Optimized Fast Filtration-Based Sampling and Extraction Enables Precise and Absolute Quantification of the Escherichia coli Central Carbon Metabolome
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Cultivation Conditions
2.2. Sampling by Conventional Cold Methanol Quenching
2.3. Fluorescence Microscopy for the Assessment of Cell Morphology, Size, and Membrane Integrity
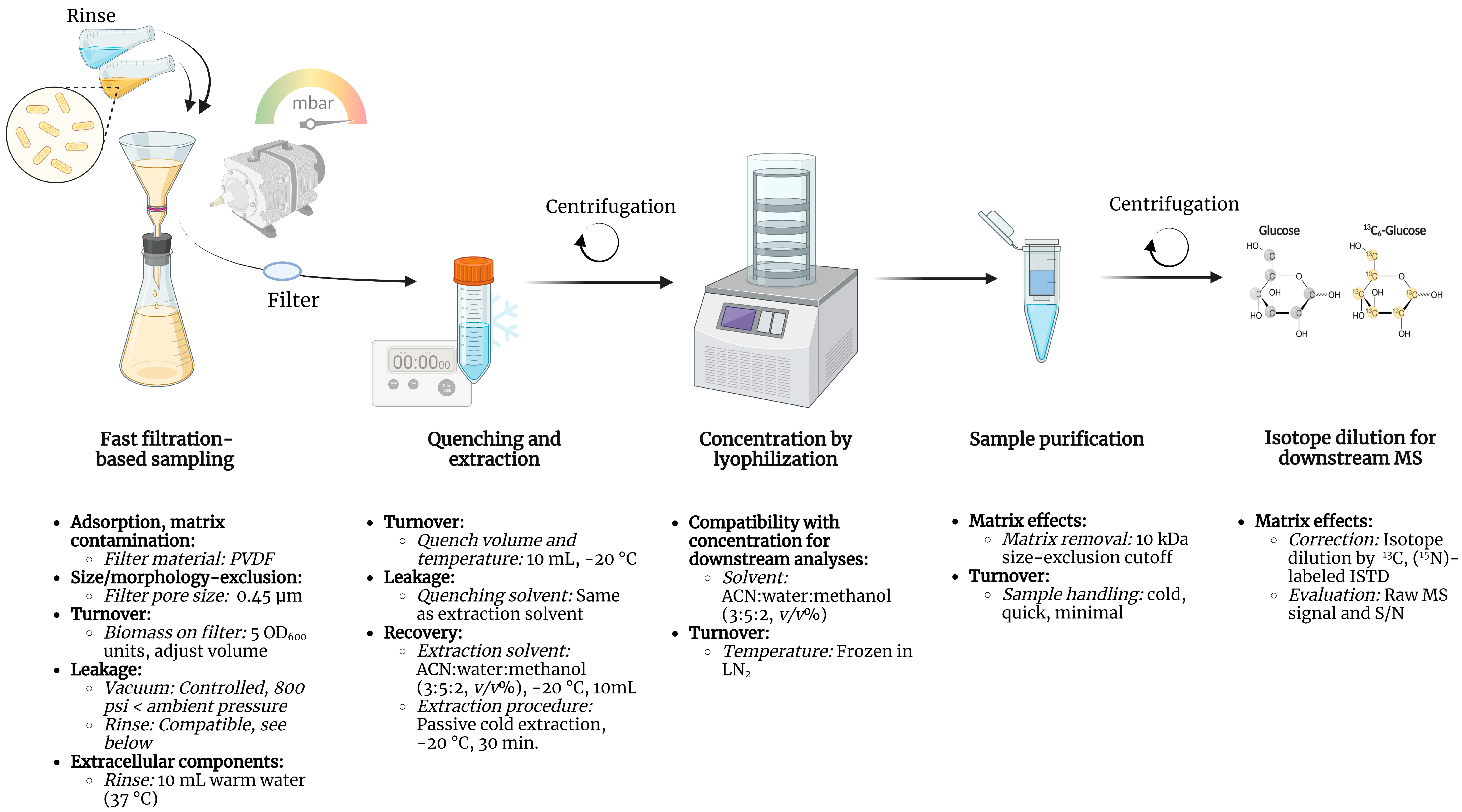
2.4. Sampling by Fast Filtration and Rinsing of Extracellular Components
2.5. Plate Colony Counts for Assessment of Bacteria in Filtered Flow-Through
2.6. Rapid Quenching of Metabolic Reactions Post Filtration
2.7. Metabolite Extraction, Concentration, and Purification
2.8. Flow Cytometric Measurements of Cell Density
2.9. Mass Spectrometric Quantification of Intracellular Amino Acids, Organic Acids, and Phosphorylated Metabolites
2.10. Mass Spectrometric Quantification of Intracellular Cyclic- and Pyridine Nucleotides
2.11. Dry-Weight Measurements
2.12. Normalization of Metabolite Extract Concentrations and Calculation of Intracellular Metabolite Concentrations
3. Results and Discussion
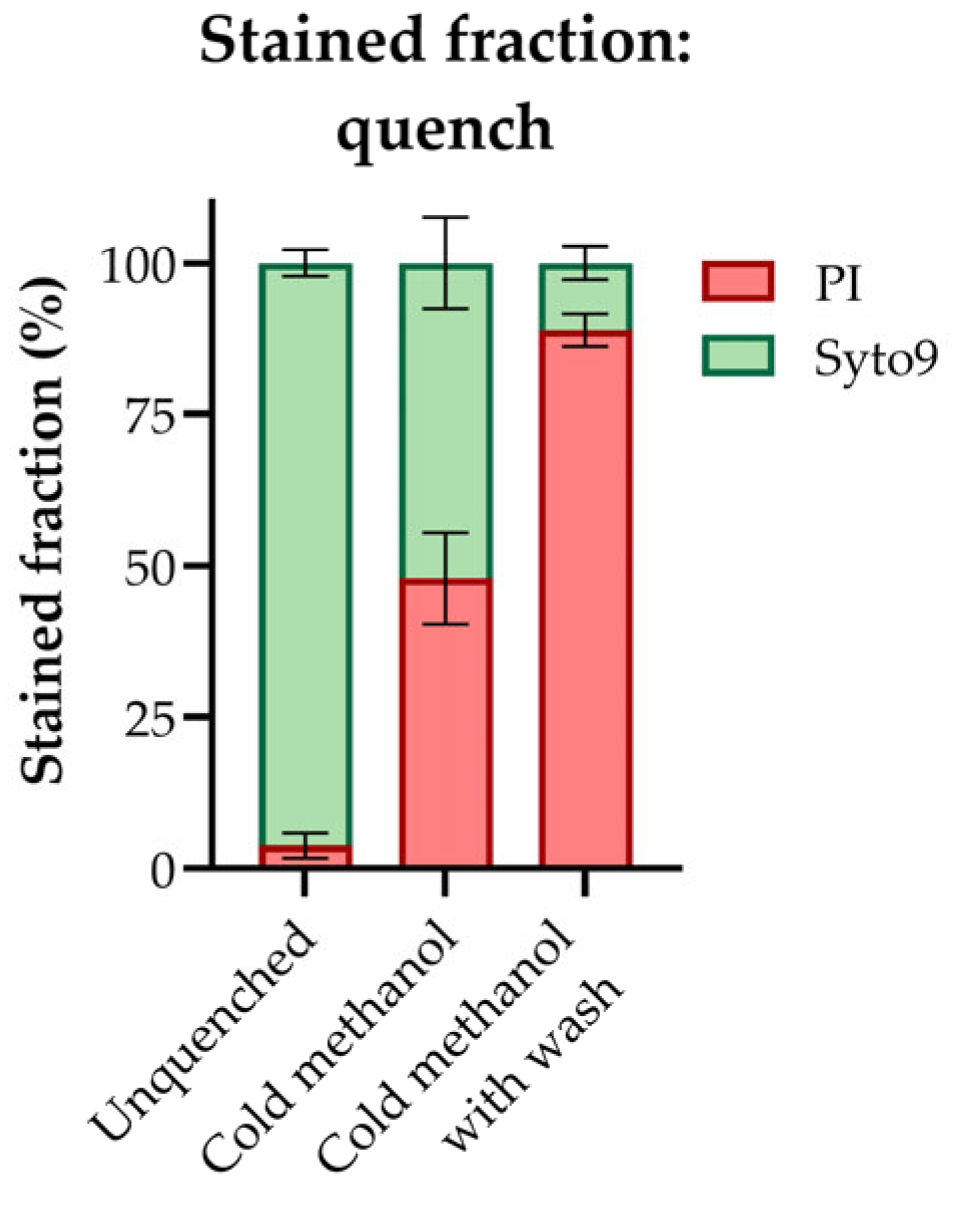
3.1. Sampling by Conventional “Cold Methanol Quenching” Permeabilizes the E. coli Cell Membrane
3.2. Filter Material and Pore Size Are Critical Parameters in Fast Filtration-Based Sampling Protocols
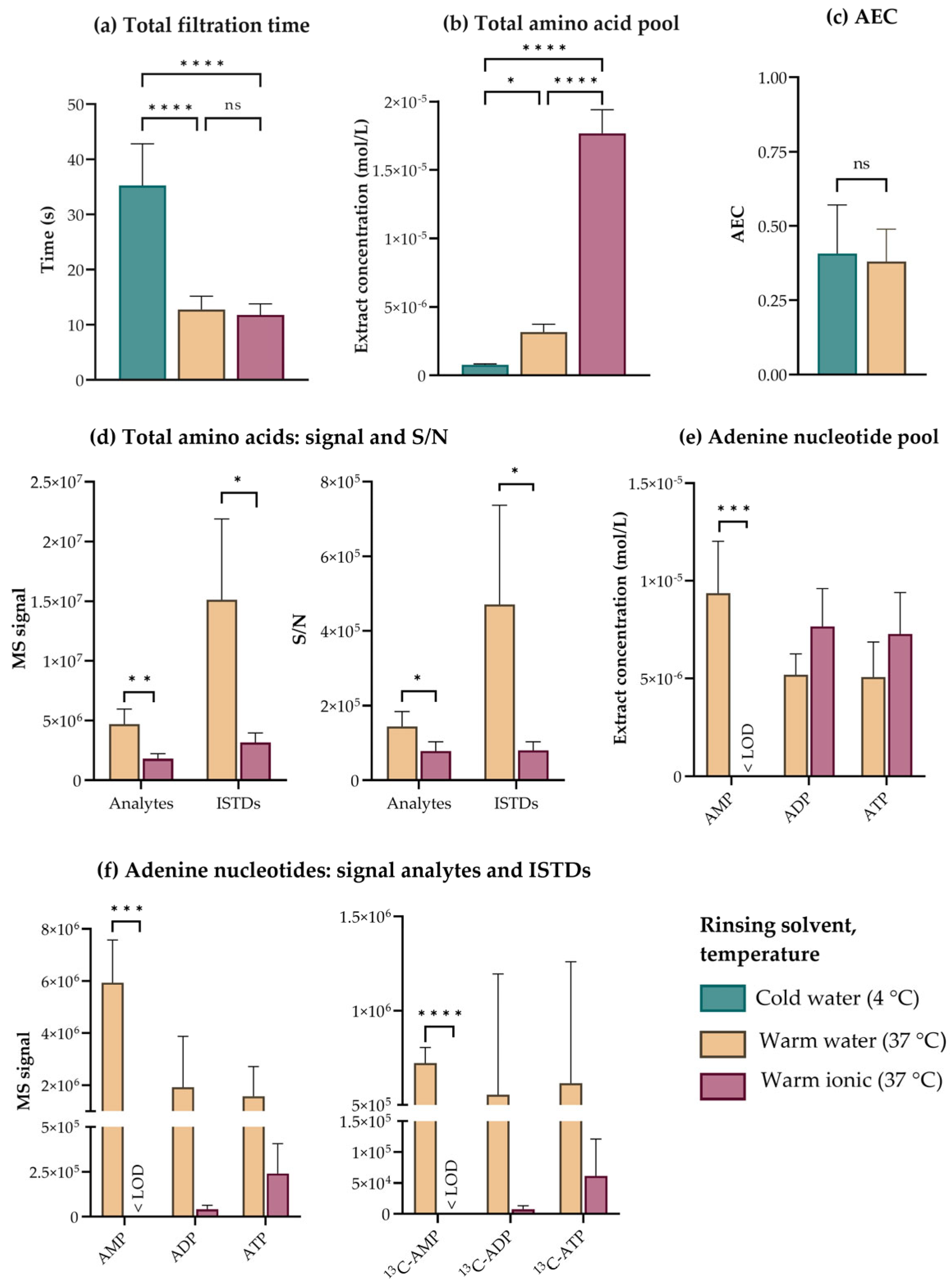
3.3. Filtered E. coli Tolerates a Quick Warm Water Rinse Serving to Remove Extracellular Components and Ensure Analytical Performance
3.4. Combined Quenching and Extraction in Cold Acetonitrile:Water:Methanol Stabilizes High Turnover Metabolites to Allow for High-Precision Metabolic Profiling of E. coli
