Several Metabolite Families Display Inflexibility during Glucose Challenge in Patients with Type 2 Diabetes: An Untargeted Metabolomics Study
Abstract
1. Introduction
2. Methods
2.1. Subjects and Study Design
2.2. Metabolites Quantification
2.3. Pathway Analysis and Statistical Analysis
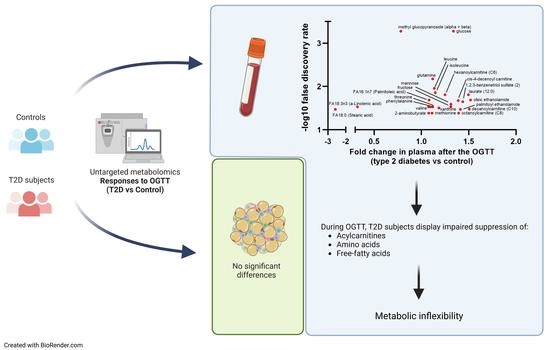
3. Results
3.1. Untargeted Metabolomics and Lipidomics during the OGTT in Controls and Type 2 Diabetes Subjects
3.2. Metabolic Pathways Associated with Abnormal Response to Oral Glucose in Type 2 Diabetes
3.3. Associations with Clinical Parameters
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fanni, G. Diabetes Is Not (Only) a Disorder of Glucose Metabolism! Diabet. Med. 2021, 38, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Tareen, S.H.K.; Kutmon, M.; Adriaens, M.E.; Mariman, E.C.M.; de Kok, T.M.; Arts, I.C.W.; Evelo, C.T. Exploring the Cellular Network of Metabolic Flexibility in the Adipose Tissue. Genes Nutr. 2018, 13, 17. [Google Scholar] [CrossRef]
- Rynders, C.A.; Blanc, S.; DeJong, N.; Bessesen, D.H.; Bergouignan, A. Sedentary Behaviour Is a Key Determinant of Metabolic Inflexibility. J. Physiol. 2018, 596, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.A.; Yu, T.; Jones, D.P.; Ramirez-Zea, M.; Stein, A.D. Metabolomic Profiling After a Meal Shows Greater Changes and Lower Metabolic Flexibility in Cardiometabolic Diseases. J. Endocr. Soc. 2020, 4, 11. [Google Scholar] [CrossRef]
- Ahola-Olli, A.V.; Mustelin, L.; Kalimeri, M.; Kettunen, J.; Jokelainen, J.; Auvinen, J.; Puukka, K.; Havulinna, A.S.; Lehtimäki, T.; Kähönen, M.; et al. Circulating Metabolites and the Risk of Type 2 Diabetes: A Prospective Study of 11,896 Young Adults from Four Finnish Cohorts. Diabetologia 2019, 62, 2298–2309. [Google Scholar] [CrossRef]
- Wittenbecher, C.; Guasch-Ferré, M.; Haslam, D.E.; Dennis, C.; Li, J.; Bhupathiraju, S.N.; Lee, C.-H.; Qi, Q.; Liang, L.; Eliassen, A.H.; et al. Changes in Metabolomics Profiles over Ten Years and Subsequent Risk of Developing Type 2 Diabetes: Results from the Nurses’ Health Study. eBioMedicine 2022, 75, 103799. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef]
- Wildberg, C.; Masuch, A.; Budde, K.; Kastenmüller, G.; Artati, A.; Rathmann, W.; Adamski, J.; Kocher, T.; Völzke, H.; Nauck, M.; et al. Plasma Metabolomics to Identify and Stratify Patients with Impaired Glucose Tolerance. J. Clin. Endocrinol. Metab. 2019, 104, 6357–6370. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martínez-González, M.A.; Salas-Salvadó, J.; Hu, F.B. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-Analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef]
- Menni, C.; Fauman, E.; Erte, I.; Perry, J.R.B.; Kastenmüller, G.; Shin, S.-Y.; Petersen, A.-K.; Hyde, C.; Psatha, M.; Ward, K.J.; et al. Biomarkers for Type 2 Diabetes and Impaired Fasting Glucose Using a Nontargeted Metabolomics Approach. Diabetes 2013, 62, 4270–4276. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite Profiles and the Risk of Developing Diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Patti, M.E.; Brambilla, E.; Luzi, L.; Landaker, E.J.; Kahn, C.R. Bidirectional Modulation of Insulin Action by Amino Acids. J. Clin. Investig. 1998, 101, 1519–1529. [Google Scholar] [CrossRef]
