Metabolic Remodeling during Early Cardiac Lineage Specification of Pluripotent Stem Cells
Abstract
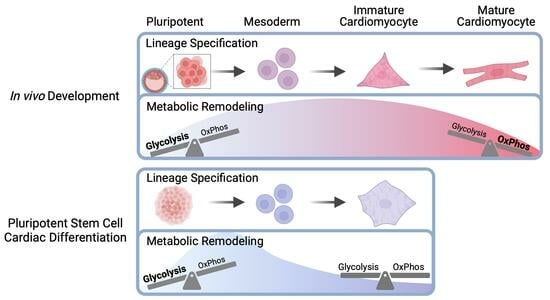
:1. Introduction
2. Materials and Methods
2.1. Human Pluripotent Stem Cell Maintenance as 3D Spheroids
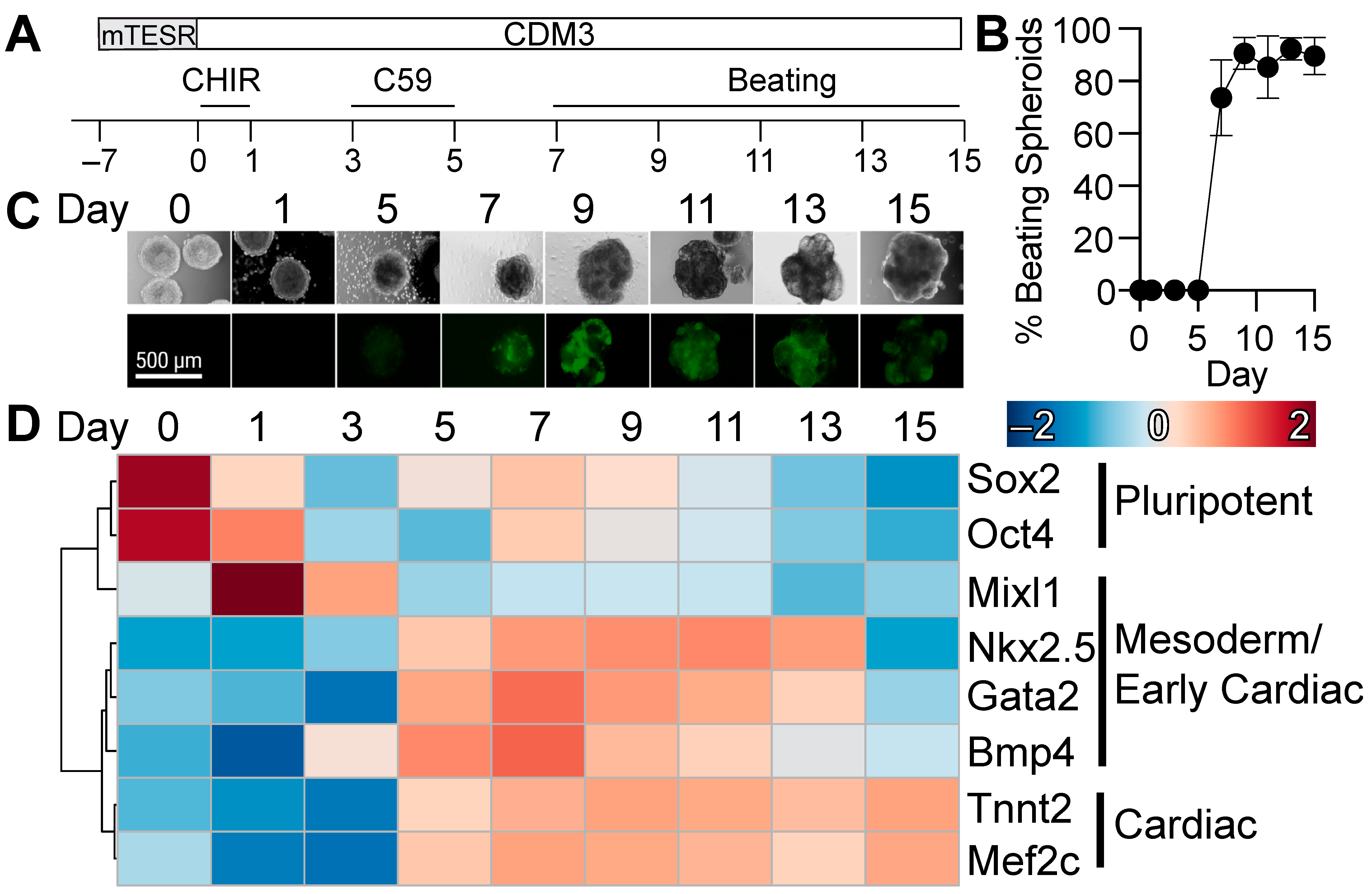
2.2. Three-Dimensional Cardiac Differentiation of Human Pluripotent Stem Cells
2.3. Expression of Lineage Specific Markers by qRT-PCR
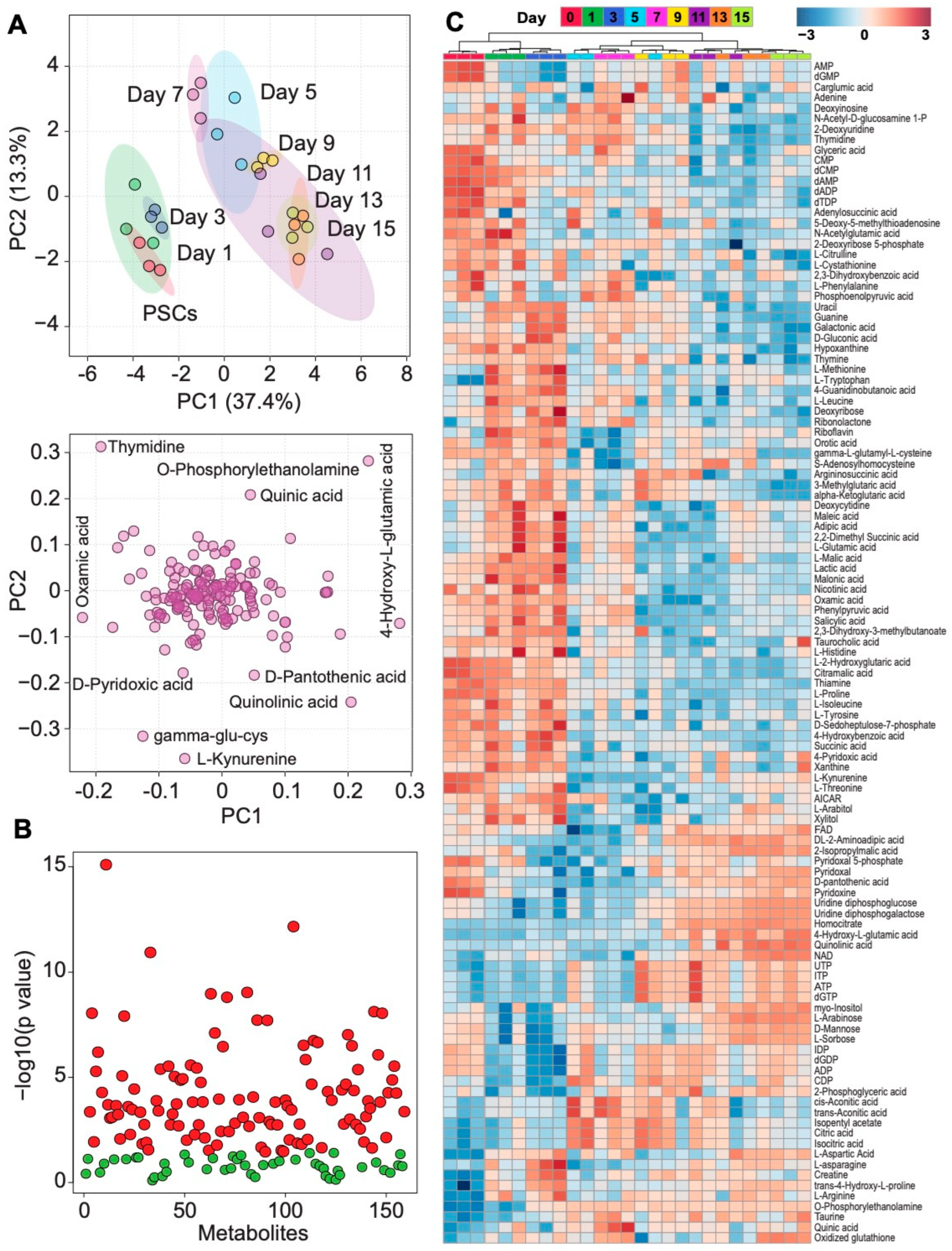
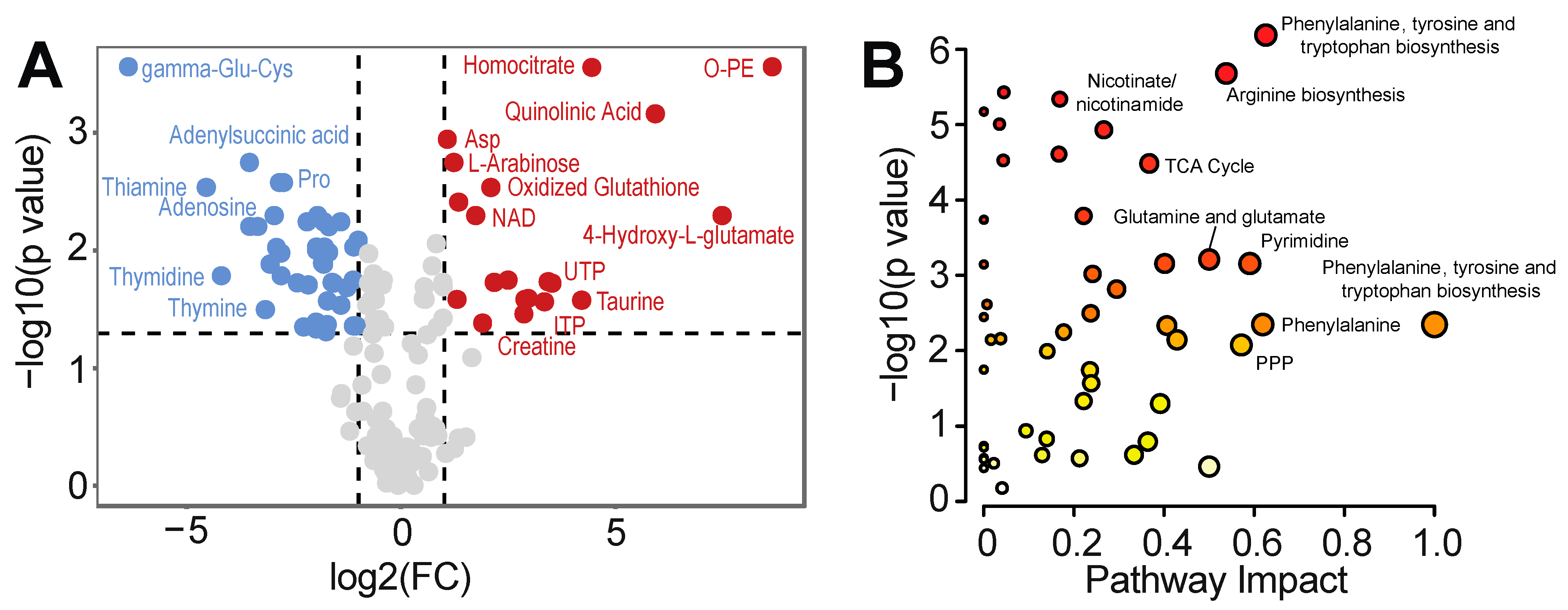
2.4. Metabolomics
2.5. Seahorse Extracellular Flux Analysis
2.6. Statistical Analysis
3. Results
3.1. Three-Dimensional Differentiation of Human Pluripotent Stem Cells
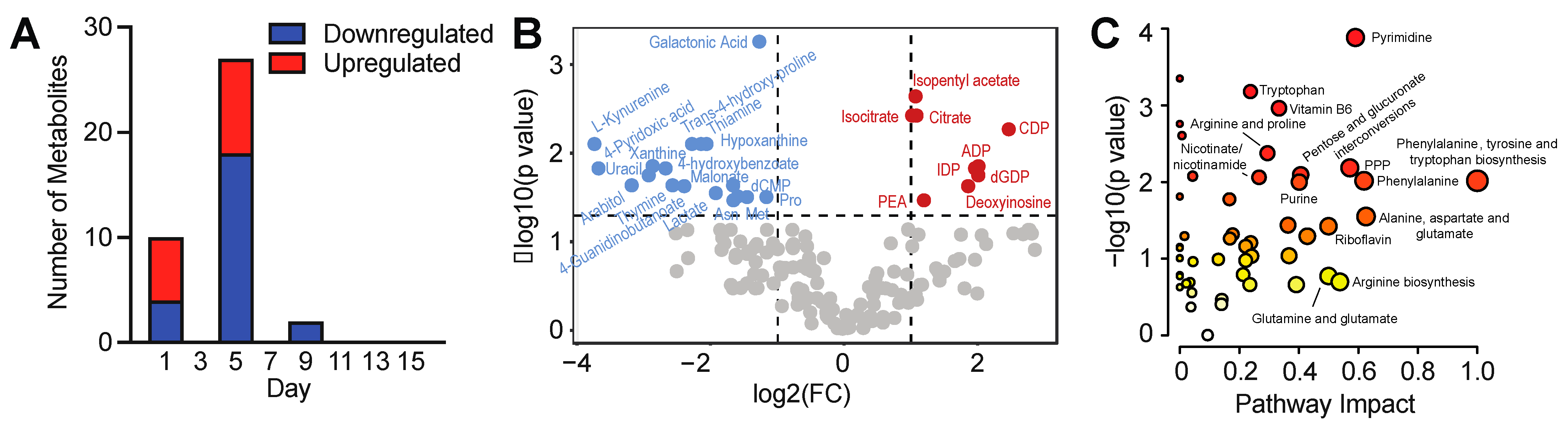
3.2. Intracellular Metabolic Remodeling during hPSC-CM Specification
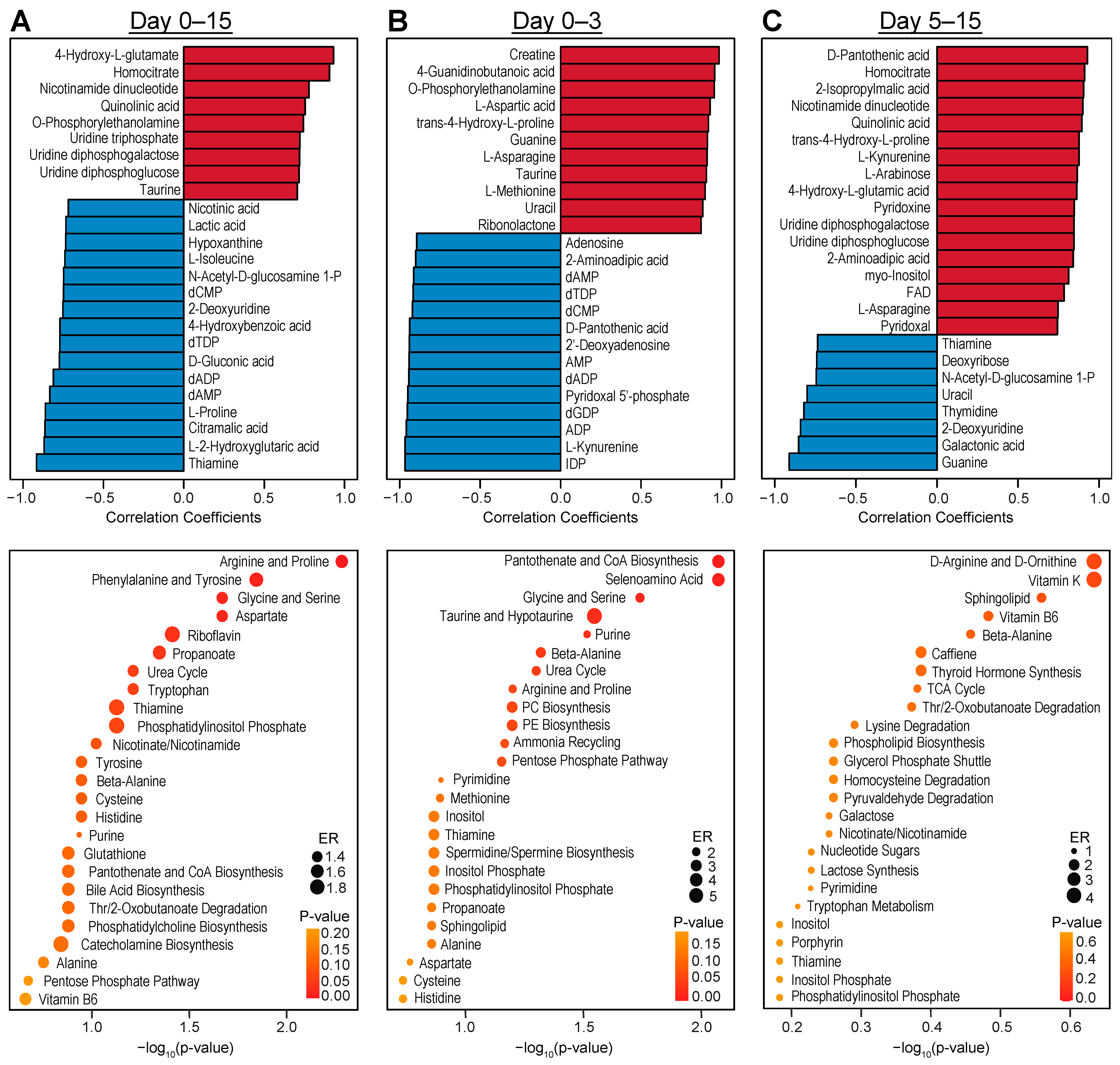
3.3. Temporal Analysis Indicates Stalled Metabolic Maturation following Cardiac Lineage Specification
3.4. Assessment of Real-Time Metabolic Function in hPSC-CM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Folmes, C.D.; Terzic, A. Metabolic determinants of embryonic development and stem cell fate. Reprod. Fertil. Dev. 2014, 27, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Dzeja, P.P.; Nelson, T.J.; Terzic, A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012, 11, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, O.A.; Pourquie, O. Exploring the Influence of Cell Metabolism on Cell Fate through Protein Post-translational Modifications. Dev. Cell 2020, 54, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Ghosh-Choudhary, S.; Liu, J.; Finkel, T. Metabolic Regulation of Cell Fate and Function. Trends Cell Biol. 2020, 30, 201–212. [Google Scholar] [CrossRef]
- Malandraki-Miller, S.; Lopez, C.A.; Al-Siddiqi, H.; Carr, C.A. Changing Metabolism in Differentiating Cardiac Progenitor Cells-Can Stem Cells Become Metabolically Flexible Cardiomyocytes? Front. Cardiovasc. Med. 2018, 5, 119. [Google Scholar] [CrossRef]
- Houghton, F.D.; Thompson, J.G.; Kennedy, C.J.; Leese, H.J. Oxygen consumption and energy metabolism of the early mouse embryo. Mol. Reprod. Dev. 1996, 44, 476–485. [Google Scholar] [CrossRef]
- Hom, J.R.; Quintanilla, R.A.; Hoffman, D.L.; de Mesy Bentley, K.L.; Molkentin, J.D.; Sheu, S.S.; Porter, G.A., Jr. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev. Cell 2011, 21, 469–478. [Google Scholar] [CrossRef]
- Folmes, C.D.; Dzeja, P.P.; Nelson, T.J.; Terzic, A. Mitochondria in control of cell fate. Circ. Res. 2012, 110, 526–529. [Google Scholar] [CrossRef]
