Comprehensive Metabolomic Fingerprinting Combined with Chemometrics Identifies Species- and Variety-Specific Variation of Medicinal Herbs: An Ocimum Study
Abstract
1. Introduction
2. Materials and Methods
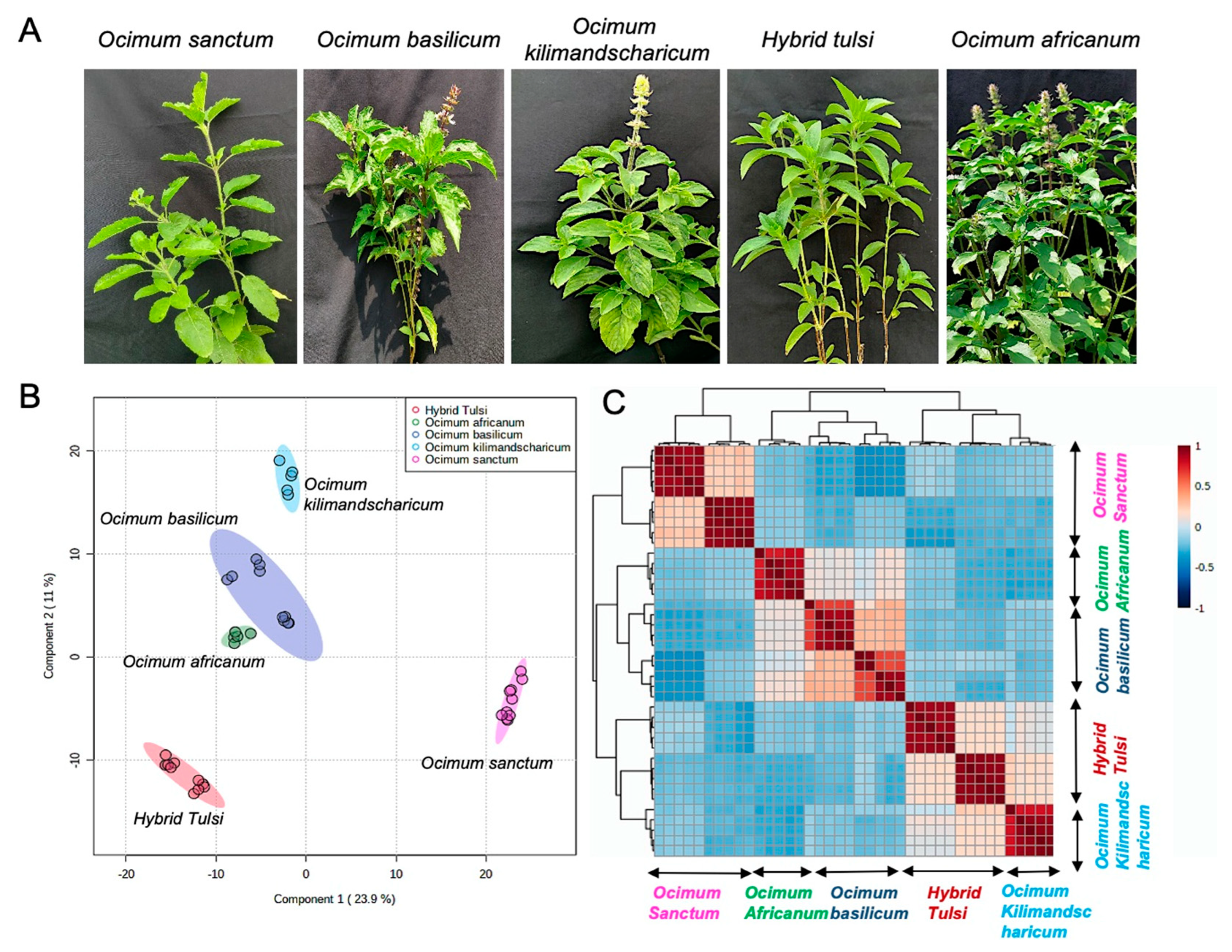
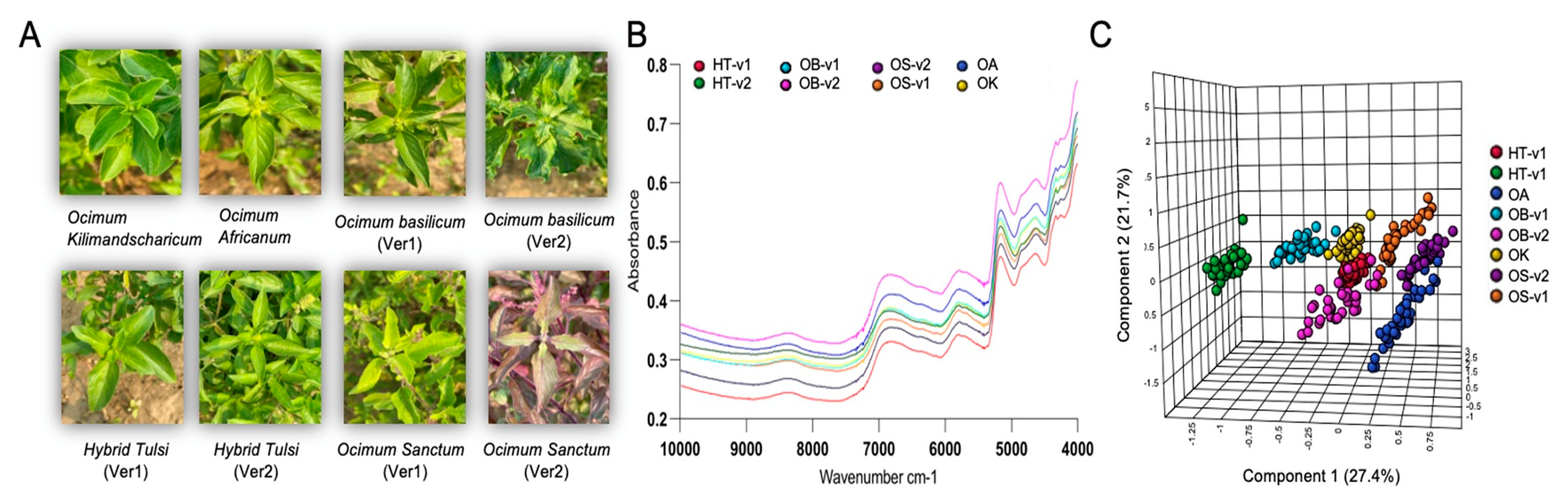
2.1. Plant Materials
2.2. Sample Preparation and LC-MS/MS Conditions