3.5. Optimized Sampling, Extraction, and Sample Purification Enable Precise Quantification of the E. coli Central Carbon Metabolome under Different Growth Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and Isotope Tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; de Mey, M.; Ras, C.; ten Pierick, A.; Seifar, R.M.; van Dam, J.C.; Heijnen, J.J.; van Gulik, W.M. Development and Application of a Differential Method for Reliable Metabolome Analysis in Escherichia coli. Anal. Biochem. 2009, 386, 9–19. [Google Scholar] [CrossRef]
- Røst, L.M.; Thorfinnsdottir, L.B.; Kumar, K.; Fuchino, K.; Langørgen, I.E.; Bartosova, Z.; Kristiansen, K.A.; Bruheim, P. Absolute Quantification of the Central Carbon Metabolome in Eight Commonly Applied Prokaryotic and Eukaryotic Model Systems. Metabolites 2020, 10, 74. [Google Scholar] [CrossRef]
- Nasution, U.; Van Gulik, W.M.; Kleijn, R.J.; Van Winden, W.A.; Proell, A.; Heijnen, J.J. Measurement of Intracellular Metabolites of Primary Metabolism and Adenine Nucleotides in Chemostat Cultivated Penicillium chrysogenum. Biotechnol. Bioeng. 2006, 94, 159–166. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Arrivault, S.; Guenther, M.; Ivakov, A.; Feil, R.; Vosloh, D.; Van Dongen, J.T.; Sulpice, R.; Stitt, M. Use of Reverse-Phase Liquid Chromatography, Linked to Tandem Mass Spectrometry, to Profile the Calvin Cycle and Other Metabolic Intermediates in Arabidopsis Rosettes at Different Carbon Dioxide Concentrations. Plant J. 2009, 59, 826–839. [Google Scholar] [CrossRef]
- Gil, A.; Siegel, D.; Permentier, H.; Reijngoud, D.J.; Dekker, F.; Bischoff, R. Stability of Energy Metabolites-An Often Overlooked Issue in Metabolomics Studies: A Review. Electrophoresis 2015, 36, 2156–2169. [Google Scholar] [CrossRef]
- Lerma-Ortiz, C.; Jeffryes, J.G.; Cooper, A.J.L.; Niehaus, T.D.; Thamm, A.M.K.; Frelin, O.; Aunins, T.; Fiehn, O.; De Crécy-Lagard, V.; Henry, C.S.; et al. ‘Nothing of Chemistry Disappears in Biology’: The Top 30 Damage-Prone Endogenous Metabolites. Biochem. Soc. Trans. 2016, 44, 961–971. [Google Scholar] [CrossRef]
