Linking Clinical Blood Metabogram and Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Mass Spectrometry Analysis of Blood Samples
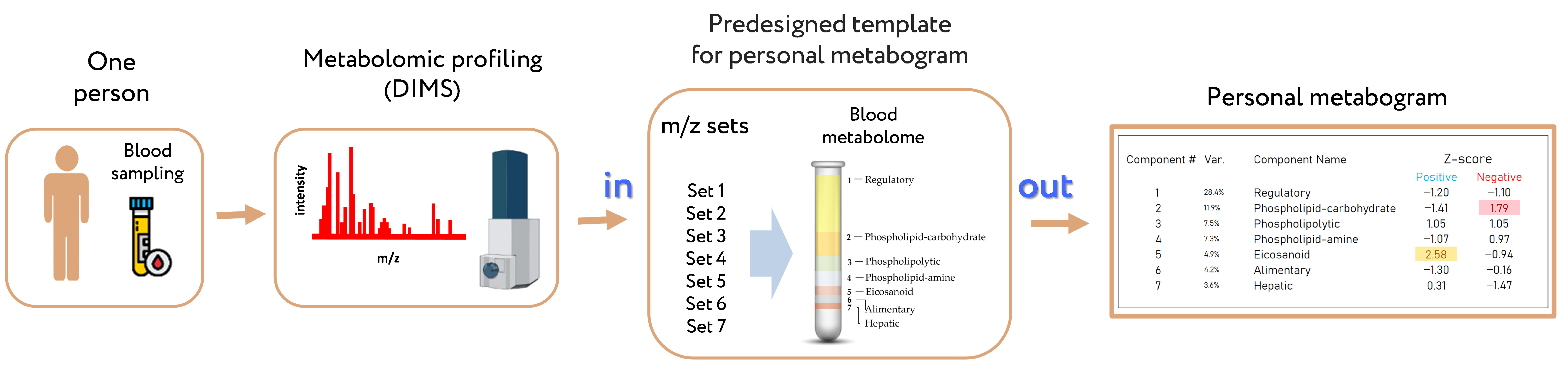
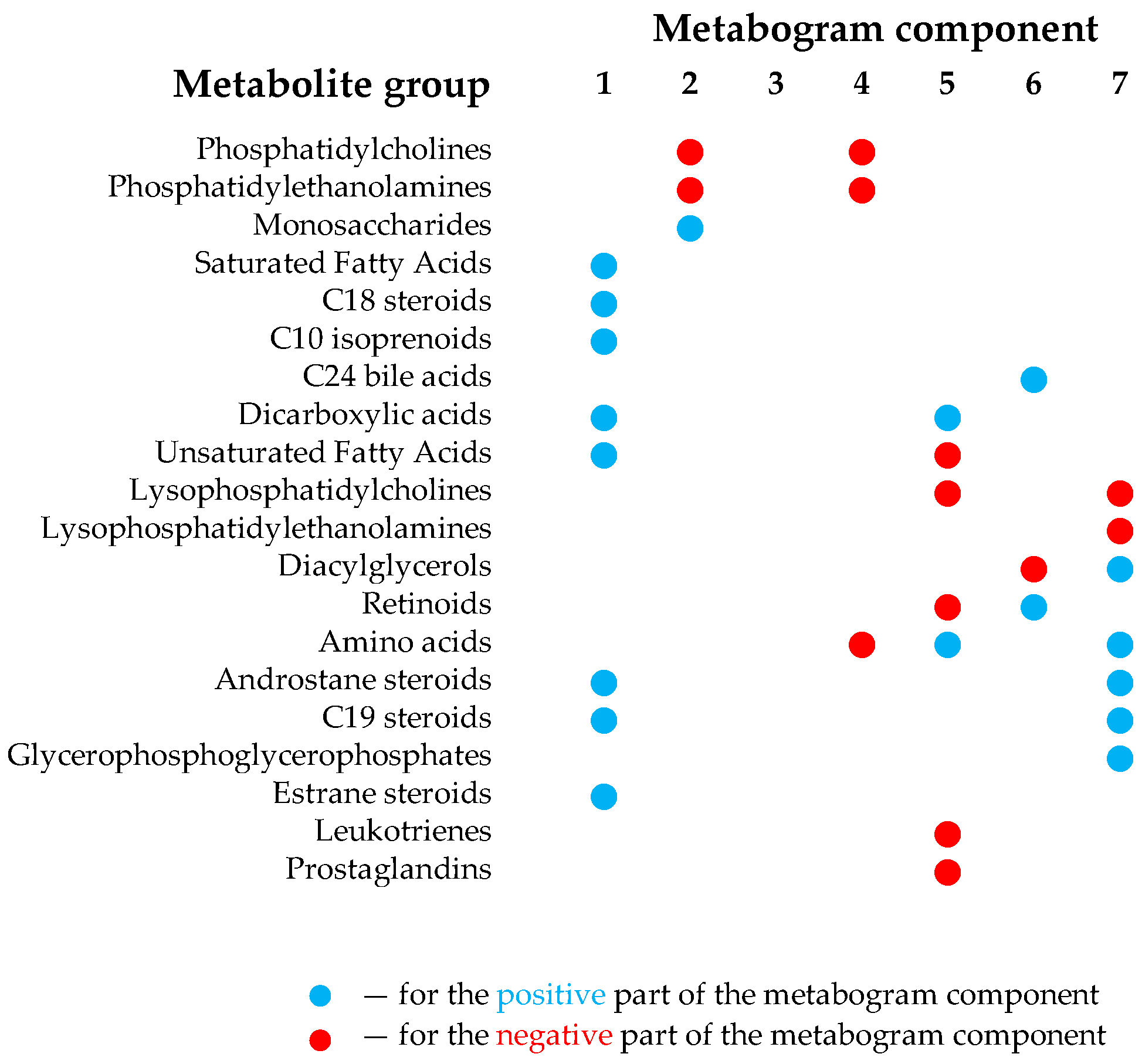
2.3. Design of Metabogram (Template for Personal Metabograms)
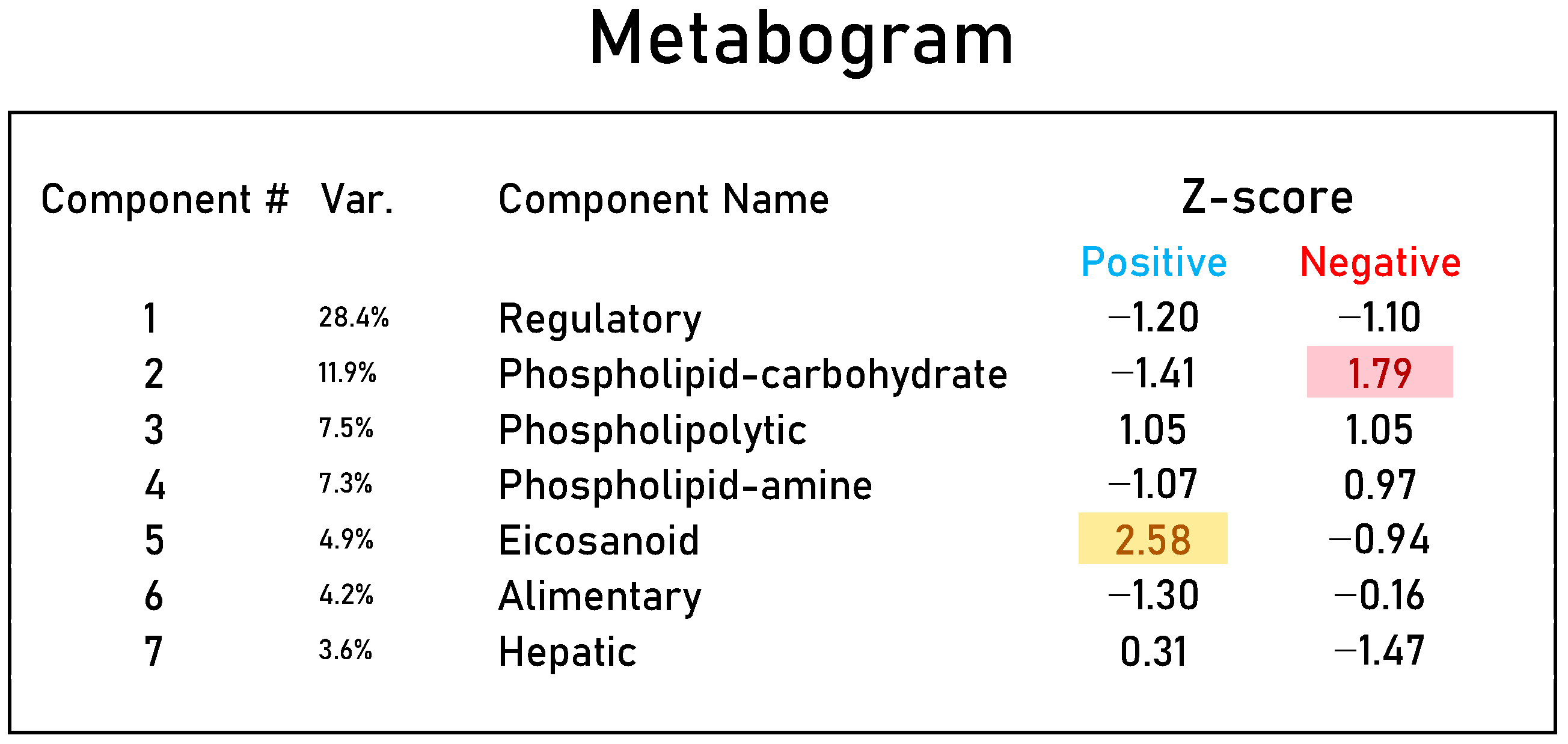
2.4. Personal Metabograms
2.5. Gut Microbiota Analysis
2.5.1. Gut Microbiota Analysis by Culture-Based Method
2.5.2. Gut Microbiota Analysis by Real-Time PCR
- Total bacteria
- Lactobacillus spp.
- Bifidobacterium spp.
- Faecalibacterium prausnitzii
- Bacteroides thetaiotaomicron
- Bacteroides spp./Faecalibacterium prausnitzii ratio
- Klebsiella pneumoniae
- Klebsiella oxytoca
- Enterobacter spp. and Citrobacter spp.