2.3. Sample Preparation and GC-MS Conditions
2.4. FT-NIR Spectroscopy Analysis
2.5. Data Processing and Analysis
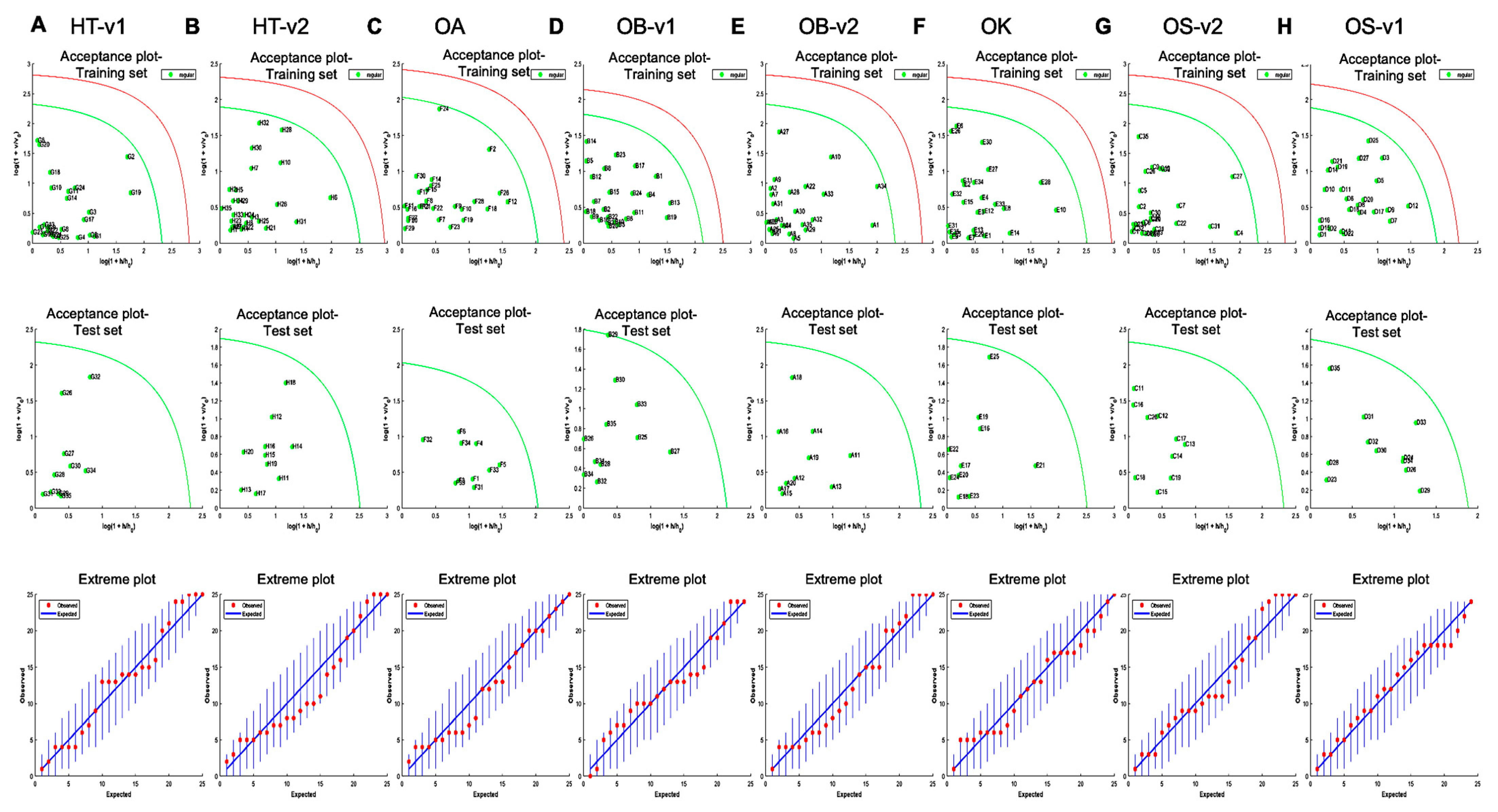
2.6. One Class Classification Model