- Lowry, H.; Carter, J.; Ward, J.B.; Glaser, L. The Effect of Carbon and Nitrogen Sources on the Level of Metabolic Intermediates in Escherichia coli. J. Biol. Chem. 1971, 246, 6511–6521. [Google Scholar] [CrossRef]
- Boer, V.M.; Crutchfield, C.A.; Bradley, P.H.; Botstein, D.; Rabinowitz, J.D. Growth-Limiting Intracellular Metabolites in Yeast Growing under Diverse Nutrient Limitations. Mol. Biol. Cell 2010, 21, 198–211. [Google Scholar] [CrossRef]
- Alipanah, L.; Winge, P.; Rohloff, J.; Najafi, J.; Brembu, T.; Bones, A.M. Molecular Adaptations to Phosphorus Deprivation and Comparison with Nitrogen Deprivation Responses in the Diatom Phaeodactylum tricornutum. PLoS ONE 2018, 13, e0193335. [Google Scholar] [CrossRef]
- Sáez, M.J.; Lagunas, R. Determination of Intermediary Metabolites in Yeast. Critical Examination of the Effect of Sampling Conditions and Recommendations for Obtaining True Levels. Mol. Cell. Biochem. 1976, 13, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Bolten, C.J.; Kiefer, P.; Letisse, F.; Portais, J.C.; Wittmann, C. Sampling for Metabolome Analysis of Microorganisms. Anal. Chem. 2007, 79, 3843–3849. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.Y.; Wohlgemuth, G.; Park, H.S.; Fiehn, O.; Kim, K.H. Evaluation and Optimization of Metabolome Sample Preparation Methods for Saccharomyces cerevisiae. Anal. Chem. 2013, 85, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Kapoore, R.V.; Vaidyanathan, S. Towards Quantitative Mass Spectrometry-Based Metabolomics in Microbial and Mammalian Systems. Philos. Trans. R. Soc. A 2016, 374, 20150363. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Kimball, E. Acidic Acetonitrile for Cellular Metabolome Extraction from Escherichia coli. Anal. Chem. 2007, 79, 6167–6173. [Google Scholar] [CrossRef]