- Clostridium difficile
- Clostridium perfringens
- Staphylococcus aureus
- Proteus vulgaris and Proteus mirabilis
- Candida spp. yeast
- Escherichia coli enteropathogenic
- Salmonella spp.
- Shigella spp.
- Fusobacterium nucleatum
- Parvimonas micra
2.6. Correlation Analysis
2.7. Plotting Links between Metabogram Components and Gut Microbiota
2.8. Diagnostic Parameters
3. Results
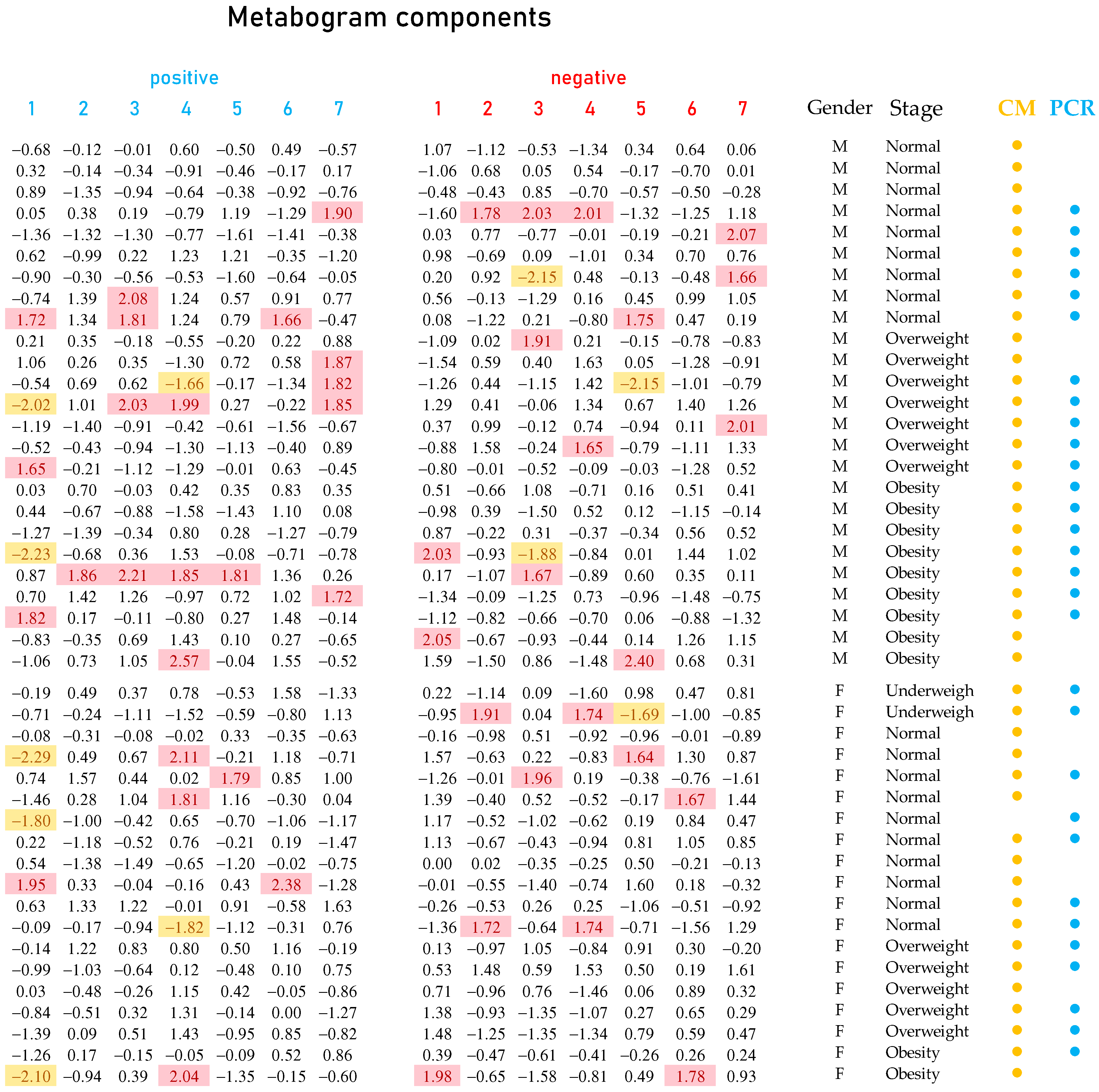
3.1. Studied Subjects
3.2. Mass Spectrometry Data for Metabograms
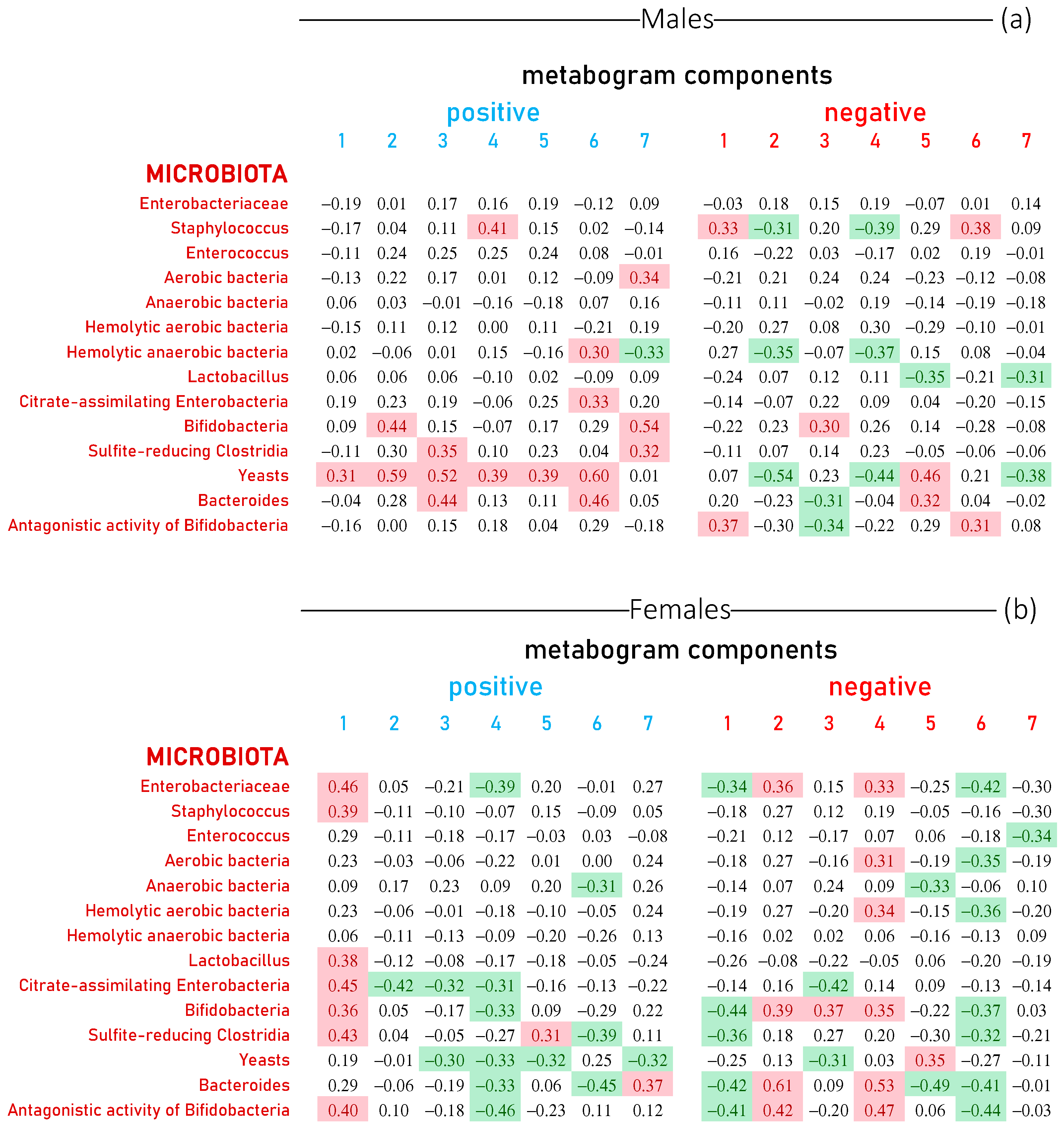