3. Results and Discussion
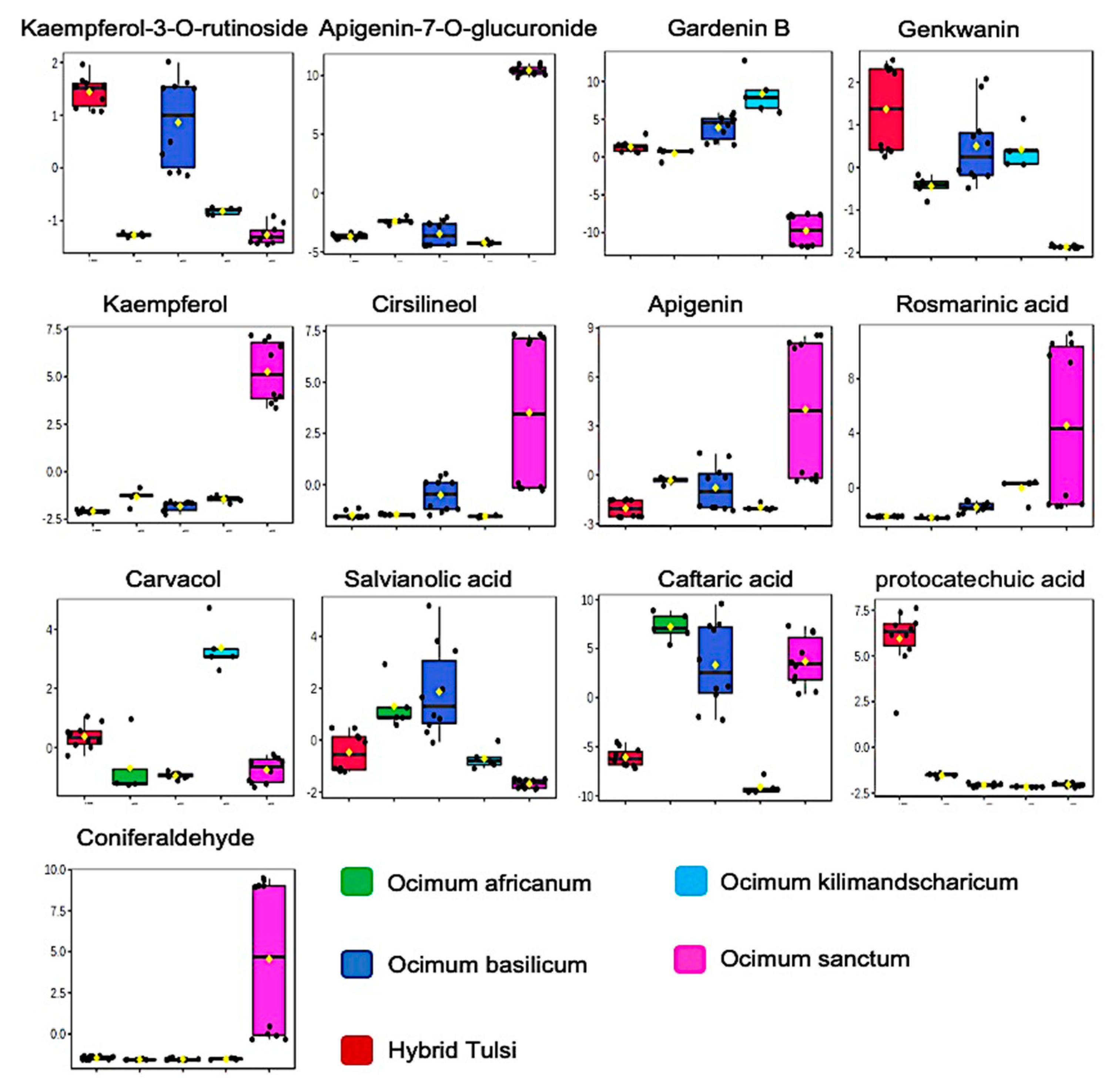
3.1. LC-MS-Based Metabolic Profiling of Ocimum Leaves Identified Species-Specific Variation
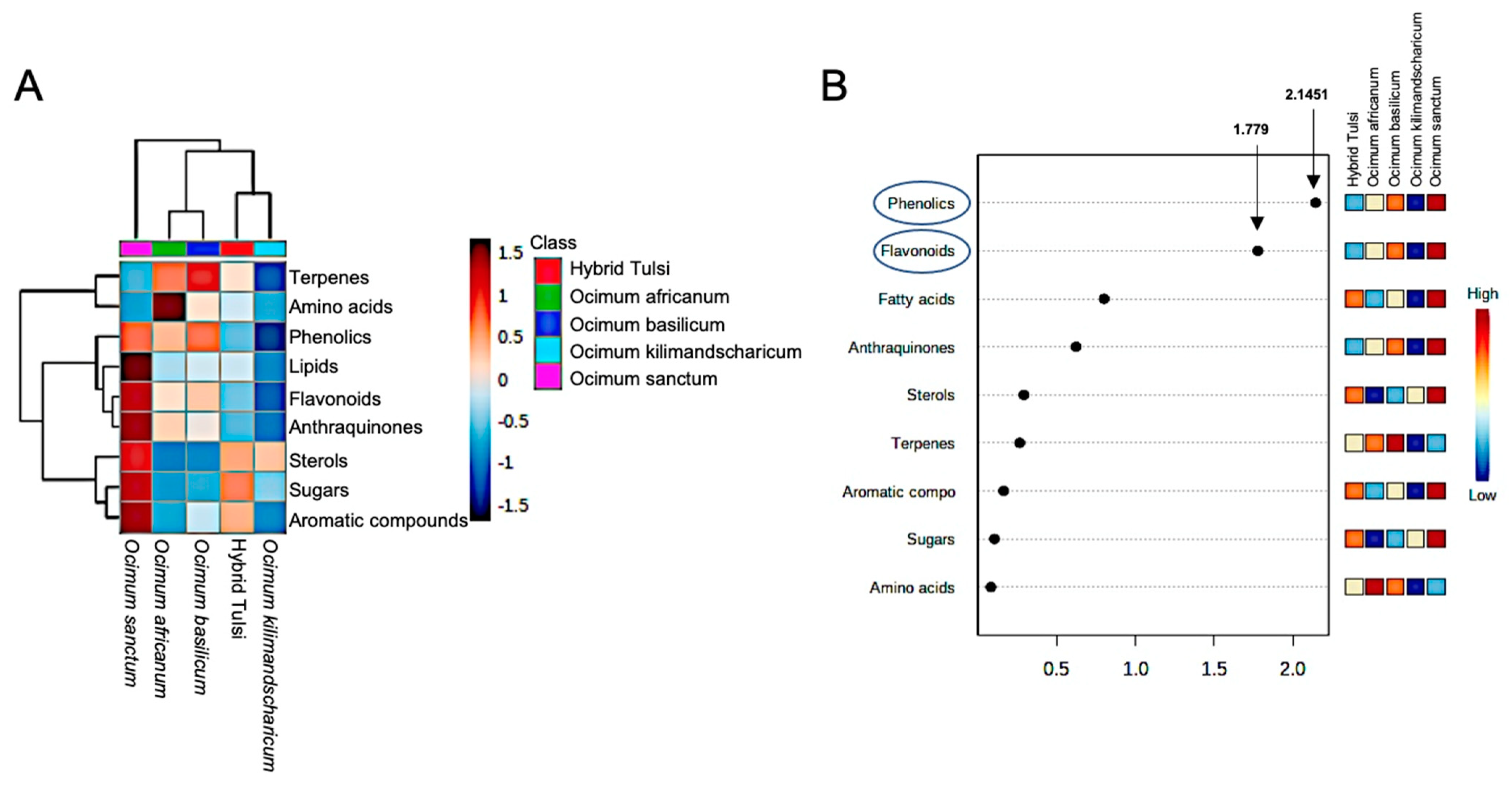