- Zhao, Q.; Sun, Q.; Zhou, L.; Liu, K.; Jiao, K. Complex Regulation of Mitochondrial Function During Cardiac Development. J. Am. Heart Assoc. 2019, 8, e012731. [Google Scholar] [CrossRef]
- Johnson, M.T.; Mahmood, S.; Patel, M.S. Intermediary metabolism and energetics during murine early embryogenesis. J. Biol. Chem. 2003, 278, 31457–31460. [Google Scholar] [CrossRef]
- Ergir, E.; Oliver-De La Cruz, J.; Fernandes, S.; Cassani, M.; Niro, F.; Pereira-Sousa, D.; Vrbsky, J.; Vinarsky, V.; Perestrelo, A.R.; Debellis, D.; et al. Generation and maturation of human iPSC-derived 3D organotypic cardiac microtissues in long-term culture. Sci. Rep. 2022, 12, 17409. [Google Scholar] [CrossRef]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H.; et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021, 39, 737–746. [Google Scholar] [CrossRef]
- Chi, F.; Sharpley, M.S.; Nagaraj, R.; Roy, S.S.; Banerjee, U. Glycolysis-Independent Glucose Metabolism Distinguishes TE from ICM Fate during Mammalian Embryogenesis. Dev. Cell 2020, 53, 9–26.e24. [Google Scholar] [CrossRef] [PubMed]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Dzeja, P.P.; Faustino, R.S.; Perez-Terzic, C.; Behfar, A.; Terzic, A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4 (Suppl. S1), S60–S67. [Google Scholar] [CrossRef]
- Morita, Y.; Tohyama, S. Metabolic Regulation of Cardiac Differentiation and Maturation in Pluripotent Stem Cells: A Lesson from Heart Development. JMA J. 2020, 3, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Minter-Dykhouse, K.; Nelson, T.J.; Folmes, C.D.L. Uncoupling of Proliferative Capacity from Developmental Stage During Directed Cardiac Differentiation of Pluripotent Stem Cells. Stem Cells Dev. 2022, 31, 521–528. [Google Scholar] [CrossRef]
- DeLaughter, D.M.; Bick, A.G.; Wakimoto, H.; McKean, D.; Gorham, J.M.; Kathiriya, I.S.; Hinson, J.T.; Homsy, J.; Gray, J.; Pu, W.; et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev. Cell 2016, 39, 480–490. [Google Scholar] [CrossRef]
- Lopez, C.A.; Al-Siddiqi, H.; Purnama, U.; Iftekhar, S.; Bruyneel, A.A.N.; Kerr, M.; Nazir, R.; da Luz Sousa Fialho, M.; Malandraki-Miller, S.; Alonaizan, R.; et al. Physiological and pharmacological stimulation for in vitro maturation of substrate metabolism in human induced pluripotent stem cell-derived cardiomyocytes. Sci. Rep. 2021, 11, 7802. [Google Scholar] [CrossRef]