3.3. Metabogram Components Connection with Gut Microbiota Studied by Culture-Based Method
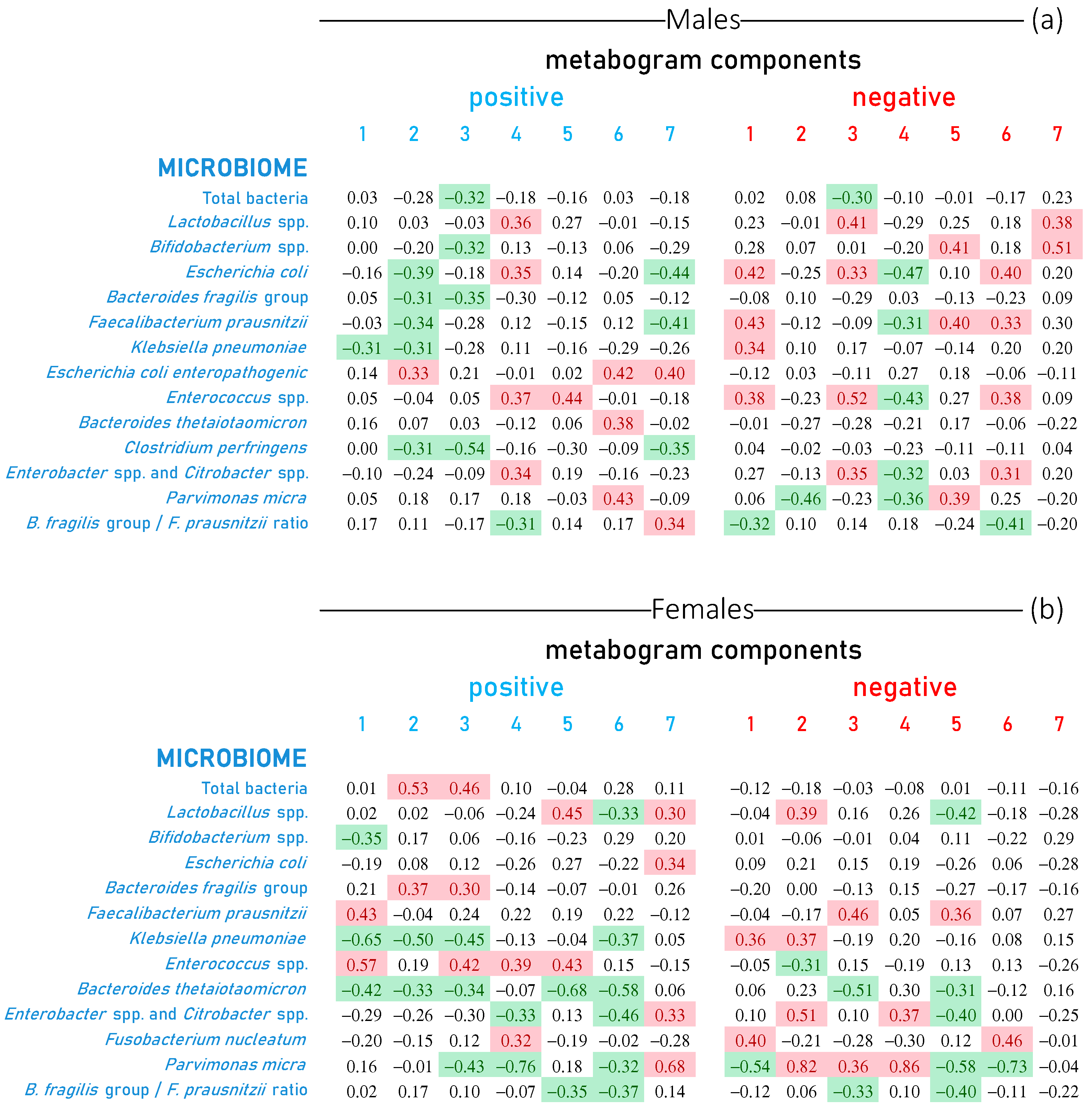
3.4. Metabogram Components Connection with Gut Microbiota Studied by Real-Time PCR
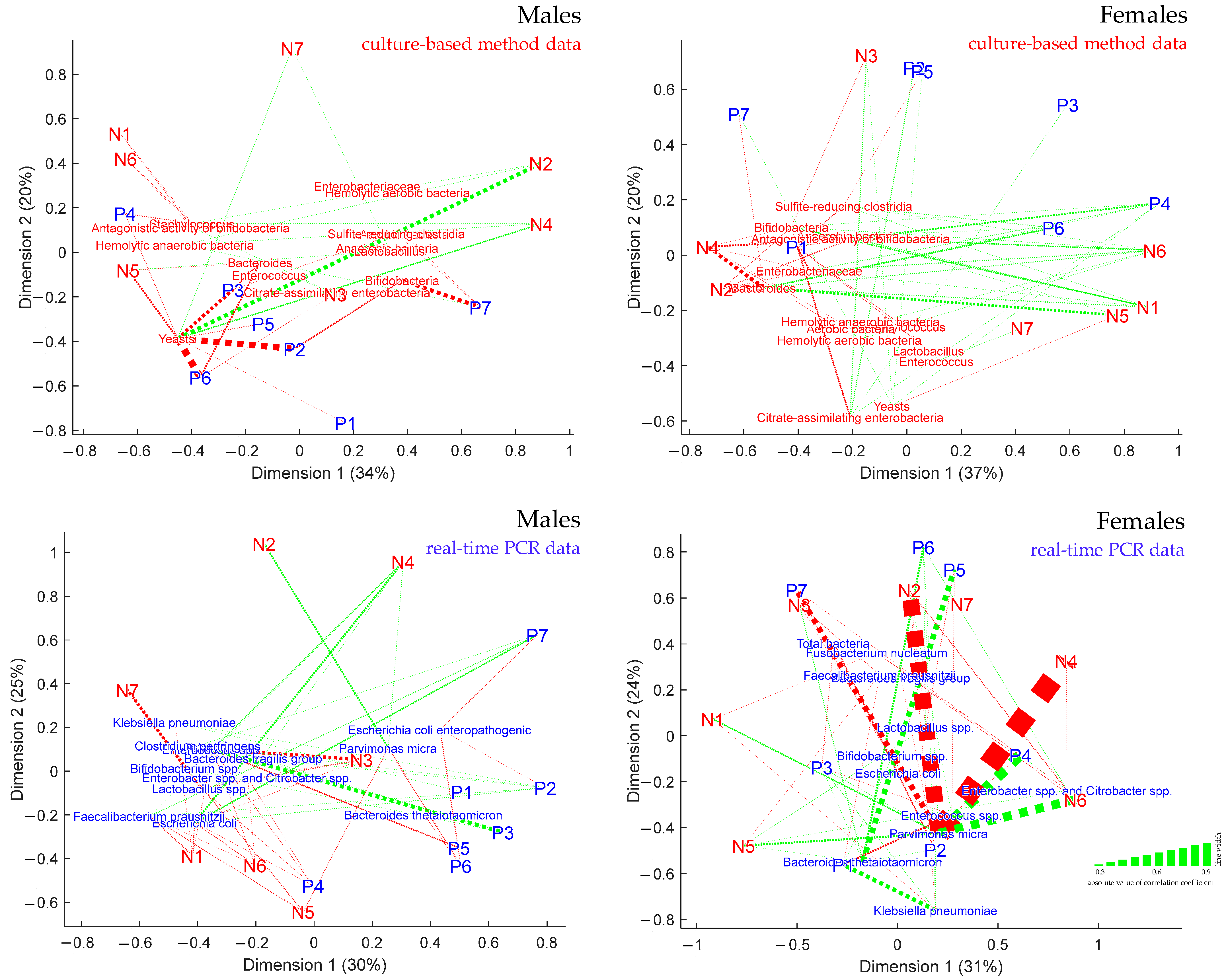