- van Gulik, W.M. Fast Sampling for Quantitative Microbial Metabolomics. Curr. Opin. Biotechnol. 2010, 21, 27–34. [Google Scholar] [CrossRef]
- Pinu, F.; Villas-Boas, S.; Aggio, R. Analysis of Intracellular Metabolites from Microorganisms: Quenching and Extraction Protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- de Koning, W.; van Dam, K. A Method for the Determination of Changes of Glycolytic Metabolites in Yeast on a Subsecond Time Scale Using Extraction at Neutral pH. Anal. Biochem. 1992, 204, 118–123. [Google Scholar] [CrossRef]
- Lange, H.C.; Eman, M.; Van Zuijlen, G.; Visser, D.; Van Dam, J.C.; Frank, J.; De Teixeira Mattos, M.J.; Heijnen, J.J. Improved Rapid Sampling for in vivo Kinetics of Intracellular Metabolites in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001, 75, 406–415. [Google Scholar] [CrossRef]
- Canelas, A.B.; Ras, C.; ten Pierick, A.; van Dam, J.C.; Heijnen, J.J.; van Gulik, W.M. Leakage-Free Rapid Quenching Technique for Yeast Metabolomics. Metabolomics 2008, 4, 226–239. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Højer-Pedersen, J.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Global Metabolite Analysis of Yeast: Evaluation of Sample Preparation Methods. Yeast 2005, 22, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Bruheim, P. Cold Glycerol-Saline: The Promising Quenching Solution for Accurate Intracellular Metabolite Analysis of Microbial Cells. Anal. Biochem. 2007, 370, 87–97. [Google Scholar] [CrossRef]
- Wittmann, C.; Krömer, J.O.; Kiefer, P.; Binz, T.; Heinzle, E. Impact of the Cold Shock Phenomenon on Quantification of Intracellular Metabolites in Bacteria. Anal. Biochem. 2004, 327, 135–139. [Google Scholar] [CrossRef]
- Mashego, M.R.; Rumbold, K.; De Mey, M.; Vandamme, E.; Soetaert, W.; Heijnen, J.J. Microbial Metabolomics: Past, Present and Future Methodologies. Biotechnol. Lett. 2007, 29, 1–16. [Google Scholar] [CrossRef]