- Feyen, D.A.M.; McKeithan, W.L.; Bruyneel, A.A.N.; Spiering, S.; Hormann, L.; Ulmer, B.; Zhang, H.; Briganti, F.; Schweizer, M.; Hegyi, B.; et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020, 32, 107925. [Google Scholar] [CrossRef]
- Dai, D.F.; Danoviz, M.E.; Wiczer, B.; Laflamme, M.A.; Tian, R. Mitochondrial Maturation in Human Pluripotent Stem Cell Derived Cardiomyocytes. Stem Cells Int. 2017, 2017, 5153625. [Google Scholar] [CrossRef] [PubMed]
- Pakzad, K.K.; Tan, J.J.; Anderson, S.; Board, M.; Clarke, K.; Carr, C.A. Metabolic maturation of differentiating cardiosphere-derived cells. Stem Cell Res. 2021, 54, 102422. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.; Joshi, A.U.; Andorf, S.; Dai, Y.; Sampathkumar, S.; Chen, H.; Li, Y.; Garg, P.; Toischer, K.; Hasenfuss, G.; et al. Proteasome-Dependent Regulation of Distinct Metabolic States During Long-Term Culture of Human iPSC-Derived Cardiomyocytes. Circ. Res. 2019, 125, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.A.; Braam, S.R.; Koutsis, K.; Ng, E.S.; Jenny, R.; Lagerqvist, E.L.; Biben, C.; Hatzistavrou, T.; Hirst, C.E.; Yu, Q.C.; et al. NKX2-5eGFP/w hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods 2011, 8, 1037–1040. [Google Scholar] [CrossRef]
- Burridge, P.W.; Holmstrom, A.; Wu, J.C. Chemically Defined Culture and Cardiomyocyte Differentiation of Human Pluripotent Stem Cells. Curr. Protoc. Hum. Genet. 2015, 87, 21.3.1–21.3.15. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kremer, D.M.; Sajjakulnukit, P.; Zhang, L.; Lyssiotis, C.A. A large-scale analysis of targeted metabolomics data from heterogeneous biological samples provides insights into metabolite dynamics. Metabolomics 2019, 15, 103. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Friedman, C.E.; Nguyen, Q.; Lukowski, S.W.; Helfer, A.; Chiu, H.S.; Miklas, J.; Levy, S.; Suo, S.; Han, J.J.; Osteil, P.; et al. Single-Cell Transcriptomic Analysis of Cardiac Differentiation from Human PSCs Reveals HOPX-Dependent Cardiomyocyte Maturation. Cell Stem Cell 2018, 23, 586–598.e8. [Google Scholar] [CrossRef]
- Itoi, T.; Lopaschuk, G.D. The contribution of glycolysis, glucose oxidation, lactate oxidation, and fatty acid oxidation to ATP production in isolated biventricular working hearts from 2-week-old rabbits. Pediatr. Res. 1993, 34, 735–741. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Spafford, M.A. Energy substrate utilization by isolated working hearts from newborn rabbits. Am. J. Physiol. 