- Zhao, X.; Peter, A.; Fritsche, J.; Elcnerova, M.; Fritsche, A.; Häring, H.U.; Schleicher, E.D.; Xu, G.; Lehmann, R. Changes of the Plasma Metabolome during an Oral Glucose Tolerance Test: Is There More than Glucose to Look At? Am. J. Physiol. Endocrinol. Metab. 2009, 296, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.E.; Larson, M.G.; Vasan, R.S.; Ghorbani, A.; Cheng, S.; Rhee, E.P.; Florez, J.C.; Clish, C.B.; Gerszten, R.E.; Wang, T.J. Metabolite Profiles During Oral Glucose Challenge. Diabetes 2013, 62, 2689–2698. [Google Scholar] [CrossRef]
- Shaham, O.; Wei, R.; Wang, T.J.; Ricciardi, C.; Lewis, G.D.; Vasan, R.S.; Carr, S.A.; Thadhani, R.; Gerszten, R.E.; Mootha, V.K. Metabolic Profiling of the Human Response to a Glucose Challenge Reveals Distinct Axes of Insulin Sensitivity. Mol. Syst. Biol. 2008, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Sun, L.; Zong, G.; Shen, X.; Zheng, H.; Jin, Q.; Li, H.; Yin, H.; Lin, X. Changes in Plasma Metabolome Profiles Following Oral Glucose Challenge among Adult Chinese. Nutrients 2021, 13, 1474. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.J.; Skrtic, S.; Katsogiannos, P.; Abrahamsson, N.; Sidibeh, C.O.; Dahgam, S.; Månsson, M.; Risérus, U.; Kullberg, J.; Eriksson, J.W. Impaired Adipose Tissue Lipid Storage, but Not Altered Lipolysis, Contributes to Elevated Levels of NEFA in Type 2 Diabetes. Degree of Hyperglycemia and Adiposity Are Important Factors. Metabolism 2016, 65, 1768–1780. [Google Scholar] [CrossRef]
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing Value Imputation Approach for Mass Spectrometry-Based Metabolomics Data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef]
- Diamanti, K.; Cavalli, M.; Pan, G.; Pereira, M.J.; Kumar, C.; Skrtic, S.; Grabherr, M.; Risérus, U.; Eriksson, J.W.; Komorowski, J.; et al. Intra- and Inter-Individual Metabolic Profiling Highlights Carnitine and Lysophosphatidylcholine Pathways as Key Molecular Defects in Type 2 Diabetes. Sci. Rep. 2019, 9, 9653. [Google Scholar] [CrossRef]
- Nowak, C.; Hetty, S.; Salihovic, S.; Castillejo-Lopez, C.; Ganna, A.; Cook, N.L.; Broeckling, C.D.; Prenni, J.E.; Shen, X.; Giedraitis, V.; et al. Glucose Challenge Metabolomics Implicates Medium-Chain Acylcarnitines in Insulin Resistance. Sci. Rep. 2018, 8, 8691. [Google Scholar] [CrossRef]
- Liu, L.; Feng, R.; Guo, F.; Li, Y.; Jiao, J.; Sun, C. Targeted Metabolomic Analysis Reveals the Association between the Postprandial Change in Palmitic Acid, Branched-Chain Amino Acids and Insulin Resistance in Young Obese Subjects. Diabetes Res. Clin. Pract. 2015, 108, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.W.; Visvanathar, R.; Kullberg, J.; Strand, R.; Skrtic, S.; Ekström, S.; Lubberink, M.; Lundqvist, M.H.; Katsogiannos, P.; Pereira, M.J.; et al. Tissue-Specific Glucose Partitioning and Fat Content in Prediabetes and Type 2 Diabetes: Whole-Body PET/MRI during Hyperinsulinemia. Eur. J. Endocrinol. 2021, 184, 879–899. [Google Scholar] [CrossRef] [PubMed]
- Galgani, J.E.; Heilbronn, L.K.; Azuma, K.; Kelley, D.E.; Albu, J.B.; Pi-Sunyer, X.; Smith, S.R.; Ravussin, E. Metabolic Flexibility in Response to Glucose Is Not Impaired in People with Type 2 Diabetes after Controlling for Glucose Disposal Rate. Diabetes 2008, 57, 841–845. [Google Scholar] [CrossRef]
- Kim, H.W. Metabolomic Approaches to Investigate the Effect of Metformin: An Overview. Int. J. Mol. Sci. 2021, 22, 10275. [Google Scholar] [CrossRef]