- de Jonge, L.P.; Douma, R.D.; Heijnen, J.J.; van Gulik, W.M. Optimization of Cold Methanol Quenching for Quantitative Metabolomics of Penicillium chrysogenum. Metabolomics 2012, 8, 727–735. [Google Scholar] [CrossRef]
- Winder, C.L.; Dunn, W.B.; Schuler, S.; Broadhurst, D.; Jarvis, R.; Stephens, G.M.; Goodacre, R. Global Metabolic Profiling of Escherichia coli Cultures: An Evaluation of Methods for Quenching and Extraction of Intracellular Metabolites. Anal. Chem. 2008, 80, 2939–2948. [Google Scholar] [CrossRef]
- Brauer, M.J.; Yuan, J.; Bennett, B.D.; Lu, W.; Kimball, E.; Botstein, D.; Rabinowitz, J.D. Conservation of the Metabolomic Response to Starvation across Two Divergent Microbes. Proc. Natl. Acad. Sci. USA 2006, 103, 19302–19307. [Google Scholar] [CrossRef]
- Kvitvang, H.F.N.; Kristiansen, K.A.; Bruheim, P. Assessment of Capillary Anion Exchange Ion Chromatography Tandem Mass Spectrometry for the Quantitative Profiling of the Phosphometabolome and Organic Acids in Biological Extracts. J. Chromatogr. A 2014, 1370, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Stafsnes, M.H.; Røst, L.M.; Bruheim, P. Improved Phosphometabolome Profiling Applying Isotope Dilution Strategy and Capillary Ion Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B 2018, 1083, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Røst, L.M.; Shafaei, A.; Fuchino, K.; Bruheim, P. Zwitterionic HILIC Tandem Mass Spectrometry with Isotope Dilution for Rapid, Sensitive and Robust Quantification of Pyridine Nucleotides in Biological Extracts. J. Chromatogr. B 2020, 1144, 122078. [Google Scholar] [CrossRef]
- Kumar, K.; Venkatraman, V.; Bruheim, P. Adaptation of Central Metabolite Pools to Variations in Growth Rate and Cultivation Conditions in Saccharomyces cerevisiae. Microb. Cell Factories 2021, 20, 64. [Google Scholar] [CrossRef]