1990, 258, H1274–H1280. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Spafford, M.A.; Marsh, D.R. Glycolysis Is Predominant Source of Myocardial Atp Production Immediately after Birth. Am. J. Physiol. 1991, 261, H1698–H1705. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Ussher, J.R.; Folmes, C.D.; Jaswal, J.S.; Stanley, W.C. Myocardial fatty acid metabolism in health and disease. Physiol. Rev. 2010, 90, 207–258. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef]
- Johnson, B.B.; Reinhold, J.; Holmes, T.L.; Moore, J.A.; Cowell, V.; Bernardo, A.S.; Rushworth, S.A.; Vassiliou, V.; Smith, J.G.W. Modelling Metabolic Shifts during Cardiomyocyte Differentiation, Iron Deficiency and Transferrin Rescue Using Human Pluripotent Stem Cells. Metabolites 2021, 12, 9. [Google Scholar] [CrossRef]
- Barison, M.J.; Pereira, I.T.; Waloski Robert, A.; Dallagiovanna, B. Reorganization of Metabolism during Cardiomyogenesis Implies Time-Specific Signaling Pathway Regulation. Int. J. Mol. Sci. 2021, 22, 1330. [Google Scholar] [CrossRef]
- Umei, T.C.; Tohyama, S.; Fukuda, K. Metabolism-based cardiomyocytes production for regenerative therapy. J. Mol. Cell. Cardiol. 2023, 176, 11–20. [Google Scholar] [CrossRef]
- Persad, K.L.; Lopaschuk, G.D. Energy Metabolism on Mitochondrial Maturation and Its Effects on Cardiomyocyte Cell Fate. Front. Cell Dev. Biol. 2022, 10, 886393. [Google Scholar] [CrossRef]
- Shi, H.; Enriquez, A.; Rapadas, M.; Martin, E.; Wang, R.; Moreau, J.; Lim, C.K.; Szot, J.O.; Ip, E.; Hughes, J.N.; et al. NAD Deficiency, Congenital Malformations, and Niacin Supplementation. N. Engl. J. Med. 2017, 377, 544–552. [Google Scholar] [CrossRef]
- Zhang, D.; Ning, J.; Ramprasath, T.; Yu, C.; Zheng, X.; Song, P.; Xie, Z.; Zou, M.H. Kynurenine promotes neonatal heart regeneration by stimulating cardiomyocyte proliferation and cardiac angiogenesis. Nat. Commun. 2022, 13, 6371. [Google Scholar] [CrossRef]
- Kropp, E.M.; Broniowska, K.A.; Waas, M.; Nycz, A.; Corbett, J.A.; Gundry, R.L. Cardiomyocyte Differentiation Promotes Cell Survival During Nicotinamide Phosphoribosyltransferase Inhibition Through Increased Maintenance of Cellular Energy Stores. Stem Cells Transl. Med. 2017, 6, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Piquereau, J.; Boitard, S.E.; Ventura-Clapier, R.; Mericskay, M. Metabolic Therapy of Heart Failure: Is There a Future for B Vitamins? Int. J. Mol. Sci. 2021, 23, 30. [Google Scholar] [CrossRef]