- Scheen, A.J. Clinical Pharmacokinetics of Metformin. Clin. Pharmacokinet. 1996, 30, 359–371. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, S.; Zhang, C.; Zhao, Y. Coordinated Modulation of Energy Metabolism and Inflammation by Branched-Chain Amino Acids and Fatty Acids. Front. Endocrinol. 2020, 11, 617. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.-S. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients 2016, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, R.J.O.; Rizo-Roca, D.; Chibalin, A.V.; Chorell, E.; Furrer, R.; Katayama, S.; Harada, J.; Karlsson, H.K.R.; Handschin, C.; Moritz, T.; et al. Branched-Chain Amino Acid Metabolism Is Regulated by ERRα in Primary Human Myotubes and Is Further Impaired by Glucose Loading in Type 2 Diabetes. Diabetologia 2021, 64, 2077–2091. [Google Scholar] [CrossRef]
- Newgard, C.B. Interplay between Lipids and Branched-Chain Amino Acids in Development of Insulin Resistance. Cell Metab. 2012, 15, 606–614. [Google Scholar] [CrossRef]
- Lynch, C.J.; Adams, S.H. Branched-Chain Amino Acids in Metabolic Signalling and Insulin Resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef]
- Vanweert, F.; Schrauwen, P.; Phielix, E. Role of Branched-Chain Amino Acid Metabolism in the Pathogenesis of Obesity and Type 2 Diabetes-Related Metabolic Disturbances BCAA Metabolism in Type 2 Diabetes. Nutr. Diabetes 2022, 12, 35. [Google Scholar] [CrossRef]
- Geidenstam, N.; Spégel, P.; Mulder, H.; Filipsson, K.; Ridderstråle, M.; Danielsson, A.P.H. Metabolite Profile Deviations in an Oral Glucose Tolerance Test-a Comparison between Lean and Obese Individuals. Obesity 2014, 22, 2388–2395. [Google Scholar] [CrossRef]
- Wang, Q.; Jokelainen, J.; Auvinen, J.; Puukka, K.; Keinänen-Kiukaanniemi, S.; Järvelin, M.R.; Kettunen, J.; Mäkinen, V.P.; Ala-Korpela, M. Insulin Resistance and Systemic Metabolic Changes in Oral Glucose Tolerance Test in 5340 Individuals: An Interventional Study. BMC Med. 2019, 17, 217. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Gao, H.Y.; Fan, Z.Y.; He, Y.; Yan, Y.X. Metabolomics Signatures in Type 2 Diabetes: A Systematic Review and Integrative Analysis. J. Clin. Endocrinol. Metab. 2020, 105, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B.; Arneth, R.; Shams, M. Metabolomics of Type 1 and Type 2 Diabetes. Int. J. Mol. Sci. 2019, 20, 2467. [Google Scholar] [CrossRef]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as Regulators of Insulin Sensitivity and Metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Lim, H.R.; Kim, J.W.; Shin, K.-H. Metabolite Changes in Risk of Type 2 Diabetes Mellitus in Cohort Studies: A Systematic Review and Meta-Analysis. Diabetes Res. Clin. Pract. 2018, 140, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, X.; Zhang, R.; Liu, Z.; Shen, N.; Di, Y.; Fang, T.; Li, H.; Tian, F. Discovery and Comparison of Serum Biomarkers for Diabetes Mellitus and Metabolic Syndrome Based on UPLC-Q-TOF/MS. Clin. Biochem. 2020, 82, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.D.; Stevens, R.D.; Antinozzi, P.A.; Anderson, A.; Bergman, R.N.; Wagenknecht, L.E.; Newgard, C.B.; Bowden, D.W. Metabolomic Profile Associated with Insulin Resistance and Conversion to Diabetes in the Insulin Resistance Atherosclerosis Study. J. Clin. Endocrinol. Metab. 2015, 100, E463–E468. [Google Scholar] [CrossRef]
- Qiu, G.; Zheng, Y.; Wang, H.; Sun, J.; Ma, H.; Xiao, Y.; Li, Y.; Yuan, Y.; Yang, H.; Li, X.; et al. Plasma Metabolomics Identified Novel Metabolites Associated with Risk of Type 2 Diabetes in Two Prospective Cohorts of Chinese Adults. Int. J. Epidemiol. 2016, 45, 1507–1516. [Google Scholar] [CrossRef]