- Kumar, K.; Bruheim, P. Large Dependency of Intracellular NAD and CoA Pools on Cultivation Conditions in Saccharomyces cerevisiae. BMC Res. Notes 2021, 14, 372. [Google Scholar] [CrossRef]
- Atkinson, D.E.; Walton, G.M. Adenosine Triphosphate Conservation in Metabolic Regulation: Rat Liver Citrate Cleavage Enzyme. J. Biol. Chem. 1967, 242, 3239–3241. [Google Scholar] [CrossRef]
- De La Fuente, I.M.; Cortés, J.M.; Valero, E.; Desroches, M.; Rodrigues, S.; Malaina, I.; Martínez, L. On the Dynamics of the Adenylate Energy System: Homeorhesis vs Homeostasis. PLoS ONE 2014, 9, e108676. [Google Scholar] [CrossRef]
- Chapman, A.G.; Fall, L.; Atkinson, D.E. Adenylate Energy Charge in Escherichia coli During Growth and Starvation. J. Bacteriol. 1971, 108, 1072–1086. [Google Scholar] [CrossRef]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Fuchino, K.; Bruheim, P. Increased Salt Tolerance in Zymomonas mobilis Strain Generated by Adaptative Evolution. Microb. Cell Factories 2020, 19, 147. [Google Scholar] [CrossRef] [PubMed]
- Skinner, S.O.; Sepúlveda, L.A.; Xu, H.; Golding, I. Measuring MRNA Copy Number in Individual Escherichia coli Cells Using Single-Molecule Fluorescent in Situ Hybridization. Nat. Protoc. 2013, 8, 1100–1113. [Google Scholar] [CrossRef]
- Volkmer, B.; Heinemann, M. Condition-Dependent Cell Volume and Concentration of Escherichia coli to Facilitate Data Conversion for Systems Biology Modeling. PLoS ONE 2011, 6, e23126. [Google Scholar] [CrossRef]
- Kvitvang, H.F.N.; Bruheim, P. Fast Filtration Sampling Protocol for Mammalian Suspension Cells Tailored for Phosphometabolome Profiling by Capillary Ion Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B 2015, 998–999, 45–49. [Google Scholar] [CrossRef]
- Molecular Probes LIVE/DEAD BacLight Bacterial Viability Kit. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fmp07007.pdf (accessed on 2 December 2022).