3.5. Visualization of the Links between Metabogram Components and Gut Microbiota
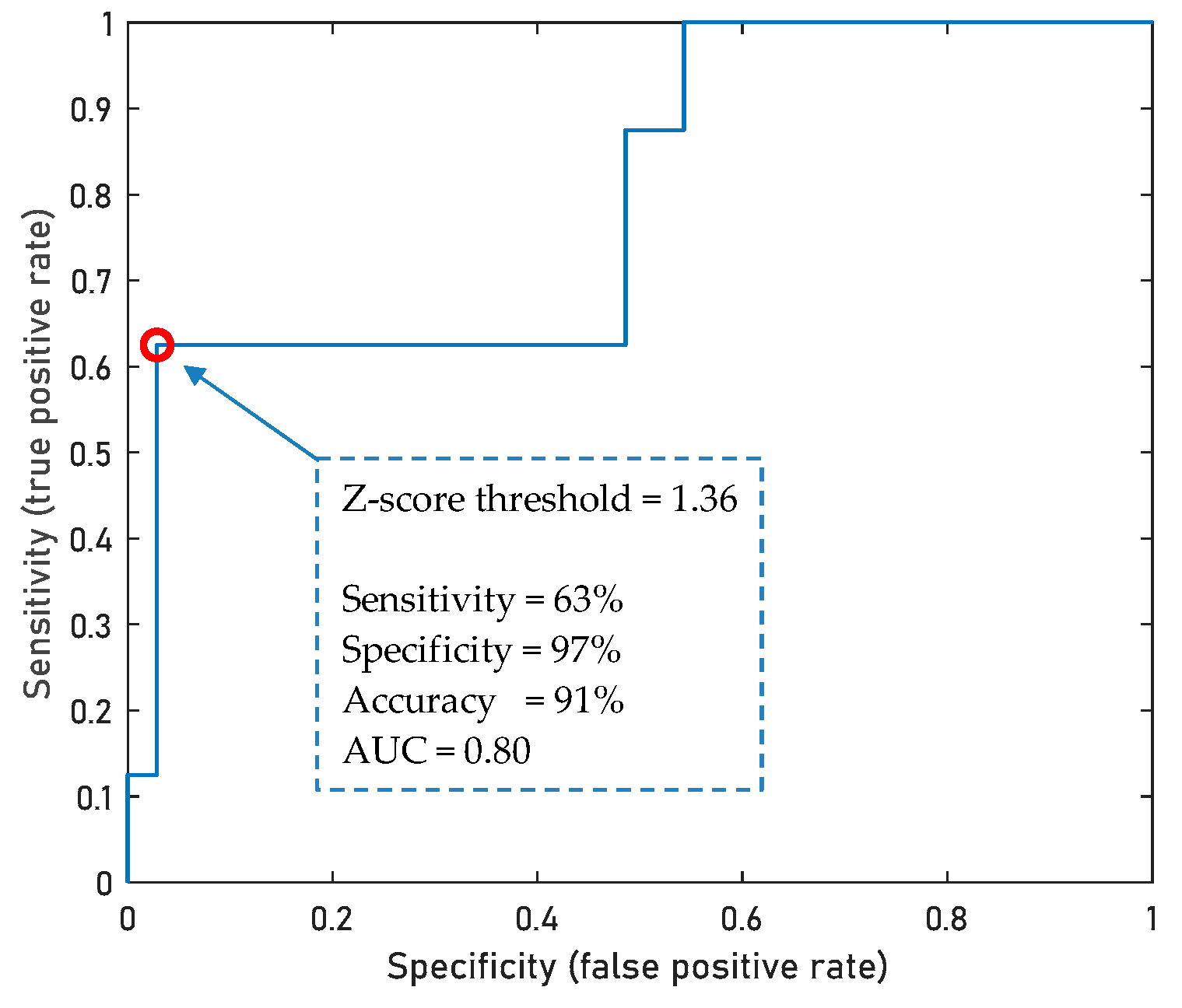
3.6. Diagnostic Potential of Metabogram Components
4. Discussion
- The difference between the sexes was clearly shown.
