection and comparison of phenolic compounds in different extracts of black currant leaves by liquid chromatography coupled with high-resolution ESI-LTQ-Orbitrap MS and high-sensitivity ESI-Qtrap MS. J. Pharm. Biomed. Anal. 2020, 179, 112926. [Google Scholar] [CrossRef]
- Russo, D.; Kenny, O.; Smyth, T.J.; Milella, L.; Hossain, M.B.; Diop, M.S.; Rai, D.K.; Brunton, N.P. Profiling of phytochemicals in tissues from Sclerocaryabirrea by HPLC-MS and their link with antioxidant activity. Int. Sch. Res. Not. 2013, 2013, 10. [Google Scholar]
- Teixeira, N.; Azevedo, J.; Mateus, N.; de Freitas, V. Proanthocyanidin screening by LC–ESI-MS of Portuguese red wines made with teinturier grapes. Food Chem. 2016, 190, 300–307. [Google Scholar] [CrossRef]
- Miketova, P.; Schram, K.H.; Whitney, J.; Li, M.; Huang, R.; Kerns, E.; Valcic, S.; Timmermann, B.N.; Rourick, R.; Klohr, S. Tandem mass spectrometry studies of green tea catechins. Identification of three minor components in the polyphenolic extract of green tea. J. Mass Spectrom. 2000, 35, 860–869. [Google Scholar] [CrossRef]
- Delcambre, A.; André, Y.; Saucier, C. Sequencing of red wine proanthocyanidins by UHPLC-ESI-QToF. J. Appl. Bioanal. 2015, 1, 46–54. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, C.; Quirantes-Piné, R.; Amessis-Ouchemoukh, N.; Madani, K.; Segura-Carretero, A.; Fernández-Gutierrez, A. A metabolite-profiling approach allows the identification of new compounds from Pistacia lentiscus leaves. J. Pharm. Biomed. Anal. 2013, 77, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Daou, M.; Elnaker, N.A.; Ochsenkühn, M.A.; Amin, S.A.; Yousef, A.F.; Yousef, L.F. In vitro α-glucosidase inhibitory activity of Tamarixnilotica shoot extracts and fractions. PLoS ONE. 2022, 17, e0264969. [Google Scholar] [CrossRef] [PubMed]
- Nokhala, A.; Siddiqui, M.J.; Ahmed, Q.U.; Ahamad Bustamam, M.S.; Zakaria, Z.A. Investigation of α-glucosidase inhibitory metabolites from Tetracera scandens leaves by GC–MS metabolite profiling and docking studies. Biomolecules. 2020, 10, 287. [Google Scholar] [CrossRef]