- Nisar, S.; Mohi, U.D.K.; Tak, S.I.; Andrabi, S.M.A.; Tanvir, M.; Muzaffer, U.; Kareem, O.; Ganie, M.A. Thiamine responsive high output heart failure of adults: An under-recognized entity. Eur. J. Clin. Nutr. 2023, 77, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Tanne, C.; Nguyen, J.; Blonde, R. Shoshin beriberi and thiamine-responsive right heart failure: A case report in Mayotte Recognition and management of infant Shoshin beriberi. Arch. Pediatr. 2022, 29, 624–625. [Google Scholar] [CrossRef] [PubMed]
- Helali, J.; Park, S.; Ziaeian, B.; Han, J.K.; Lankarani-Fard, A. Thiamine and Heart Failure: Challenging Cases of Modern-Day Cardiac Beriberi. Mayo Clin. Proc. Innov. Qual. Outcomes 2019, 3, 221–225. [Google Scholar] [CrossRef]
- Dinicolantonio, J.J.; Lavie, C.J.; Niazi, A.K.; O’Keefe, J.H.; Hu, T. Effects of thiamine on cardiac function in patients with systolic heart failure: Systematic review and metaanalysis of randomized, double-blind, placebo-controlled trials. Ochsner J. 2013, 13, 495–499. [Google Scholar]
- Wang, J.; Xue, Z.; Lin, J.; Wang, Y.; Ying, H.; Lv, Q.; Hua, C.; Wang, M.; Chen, S.; Zhou, B. Proline improves cardiac remodeling following myocardial infarction and attenuates cardiomyocyte apoptosis via redox regulation. Biochem. Pharmacol. 2020, 178, 114065. [Google Scholar] [CrossRef]
- Patriarca, E.J.; Cermola, F.; D’Aniello, C.; Fico, A.; Guardiola, O.; De Cesare, D.; Minchiotti, G. The Multifaceted Roles of Proline in Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 728576. [Google Scholar] [CrossRef]
- Washington, J.M.; Rathjen, J.; Felquer, F.; Lonic, A.; Bettess, M.D.; Hamra, N.; Semendric, L.; Tan, B.S.; Lake, J.A.; Keough, R.A.; et al. L-Proline induces differentiation of ES cells: A novel role for an amino acid in the regulation of pluripotent cells in culture. Am. J. Physiol. Cell Physiol. 2010, 298, C982–C992. [Google Scholar] [CrossRef]
- Tanner, J.J.; Fendt, S.M.; Becker, D.F. The Proline Cycle as a Potential Cancer Therapy Target. Biochemistry 2018, 57, 3433–3444. [Google Scholar] [CrossRef]
- Poluektov, Y.M.; Petrushanko, I.Y.; Undrovinas, N.A.; Lakunina, V.A.; Khapchaev, A.Y.; Kapelko, V.I.; Abramov, A.A.; Lakomkin, V.L.; Novikov, M.S.; Shirinsky, V.P.; et al. Glutathione-related substances maintain cardiomyocyte contractile function in hypoxic conditions. Sci. Rep. 2019, 9, 4872. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Yin, Y.; Ma, X.; Zhang, J.; Pan, W.; Tan, M.; Zhao, Y.; Yang, T.; Jiang, T.; Li, H. Glutathione system enhancement for cardiac protection: Pharmacological options against oxidative stress and ferroptosis. Cell Death Dis. 2023, 14, 131. [Google Scholar] [CrossRef]