- Porcu, E.; Gilardi, F.; Darrous, L.; Yengo, L.; Bararpour, N.; Gasser, M.; Marques-Vidal, P.; Froguel, P.; Waeber, G.; Thomas, A.; et al. Triangulating Evidence from Longitudinal and Mendelian Randomization Studies of Metabolomic Biomarkers for Type 2 Diabetes. Sci. Rep. 2021, 11, 6197. [Google Scholar] [CrossRef]
- Papandreou, C.; Hernández-Alonso, P.; Bulló, M.; Ruiz-Canela, M.; Li, J.; Guasch-Ferré, M.; Toledo, E.; Clish, C.; Corella, D.; Estruch, R.; et al. High Plasma Glutamate and a Low Glutamine-to-Glutamate Ratio Are Associated with Increased Risk of Heart Failure but Not Atrial Fibrillation in the Prevención Con Dieta Mediterránea (PREDIMED) Study. J. Nutr. 2020, 150, 2882–2889. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.R.; de la Rosa, M.V.G.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Makarova, E.; Makrecka-Kuka, M.; Vilks, K.; Volska, K.; Sevostjanovs, E.; Grinberga, S.; Zarkova-Malkova, O.; Dambrova, M.; Liepinsh, E. Decreases in Circulating Concentrations of Long-Chain Acylcarnitines and Free Fatty Acids During the Glucose Tolerance Test Represent Tissue-Specific Insulin Sensitivity. Front. Endocrinol. 2019, 10, 870. [Google Scholar] [CrossRef]
- Yoshimura, K.; Hirano, S.; Takata, H.; Funakoshi, S.; Ohmi, S.; Amano, E.; Nishi, Y.; Inoue, M.; Fukuda, Y.; Hayashi, H.; et al. Plasma Mannose Level, a Putative Indicator of Glycogenolysis, and Glucose Tolerance in Japanese Individuals. J. Diabetes Investig. 2017, 8, 489. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, T.; Yamashita, E.; Mizutani, T.; Nakajima, H.; Yabuuchi, M.; Asano, N.; Miwa, I. Hepatic Glycogen Breakdown Is Implicated in the Maintenance of Plasma Mannose Concentration. Am. J. Physiol.-Endocrinol. Metab. 2005, 288, E534–E540. [Google Scholar] [CrossRef]
- Gabbay, K.H. Hyperglycemia, Polyol Metabolism, and Complications of Diabetes Mellitus. Annu. Rev. Med. 1975, 26, 521–536. [Google Scholar] [CrossRef]
- Lanaspa, M.A.; Ishimoto, T.; Li, N.; Cicerchi, C.; Orlicky, D.J.; Ruzycki, P.; Rivard, C.; Inaba, S.; Roncal-Jimenez, C.A.; Bales, E.S.; et al. Endogenous Fructose Production and Metabolism in the Liver Contributes to the Development of Metabolic Syndrome. Nat. Commun. 2013, 4, 2434. [Google Scholar] [CrossRef]
- Burns, T.W.; Terry, B.E.; Langley, P.E.; Robison, G.A. Insulin Inhibition of Lipolysis of Human Adipocytes the Role of Cyclic Adenosine Monophosphate. Diabetes 1979, 28, 957–961. [Google Scholar] [CrossRef]
- Frape, D.L.; Williams, N.R.; Carpenter, K.L.H.; Freeman, M.A.; Palmer, C.R.; Fletcher, R.J. Insulin Response and Changes in Composition of Non-Esterified Fatty Acids in Blood Plasma of Middle-Aged Men Following Isoenergetic Fatty and Carbohydrate Breakfasts. Br. J. Nutr. 2000, 84, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Faludi, G.; Spitzer, J.J. Changes of Individual Free Fatty Acids during Glucose Tolerance Test. Diabetes 1967, 16, 175–180. [Google Scholar] [CrossRef]
- Gilbert, C.H.; Kaye, J.; Galton, D.J. The Effect of a Glucose Load on Plasma Fatty Acids and Lipolysis in Adipose Tissue of Obese Diabetic and Non-Diabetic Patients. Diabetologia 1974, 10, 135–138. [Google Scholar] [CrossRef]