- da Luz, J.A.; Hans, E.; Zeng, A.-P. Automated Fast Filtration and On-Filter Quenching Improve the Intracellular Metabolite Analysis of Microorganisms. Eng. Life Sci. 2014, 14, 135–142. [Google Scholar] [CrossRef]
- Furey, A.; Moriarty, M.; Bane, V.; Kinsella, B.; Lehane, M. Ion Suppression; A Critical Review on Causes, Evaluation, Prevention and Applications. Talanta 2013, 115, 104–122. [Google Scholar] [CrossRef]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms—Global Edition, 16th ed.; Pearson Education: London, UK, 2022; ISBN 978-0-13-487440-1. [Google Scholar]
- Bordag, N.; Janakiraman, V.; Nachtigall, J.; González Maldonado, S.; Bethan, B.; Laine, J.-P.; Fux, E. Fast Filtration of Bacterial or Mammalian Suspension Cell Cultures for Optimal Metabolomics Results. PLoS ONE 2016, 11, e0159389. [Google Scholar] [CrossRef]
- Merck Millipore Binding Properties of Filters. Available online: https://www.merckmillipore.com/NO/en/life-science-research/chromatography-sample-preparation/membrane-learning-center/Binding-Properties-of-Filters/596b.qB.Hj0AAAFM5FB88eJw,nav (accessed on 2 December 2022).
- Wu, L.; Mashego, M.R.; Van Dam, J.C.; Proell, A.M.; Vinke, J.L.; Ras, C.; Van Winden, W.A.; Van Gulik, W.M.; Heijnen, J.J. Quantitative Analysis of the Microbial Metabolome by Isotope Dilution Mass Spectrometry Using Uniformly 13C-Labeled Cell Extracts as Internal Standards. Anal. Biochem. 2005, 336, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Vuckovic, D. Current Trends and Challenges in Sample Preparation for Global Metabolomics Using Liquid Chromatography-Mass Spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef]
- Dietmair, S.; Timmins, N.E.; Gray, P.P.; Nielsen, L.K.; Krömer, J.O. Towards Quantitative Metabolomics of Mammalian Cells: Development of a Metabolite Extraction Protocol. Anal. Biochem. 2010, 404, 155–164. [Google Scholar] [CrossRef]
- McCloskey, D.; Utrilla, J.; Naviaux, R.K.; Palsson, B.O.; Feist, A.M. Fast Swinnex Filtration (FSF): A Fast and Robust Sampling and Extraction Method Suitable for Metabolomics Analysis of Cultures Grown in Complex Media. Metabolomics 2015, 11, 198–209. [Google Scholar] [CrossRef]
- Vadia, S.; Levin, P.A. Growth Rate and Cell Size: A Re-Examination of the Growth Law. Curr. Opin. Microbiol. 2015, 24, 96–103. [Google Scholar] [CrossRef]
- Bennett, B.D.; Yuan, J.; Kimball, E.H.; Rabinowitz, J.D. Absolute Quantitation of Intracellular Metabolite Concentrations by an Isotope Ratio-Based Approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar] [CrossRef]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; Van Dien, S.J.; Rabinowitz, J.D. Absolute Metabolite Concentrations and Implied Enzyme Active Site Occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Fell, D.A. Understanding the Control of Metabolism, 1st ed.; Portland Press: London, UK, 1997; ISBN 1-85578-047-X. [Google Scholar]