- Almeida, H.V.; Tenreiro, M.F.; Louro, A.F.; Abecasis, B.; Santinha, D.; Calmeiro, T.; Fortunato, E.; Ferreira, L.; Alves, P.M.; Serra, M. Human Extracellular-Matrix Functionalization of 3D hiPSC-Based Cardiac Tissues Improves Cardiomyocyte Maturation. ACS Appl. Bio Mater. 2021, 4, 1888–1899. [Google Scholar] [CrossRef]
- Branco, M.A.; Cotovio, J.P.; Rodrigues, C.A.V.; Vaz, S.H.; Fernandes, T.G.; Moreira, L.M.; Cabral, J.M.S.; Diogo, M.M. Transcriptomic analysis of 3D Cardiac Differentiation of Human Induced Pluripotent Stem Cells Reveals Faster Cardiomyocyte Maturation Compared to 2D Culture. Sci. Rep. 2019, 9, 9229. [Google Scholar] [CrossRef]
- Yang, X.; Pabon, L.; Murry, C.E. Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014, 114, 511–523. [Google Scholar] [CrossRef]
- Malandraki-Miller, S.; Lopez, C.A.; Alonaizan, R.; Purnama, U.; Perbellini, F.; Pakzad, K.; Carr, C.A. Metabolic flux analyses to assess the differentiation of adult cardiac progenitors after fatty acid supplementation. Stem Cell Res. 2019, 38, 101458. [Google Scholar] [CrossRef]
- Murphy, S.A.; Chen, E.Z.; Tung, L.; Boheler, K.R.; Kwon, C. Maturing heart muscle cells: Mechanisms and transcriptomic insights. Semin. Cell Dev. Biol. 2021, 119, 49–60. [Google Scholar] [CrossRef]
- Emanuelli, G.; Zoccarato, A.; Reumiller, C.M.; Papadopoulos, A.; Chong, M.; Rebs, S.; Betteridge, K.; Beretta, M.; Streckfuss-Bomeke, K.; Shah, A.M. A roadmap for the characterization of energy metabolism in human cardiomyocytes derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2022, 164, 136–147. [Google Scholar] [CrossRef]
- Tohyama, S.; Hattori, F.; Sano, M.; Hishiki, T.; Nagahata, Y.; Matsuura, T.; Hashimoto, H.; Suzuki, T.; Yamashita, H.; Satoh, Y.; et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 2013, 12, 127–137. [Google Scholar] [CrossRef]
- Paredes, A.; Justo-Mendez, R.; Jimenez-Blasco, D.; Nunez, V.; Calero, I.; Villalba-Orero, M.; Alegre-Marti, A.; Fischer, T.; Gradillas, A.; Sant’Anna, V.A.R.; et al. gamma-Linolenic acid in maternal milk drives cardiac metabolic maturation. Nature 2023, 618, 365–373. [Google Scholar] [CrossRef]