- Wan-Nadilah, W.A.; Akhtar, M.T.; Shaari, K.; Khatib, A.; Hamid, A.A.; Hamid, M. Variation in the metabolites and α-glucosidase inhibitory activity of Cosmos caudatus at different growth stages. BMC Complement. Altern. Med. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Saleh, M.S.; Jalil, J.; Mustafa, N.H.; Ramli, F.F.; Asmadi, A.Y.; Kamisah, Y. UPLC-MS-based metabolomics profiling for α-glucosidase inhibiting property of Parkiaspeciosa pods. Life. 2021, 11, 78. [Google Scholar] [CrossRef]
- William, J.; John, P.; Mumtaz, M.W.; Ch, A.R.; Adnan, A.; Mukhtar, H.; Sharif, S.; Raza, S.A.; Akhtar, M.T. Antioxidant activity, α-glucosidase inhibition and phytochemical profiling of Hyophorbelagenicaulis leaf extracts. PeerJ. 2019, 7, e7022. [Google Scholar] [CrossRef]
- Abo-Salem, O.M.; Ali, T.M.; Harisa, G.I.; Mehanna, O.M.; Younos, I.H.; Almalki, W.H. Beneficial effects of (−)-epigallocatechin-3-O-gallate on diabetic peripheral neuropathy in the rat model. J. Biochem. Mol. Toxicol. 2020, 34, e22508. [Google Scholar] [CrossRef]
- Kamiyama, O.; Sanae, F.; Ikeda, K.; Higashi, Y.; Minami, Y.; Asano, N.; Adachi, I.; Kato, A. In vitro inhibition of α-glucosidases and glycogen phosphorylase by catechingallates in green tea. Food Chem. 2010, 122, 1061–1066. [Google Scholar] [CrossRef]
- Gamberucci, A.; Konta, L.; Colucci, A.; Giunti, R.; Magyar, J.E.; Mandl, J.; Banhegyi, G.; Bendetti, A.; Csala, M. Green tea flavonols inhibit glucosidase II. Biochem. Pharmacol. 2006, 72, 640–646. [Google Scholar] [CrossRef]
- Tadera, K.; Minami, Y.; Takamatsu, K.; Matsuoka, T. Inhibition of α-glucosidase and α-amylase by flavonoids. J. Nutr. Sci. Vitaminol. 2006, 52, 149–153. [Google Scholar] [CrossRef]
- Cai, Y.; Wu, L.; Lin, X.; Hu, X.; Wang, L. Phenolic profiles and screening of potential α-glucosidase inhibitors from Polygonumaviculare L. leaves using ultra-filtration combined with HPLC-ESI-qTOF-MS/MS and molecular docking analysis. Ind. Crops Prod. 2020, 154, 112673. [Google Scholar] [CrossRef]
- Quaresma, D.M.; Justino, A.B.; Sousa, R.M.; Munoz, R.A.; de Aquino, F.J.; Martins, M.M.; Goulart, L.R.; Pivatto, M.; Espindola, F.S.; de Oliveira, A. Antioxidant compounds from Banisteriopsis argyrophylla leaves as α-amylase, αglucosidase, lipase, and glycation inhibitors. Bioorganic Chem. 2020, 105, 104335. [Google Scholar] [CrossRef] [PubMed]