- Rahman, S.M.K.; Uyama, T.; Hussain, Z.; Ueda, N. Roles of Endocannabinoids and Endocannabinoid-Like Molecules in Energy Homeostasis and Metabolic Regulation: A Nutritional Perspective. Annu. Rev. Nutr. 2021, 41, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Chiazza, F.; Collino, M. Peroxisome Proliferator-Activated Receptors (PPARs) in Glucose Control. In Molecular Nutrition and Diabetes: A Volume in the Molecular Nutrition Series; Academic Press: Cambridge, MA, USA, 2016; pp. 105–114. [Google Scholar] [CrossRef]
- Diamanti, K.; Cavalli, M.; Pereira, M.J.; Pan, G.; Castillejo-López, C.; Kumar, C.; Mundt, F.O.; Komorowski, J.; Deshmukh, A.S.; Mann, M.; et al. Organ-specific metabolic pathways distinguish prediabetes, type 2 diabetes, and normal tissues. Cell Rep. Med. 2022, 3, 100763. [Google Scholar] [CrossRef]
- Boden, G.; Lebed, B.; Schatz, M.; Homko, C.; Lemieux, S. Effects of Acute Changes of Plasma Free Fatty Acids on Intramyocellular Fat Content and Insulin Resistance in Healthy Subjects. Diabetes 2001, 50, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Muoio, D.M. Metabolic Inflexibility: When Mitochondrial Indecision Leads to Metabolic Gridlock. Cell 2014, 159, 1253. [Google Scholar] [CrossRef] [PubMed]
- Lackey, D.E.; Lynch, C.J.; Olson, K.C.; Mostaedi, R.; Ali, M.; Smith, W.H.; Karpe, F.; Humphreys, S.; Bedinger, D.H.; Dunn, T.N.; et al. Regulation of Adipose Branched-Chain Amino Acid Catabolism Enzyme Expression and Cross-Adipose Amino Acid Flux in Human Obesity. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E1175–E1187. [Google Scholar] [CrossRef]




| Control | T2D | |
|---|---|---|
| N (F/M) | 10/10 | 10/10 |
| Age (years) | 58 (11) | 58 (9) |
| Diabetes duration (years) | N/A | 4 (3) |
| BMI (kg/m2) | 30.8 (4.6) | 30.7 (4.9) |
| Waist-to-hip ratio | 0.96 (0.07) | 0.99 (0.05) |
| Plasma glucose (mmol/L) | 6.0 (0.7) | 8.2 (1.5) * |
| Serum insulin (mU/L) | 11.5 (5.2) | 15.5 (7.0) * |
| HbA1c (mmol/mol) | 37.3 (3.7) | 48.8 (8.6) *** |
| HOMA-IR | 3.08 (1.58) | 5.27 (2.86) ** |
| Standardized Beta | p-Value | 95% CI | R2 | |
|---|---|---|---|---|
| Model 1 | ||||
| GluAUC | −0.736 | <0.001 | [−0.30, −0.13] | |
| HOMA-IR | −0.010 | 0.950 | [−0.46, 0.43] | |
| BMI | −0.222 | 0.098 | [−0.36, 0.03] | |
| 0.61 *** | ||||
| Model 2 | ||||
| HbA1c | −0.463 | 0.003 | [−0.30, −0.07] | |
| HOMA-IR | −0.289 | 0.068 | [−0.85, 0.03] | |
| BMI | −0.132 | 0.379 | [−0.32, 0.12] | |
| 0.47 *** | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanni, G.; Eriksson, J.W.; Pereira, M.J. Several Metabolite Families Display Inflexibility during Glucose Challenge in Patients with Type 2 Diabetes: An Untargeted Metabolomics Study. Metabolites 2023, 13, 131. https://doi.org/10.3390/metabo13010131
Fanni G, Eriksson JW, Pereira MJ. Several Metabolite Families Display Inflexibility during Glucose Challenge in Patients with Type 2 Diabetes: An Untargeted Metabolomics Study. Metabolites. 2023; 13(1):131. https://doi.org/10.3390/metabo13010131
Chicago/Turabian StyleFanni, Giovanni, Jan W. Eriksson, and Maria J. Pereira. 2023. "Several Metabolite Families Display Inflexibility during Glucose Challenge in Patients with Type 2 Diabetes: An Untargeted Metabolomics Study" Metabolites 13, no. 1: 131. https://doi.org/10.3390/metabo13010131
APA StyleFanni, G., Eriksson, J. W., & Pereira, M. J. (2023). Several Metabolite Families Display Inflexibility during Glucose Challenge in Patients with Type 2 Diabetes: An Untargeted Metabolomics Study. Metabolites, 13(1), 131. https://doi.org/10.3390/metabo13010131