- Yang, X.; Rodriguez, M.L.; Leonard, A.; Sun, L.; Fischer, K.A.; Wang, Y.; Ritterhoff, J.; Zhao, L.; Kolwicz, S.C., Jr.; Pabon, L.; et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2019, 13, 657–668. [Google Scholar] [CrossRef]
- Hu, D.; Linders, A.; Yamak, A.; Correia, C.; Kijlstra, J.D.; Garakani, A.; Xiao, L.; Milan, D.J.; van der Meer, P.; Serra, M.; et al. Metabolic Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes by Inhibition of HIF1alpha and LDHA. Circ. Res. 2018, 123, 1066–1079. [Google Scholar] [CrossRef]
- Kadota, S.; Pabon, L.; Reinecke, H.; Murry, C.E. In Vivo Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Neonatal and Adult Rat Hearts. Stem Cell Rep. 2017, 8, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Cho, G.S.; Lee, D.I.; Tampakakis, E.; Murphy, S.; Andersen, P.; Uosaki, H.; Chelko, S.; Chakir, K.; Hong, I.; Seo, K.; et al. Neonatal Transplantation Confers Maturation of PSC-Derived Cardiomyocytes Conducive to Modeling Cardiomyopathy. Cell Rep. 2017, 18, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.; Kwon, C. Regulation of cardiomyocyte maturation during critical perinatal window. J. Physiol. 2019, 598, 2941–2956. [Google Scholar] [CrossRef]
- Cantor, J.R. The Rise of Physiologic Media. Trends Cell Biol. 2019, 29, 854–861. [Google Scholar] [CrossRef]
- Eagle, H. Nutrition needs of mammalian cells in tissue culture. Science 1955, 122, 501–514. [Google Scholar] [CrossRef]
- Dulbecco, R.; Freeman, G. Plaque production by the polyoma virus. Virology 1959, 8, 396–397. [Google Scholar] [CrossRef]
- Claycomb, W.C.; Palazzo, M.C. Culture of the terminally differentiated adult cardiac muscle cell: A light and scanning electron microscope study. Dev. Biol. 1980, 80, 466–482. [Google Scholar] [CrossRef]
- Horackova, M.; Byczko, Z. Differences in the structural characteristics of adult guinea pig and rat cardiomyocytes during their adaptation and maintenance in long-term cultures: Confocal microscopy study. Exp. Cell Res. 1997, 237, 158–175. [Google Scholar] [CrossRef]
- Poindexter, B.J.; Smith, J.R.; Buja, L.M.; Bick, R.J. Calcium signaling mechanisms in dedifferentiated cardiac myocytes: Comparison with neonatal and adult cardiomyocytes. Cell Calcium 2001, 30, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Banyasz, T.; Lozinskiy, I.; Payne, C.E.; Edelmann, S.; Norton, B.; Chen, B.; Chen-Izu, Y.; Izu, L.T.; Balke, C.W. Transformation of adult rat cardiac myocytes in primary culture. Exp. Physiol. 2008, 93, 370–382. [Google Scholar] [CrossRef]