- Amel, Z.; Nabila, B.B.; Nacéra, G.; Fethi, T.; Fawzia, A.B. Assessment of phytochemical composition and antioxidant properties of extracts from the leaf, stem, fruit and root of Pistacia lentiscus L. Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 627–633. [Google Scholar]
- ElezGarofulić, I.; Kruk, V.; Martić, A.; Martić, I.; Zorić, Z.; Pedisić, S.; Dragović, S.; Dragović-Uzelac, V. Evaluation of polyphenolic profile and antioxidant activity of Pistacia lentiscus L. leaves and fruit extract obtained by optimized microwave-assisted extraction. Foods 2020, 9, 1556. [Google Scholar] [CrossRef]
- Aissat, A.K.; Chaher-Bazizi, N.; Richard, T.; Kilani-Atmani, D.; Pedrot, E.; Renouf, E.; Atmani, D.; Fonayet, J.V. Analysis of individual anthocyanins, flavanols, flavonols and other polyphenols in Pistacia lentiscus L. fruits during ripening. J. Food Compos. Anal. 2020, 106, 104286. [Google Scholar] [CrossRef]
- Sehaki, C.; Jullian, N.; Choque, E.; Dauwe, R.; Fontaine, J.X.; Molinie, R.; Ayati, F.; Fernane, F.; Gontier, E. Profiling of Essential Oils from the Leaves of Pistacia lentiscus Collected in the Algerian Region of Tizi-Ouzou: Evidence of Chemical Variations Associated with Climatic Contrasts between Littoral and Mountain Samples. Molecules 2022, 27, 4148. [Google Scholar] [CrossRef]

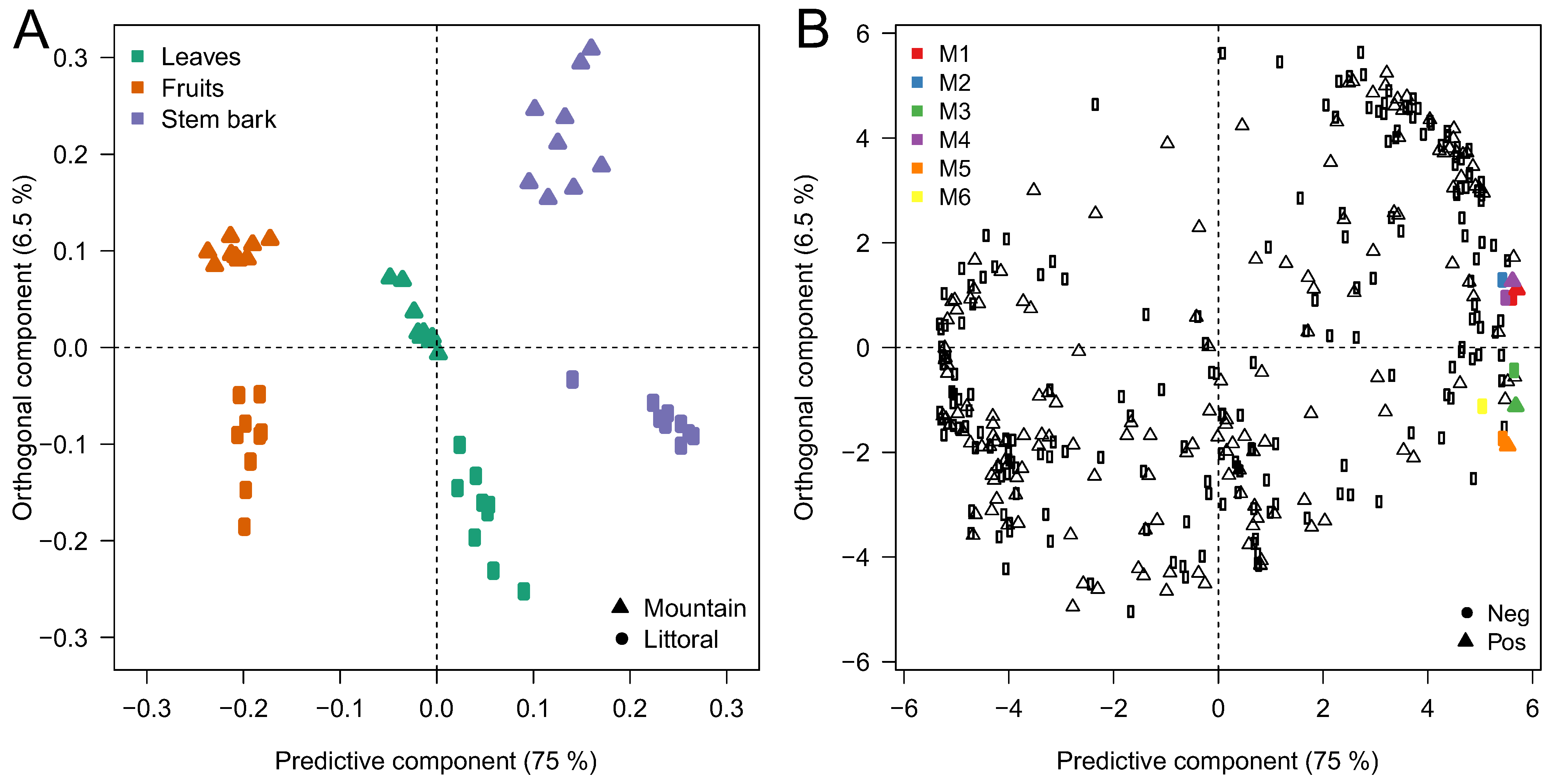



| Sites | Collected Date | 54 Samples (9 Trees × 3 Organs × 2 Sites) | Morphological Variations | Longitude | Latitude | Altitude | Annual Temperature | |
|---|---|---|---|---|---|---|---|---|
| Site 1 Ait-Irane Mountain | Mid-October 2019 | 9 lentisk trees (3 organs: leaves, stem bark and fruit) = 27 samples | Unremarkable for the same organ type | 36°29′58.3″ N | 4°04′43.4″ E | 876 m | Min | Max |
| 0.5 °C | 31.9 °C | |||||||
| Site 2 Tigzirt Littoral | Mid-October 2019 | 9 lentisk trees (3 organs: leaves, stem bark and fruit) = 27 samples | Unremarkable for the same organ type | 36°53′43.0″ N | 4°11′00.4″ E | 13 m | Min | Max |
| 7.4 °C | 27.9 °C | |||||||
| N° Metabolite | RT (min) | m/z (obs) [M-H]− | Error (ppm) | HDMSE Fragment Ions (Intensity, %) | Molecular Formula | Suggested Compound | Ref |
|---|---|---|---|---|---|---|---|