- Vasilakou, E.; Van Loosdrecht, M.C.M.; Wahl, S.A. Escherichia coli Metabolism under Short-Term Repetitive Substrate Dynamics: Adaptation and Trade-Offs. Microb. Cell Factories 2020, 19, 116. [Google Scholar] [CrossRef]
- Álvarez-Añorve, L.I.; Gaugué, I.; Link, H.; Marcos-Viquez, J.; Díaz-Jiménez, D.M.; Zonszein, S.; Bustos-Jaimes, I.; Schmitz-Afonso, I.; Calcagno, M.L.; Plumbridge, J. Allosteric Activation of Escherichia coli Glucosamine-6-Phosphate Deaminase (NagB) in Vivo Justified by Intracellular Amino Sugar Metabolite Concentrations. J. Bacteriol. 2016, 198, 1610–1620. [Google Scholar] [CrossRef]
- Kim, J.; Kim, K.H. Effects of Minimal Media vs. Complex Media on the Metabolite Profiles of Escherichia coli and Saccharomyces cerevisiae. Process Biochem. 2017, 57, 64–71. [Google Scholar] [CrossRef]
- Radoš, D.; Donati, S.; Lempp, M.; Rapp, J.; Link, H. Homeostasis of the Biosynthetic E. coli Metabolome. iScience 2022, 25, 104503. [Google Scholar] [CrossRef]




| Brand Name | Filter Material | Pore Size (µm) | Supplier | Product Number |
|---|---|---|---|---|
| Durapore | PVDF | 0.22 | Sigma-Aldrich | GVWP04700 |
| Durapore | PVDF | 0.45 | Sigma-Aldrich | HVLP04700 |
| Durapore | PVDF | 0.65 | Sigma-Aldrich | DVPP04700 |
| Magna Nylon Filter | Nylon | 0.22 | GVS Life Sciences | 1213769 |
| Nylon Net Filter | Nylon | 0.45 | Sigma-Aldrich | HNWP04700 |
| Omnipore | Hydrophilic PTFE | 0.20 | Sigma-Aldrich | JGWP04700 |
| Omnipore | Hydrophilic PTFE | 0.45 | Sigma-Aldrich | JHWP04700 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thorfinnsdottir, L.B.; García-Calvo, L.; Bø, G.H.; Bruheim, P.; Røst, L.M. Optimized Fast Filtration-Based Sampling and Extraction Enables Precise and Absolute Quantification of the Escherichia coli Central Carbon Metabolome. Metabolites 2023, 13, 150. https://doi.org/10.3390/metabo13020150
Thorfinnsdottir LB, García-Calvo L, Bø GH, Bruheim P, Røst LM. Optimized Fast Filtration-Based Sampling and Extraction Enables Precise and Absolute Quantification of the Escherichia coli Central Carbon Metabolome. Metabolites. 2023; 13(2):150. https://doi.org/10.3390/metabo13020150
Chicago/Turabian StyleThorfinnsdottir, Lilja Brekke, Laura García-Calvo, Gaute Hovde Bø, Per Bruheim, and Lisa Marie Røst. 2023. "Optimized Fast Filtration-Based Sampling and Extraction Enables Precise and Absolute Quantification of the Escherichia coli Central Carbon Metabolome" Metabolites 13, no. 2: 150. https://doi.org/10.3390/metabo13020150
APA StyleThorfinnsdottir, L. B., García-Calvo, L., Bø, G. H., Bruheim, P., & Røst, L. M. (2023). Optimized Fast Filtration-Based Sampling and Extraction Enables Precise and Absolute Quantification of the Escherichia coli Central Carbon Metabolome. Metabolites, 13(2), 150. https://doi.org/10.3390/metabo13020150