- A precise metric of the blood metabolome/gut microbiota relationship was provided (the portion of the metabolome covered by each component of the metabogram is indicated in the metabogram, and the strength of connection is expressed by the correlation coefficient).
- New data about blood metabolome/gut microorganism relations were revealed (e.g., the strong connection of the metabolome with the yeast levels).
- A high diagnostic capacity of blood metabolites (by means of a metabogram) in relation to gut microbiota was demonstrated.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Micheel, C.M.; Nass, S.J.; Omenn, G.S. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine. In Evolution of Translational Omics : Lessons Learned and the Path Forward; Micheel, C.M., Sharyl, N.J., Omenn, G.S., Eds.; National Academies Press (US): Washington, DC, USA, 2012; ISBN 9780309224185. [Google Scholar]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Nalbantoglu, S. Metabolomics: Basic Principles and Strategies; Nalbantoglu, S., Amri, H., Eds.; IntechOpen: Rijeka, Croatia, 2019; p. 8. ISBN 978-1-83962-760-6. [Google Scholar]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Lichtenberg, S.; Balashova, E.E. Personal Metabolomics: A Global Challenge. Metabolites 2021, 11, 715. [Google Scholar] [CrossRef]
- Lichtenberg, S.; Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Metabolomic Laboratory-Developed Tests: Current Statusand Perspectives. Metabolites 2021, 11, 423. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Grigoriev, A.I.; Ponomarenko, E.A.; Archakov, A.I. Mass Spectrometric Blood Metabogram: Acquisition, Characterization, and Prospects for Application. Int. J. Mol. Sci. 2023, 24, 1736. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Plotnikova, O.A.; Sharafetdinov, K.K.; Nikityuk, D.B.; Tutelyan, V.A.; Ponomarenko, E.A.; Archakov, A.I. Clinical Blood Metabogram: Application to Overweight and Obese Patients. Metabolites 2023, 13, 798. [Google Scholar] [CrossRef]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.A.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; Roy, C.I.L.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef]
- Coelho, G.D.P.; Ayres, L.F.A.; Barreto, D.S.; Henriques, B.D.; Prado, M.R.M.C.; Passos, C.M. Dos Acquisition of microbiota according to the type of birth: An integrative review. Rev. Lat. Am. Enferm. 2021, 29, e3446. [Google Scholar] [CrossRef]
- Lif Holgerson, P.; Harnevik, L.; Hernell, O.; Tanner, A.C.R.; Johansson, I. Mode of birth delivery affects oral microbiota in infants. J. Dent. Res. 2011, 90, 1183–1188. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. Msystems 2019, 4, e00261-19. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Unno, T.; Kim, B.Y.; Park, M.S. Sex Differences in Gut Microbiota. World J. Mens. Health 2020, 38, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Ponomarenko, E.A.; Archakov, A.I. Mass Spectrometry-Based Metabolomics Analysis of Obese Patients’ Blood Plasma. Int. J. Mol. Sci. 2020, 21, 568. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Kharybin, O.N.; Archakov, A.I. Diagnosis of lung cancer based on direct-infusion electrospray mass spectrometry of blood plasma metabolites. Int. J. Mass Spectrom. 2011, 309, 200–205. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Differences between correlation coefficients. In Statistical Power Analysis for the Behavioral Science, 2nd ed.; Lawrence Erlbaum Associates: Hillsdale, NJ, USA, 1988; pp. 109–145. [Google Scholar]
- Crews, B.; Wikoff, W.R.; Patti, G.J.; Woo, H.-K.; Kalisiak, E.; Heideker, J.; Siuzdak, G. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal. Chem. 2009, 81, 8538–8544. [Google Scholar] [CrossRef]
- Liu, B.N.; Liu, X.T.; Liang, Z.H.; Wang, J.H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; Bäckhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Shin, J.-H.; Park, Y.-H.; Sim, M.; Kim, S.-A.; Joung, H.; Shin, D.-M. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res. Microbiol. 2019, 170, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Stilling, R.M.; Stanton, C.; Cryan, J.F. Collective unconscious: How gut microbes shape human behavior. J. Psychiatr. Res. 2015, 63, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Borre, Y.E.; Moloney, R.D.; Clarke, G.; Dinan, T.G.; Cryan, J.F. The impact of microbiota on brain and behavior: Mechanisms & therapeutic potential. Adv. Exp. Med. Biol. 2014, 817, 373–403. [Google Scholar] [CrossRef]