| 1 | 0.45 | 609.1242 | −1.26 | 441.0825 (2.99), 423.0722 (100), 305.0667 (29.49), 261.0397 (15.71), 219.0659 (14.46), 177.0186 (26.94), 125.0241(18.09) | C30H26O14 | Epigallocatechin(4a>8)epigallocatechin | [55,56,57] |
| 2 | 0.55 | 495.0774 | −2.03 | 343.0661(100), 325.0550(15.74), 191.0551(87.47), 169.0136(37.71) | C21H20O14 | 3,5-O-Digalloylquinic acid | [43] |
| 3 | 0.56 | 761.1339 | −3.46 | 609.1228(12.42), 591.1134(5.98), 465.0820 (12.76), 423.0820(100), 305.0658(46.73) | C37H30O18 | (Epi)gallocatechin-3′-Ogalloyl-(epi)gallocatechin | [58] |
| 4 | 0.61 | 609.1233 | −1.51 | 423.0716 (100) | C30H26O14 | Epigallocatechin(4a->8)epigallocatechi-n (isomer) | [57,58,59] |
| 5 | 0.72 | 761.1344 | −2.28 | 423.0715 (100) | C37H30O18 | (Epi)gallocatechin-3′-Ogalloyl-(epi)gallocatechin (isomer) | [58] |
| 6 | 0.73 | 285.0608 | −2.76 | 108.0209 (100) | C12H14O8 | Dihydroxy benzoic acid pentoside | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sehaki, C.; Molinie, R.; Mathiron, D.; Fontaine, J.-X.; Jullian, N.; Ayati, F.; Fernane, F.; Gontier, E. Metabolomics-Based Profiling via a Chemometric Approach to Investigate the Antidiabetic Property of Different Parts and Origins of Pistacia lentiscus L. Metabolites 2023, 13, 275. https://doi.org/10.3390/metabo13020275
Sehaki C, Molinie R, Mathiron D, Fontaine J-X, Jullian N, Ayati F, Fernane F, Gontier E. Metabolomics-Based Profiling via a Chemometric Approach to Investigate the Antidiabetic Property of Different Parts and Origins of Pistacia lentiscus L. Metabolites. 2023; 13(2):275. https://doi.org/10.3390/metabo13020275
Chicago/Turabian StyleSehaki, Chabha, Roland Molinie, David Mathiron, Jean-Xavier Fontaine, Nathalie Jullian, Fadila Ayati, Farida Fernane, and Eric Gontier. 2023. "Metabolomics-Based Profiling via a Chemometric Approach to Investigate the Antidiabetic Property of Different Parts and Origins of Pistacia lentiscus L." Metabolites 13, no. 2: 275. https://doi.org/10.3390/metabo13020275
APA StyleSehaki, C., Molinie, R., Mathiron, D., Fontaine, J.-X., Jullian, N., Ayati, F., Fernane, F., & Gontier, E. (2023). Metabolomics-Based Profiling via a Chemometric Approach to Investigate the Antidiabetic Property of Different Parts and Origins of Pistacia lentiscus L. Metabolites, 13(2), 275. https://doi.org/10.3390/metabo13020275