- Zhao, J.; Yao, K.; Yu, H.; Zhang, L.; Xu, Y.; Chen, L.; Sun, Z.; Zhu, Y.; Zhang, C.; Qian, Y.; et al. Metabolic remodelling during early mouse embryo development. Nat. Metab. 2021, 3, 1372–1384. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.R.; Abu-Remaileh, M.; Kanarek, N.; Freinkman, E.; Gao, X.; Louissaint, A., Jr.; Lewis, C.A.; Sabatini, D.M. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 2017, 169, 258–272.e217. [Google Scholar] [CrossRef] [PubMed]
- Vande Voorde, J.; Ackermann, T.; Pfetzer, N.; Sumpton, D.; Mackay, G.; Kalna, G.; Nixon, C.; Blyth, K.; Gottlieb, E.; Tardito, S. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv. 2019, 5, eaau7314. [Google Scholar] [CrossRef]

| Gene | Reverse | Forward |
|---|---|---|
| MEF2C | 5′-CCC AAG GAC TAA TCT GAT CGG-3′ | 5′-CTT TCT CTT TCC TGT TTC CTC CA-3′ |
| NKX 2.5 | 5′-CAC TCA GCA TTT GTA GAA AGT CAG-3′ | 5′-ACC CTA GAG CCG AAA AGA AAG-3′ |
| TNNT2 | 5′-TCT TCG TCC TCT CTC CAG TC-3′ | 5′-AGA AGA GGT GGT GGA AGA GTA-3′ |
| RPS29 | 5′-AAT ATG TGC CGC CAG TGT TT-3′ | 5′-CCC GGA TAA TCC TCT GAA GG-3′ |
| GATA2 | 5′-CTG TCT GCA ACG CCT GTG-3′ | 5′-GTT CCG AGT CTG GAT CCC TT-3′ |
| BMP4 | 5′-GCA CTG GTC TTG AGT ATC CTG-3′ | 5′-TGC TGA GGT TAA AGA GGA AAC G-3′ |
| OCT4 | 5′-AGT TTG TGC CAG GGT TTT TG-3′ | 5′-ACT TCA CCT TCC CTC CAA CC-3′ |
| SOX2 | 5′-CTT GAC CAC CGA ACC CAT-3′ | 5′-GTA CAA CTC CAT GAC CAG CTC-3′ |
| MIXL1 | 5′-GAA GGA TTT CCC ACT CTG ACG-3′ | 5′-GTA CCC CGA CAT CCA CTT G-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobori, S.N.; Zhu, Y.; Saarinen, A.; Liuzzo, A.J.; Folmes, C.D.L. Metabolic Remodeling during Early Cardiac Lineage Specification of Pluripotent Stem Cells. Metabolites 2023, 13, 1086. https://doi.org/10.3390/metabo13101086
Bobori SN, Zhu Y, Saarinen A, Liuzzo AJ, Folmes CDL. Metabolic Remodeling during Early Cardiac Lineage Specification of Pluripotent Stem Cells. Metabolites. 2023; 13(10):1086. https://doi.org/10.3390/metabo13101086
Chicago/Turabian StyleBobori, Sunday Ndoma, Yuxiang Zhu, Alicia Saarinen, Alexis Josephine Liuzzo, and Clifford D. L. Folmes. 2023. "Metabolic Remodeling during Early Cardiac Lineage Specification of Pluripotent Stem Cells" Metabolites 13, no. 10: 1086. https://doi.org/10.3390/metabo13101086
APA StyleBobori, S. N., Zhu, Y., Saarinen, A., Liuzzo, A. J., & Folmes, C. D. L. (2023). Metabolic Remodeling during Early Cardiac Lineage Specification of Pluripotent Stem Cells. Metabolites, 13(10), 1086. https://doi.org/10.3390/metabo13101086