- Pennisi, E. Meet the psychobiome. Science 2020, 368, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Sanmiguel, C.; Gupta, A.; Mayer, E.A. Gut Microbiome and Obesity: A Plausible Explanation for Obesity. Curr. Obes. Rep. 2015, 4, 250–261. [Google Scholar] [CrossRef]
- Shen, J.; Obin, M.S.; Zhao, L. The gut microbiota, obesity and insulin resistance. Mol. Aspects Med. 2013, 34, 39–58. [Google Scholar] [CrossRef]
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Chen, M.X.; Wang, S.-Y.; Kuo, C.-H.; Tsai, I.-L. Metabolome analysis for investigating host-gut microbiota interactions. J. Formos. Med. Assoc. 2019, 118, S10–S22. [Google Scholar] [CrossRef]
- Stephens, R.W.; Arhire, L.; Covasa, M. Gut Microbiota: From Microorganisms to Metabolic Organ Influencing Obesity. Obesity 2018, 26, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
| Groups of Microorganisms | Culture Media | Dilutions | Incubation |
|---|---|---|---|
| Enterobacteria | Endo agar (Biokompas—S, Moscow, Russia) | 10−3, 10−4, 10−5, 10−6 50 μL | 37 °C, 24 h |
| Enterobacteria utilizing citrate | Simmons Citrate Agar (Biokompas—S, Russia) | 10−4, 10−5 50 μL | 37 °C, 4 days |
| Bacteroides | Bacteroides Bile Esculin Agar with Bacteroides Selective Supplement (FD062) (HiMedia, Mumbai, India) | 10−4, 10−5, 10−6, 10−7 50 μL | 37 °C, 48 h, anaerobic conditions |
| Total number of aerobic microorganisms, hemolytic microorganisms | Columbia Blood Agar (HiMedia) with 5% v/v sterile defibrinated sheep blood | 10−5, 10−6 50 μL | 37 °C, 48 h |
| Total number of anaerobic microorganisms, hemolytic anaerobes | Columbia Blood Agar (HiMedia) with 7% v/v sterile defibrinated sheep blood | 10−6, 10−8 50 μL | 37 °C up to 7 days |
| Lactic acid bacteria | MRS agar with sorbic acid additive (Biokompas—S) | 10−4, 10−5, 10−6, 10−7 50 μL | 37 °C, 3 days |
| Enterococci | Kanamycin esculin azide agar (105222) (Merck) | 10−4, 10−6 50 μL | 37 °C, 48 h |
| Staphylococci | Baird-Parker Agar with egg yolk and tellurite additive (Biokompas—S) | 10−3, 10−5 50 μL | 37 °C, 48 h |
| Bifidobacteria | Corn–lactose medium (Biokompas—S) | 10−7, 10−8, 10−9, 10−10 1 mL | 37 °C, 5 days anaerobic conditions |
| Sulfite-reducing clostridia | Iron–sulfite medium (Biokompas—S) | 10−6, 10−7, 10−8, 10−9 1 mL | 37 °C, 5 days anaerobic conditions |
| Yeasts and molds | Sabouraud agar (Biokompas—S) with streptomycin | 10−1, 10−2, 10−3 50 μL | 30°C, 5 days |
| Subjects | Age (Years) | Body Mass Index (kg/m2) | Gender (Number) |
|---|---|---|---|
| Cohort for culture-based method testing of gut microbiota | |||
| Males (Normal—9, Overweight—7, Obesity—9) | Males 30.4 ± 6.8 1 | Males 27.7 ± 5.3 | Males—25 |
| Females (Underweight—2, Normal—9, Overweight—5, Obesity—2) | Females 30.3 ± 5.2 | Females 24.9 ± 7.1 | Females—18 |
| Cohort for real-time PCR testing of gut microbiota | |||
| Males (Normal—6, Overweight—5, Obesity—7) | Males 30.9 ± 7.0 | Males 27.7 ± 4.4 | Males—18 |
| Females (Underweight—2, Normal—5, Overweight—4, Obesity—1) | Females 31.6 ± 5.0 | Females 24.4 ± 6.5 | Females—12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lokhov, P.G.; Balashova, E.E.; Maslov, D.L.; Trifonova, O.P.; Lisitsa, A.V.; Markova, Y.M.; Stetsenko, V.V.; Polyanina, A.S.; Sheveleva, S.A.; Sharafetdinov, K.K.; et al. Linking Clinical Blood Metabogram and Gut Microbiota. Metabolites 2023, 13, 1095. https://doi.org/10.3390/metabo13101095
Lokhov PG, Balashova EE, Maslov DL, Trifonova OP, Lisitsa AV, Markova YM, Stetsenko VV, Polyanina AS, Sheveleva SA, Sharafetdinov KK, et al. Linking Clinical Blood Metabogram and Gut Microbiota. Metabolites. 2023; 13(10):1095. https://doi.org/10.3390/metabo13101095
Chicago/Turabian StyleLokhov, Petr G., Elena E. Balashova, Dmitry L. Maslov, Oxana P. Trifonova, Andrey V. Lisitsa, Yulia M. Markova, Valentina V. Stetsenko, Anna S. Polyanina, Svetlana A. Sheveleva, Khaider K. Sharafetdinov, and et al. 2023. "Linking Clinical Blood Metabogram and Gut Microbiota" Metabolites 13, no. 10: 1095. https://doi.org/10.3390/metabo13101095
APA StyleLokhov, P. G., Balashova, E. E., Maslov, D. L., Trifonova, O. P., Lisitsa, A. V., Markova, Y. M., Stetsenko, V. V., Polyanina, A. S., Sheveleva, S. A., Sharafetdinov, K. K., Nikityuk, D. B., Tutelyan, V. A., & Archakov, A. I. (2023). Linking Clinical Blood Metabogram and Gut Microbiota. Metabolites, 13(10), 1095. https://doi.org/10.3390/metabo13101095