3.2. GC-MS-Based Metabolic Fingerprinting Identified Ocimum Species-Specific Variation
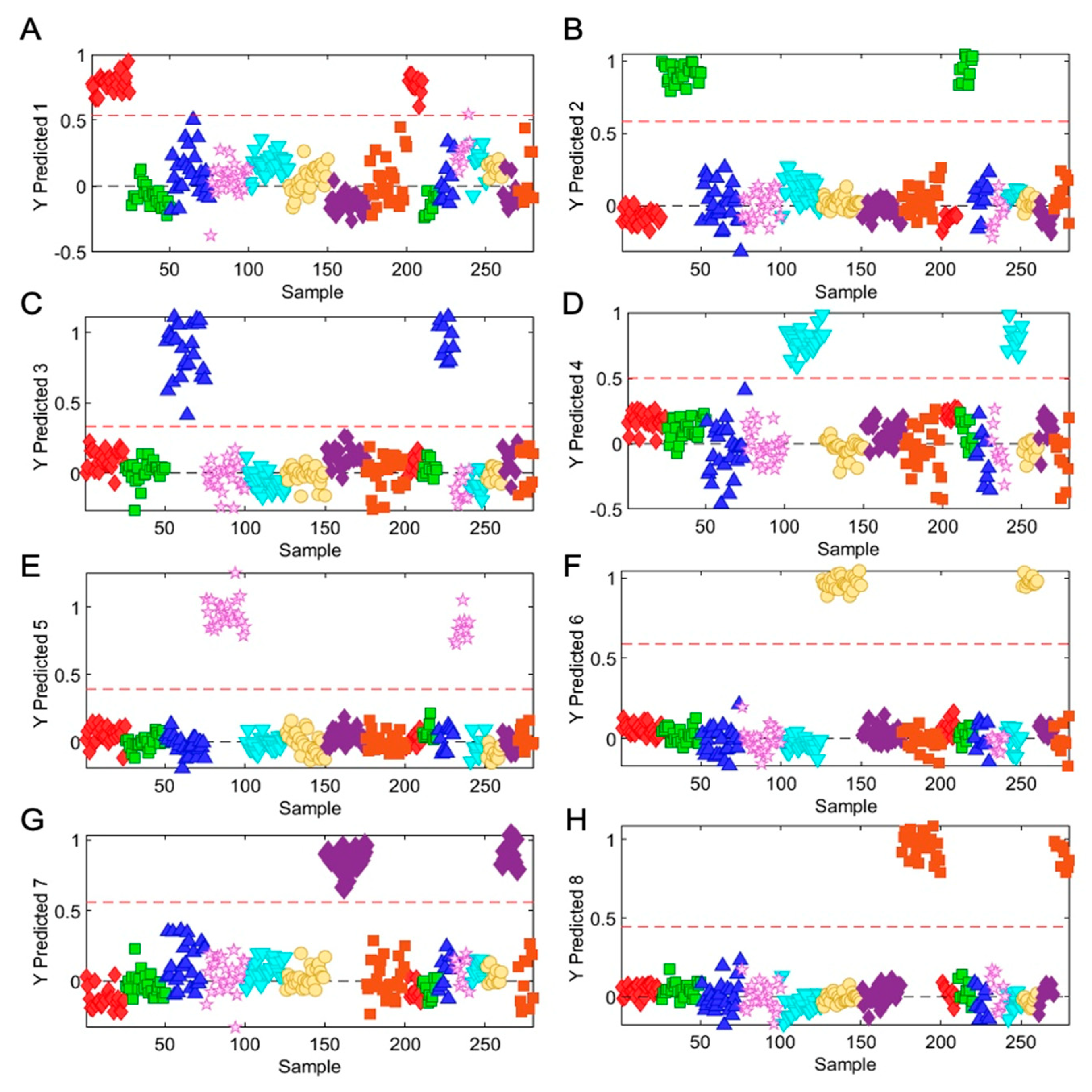
3.3. Rapid Metabolic Fingerprinting with FT-NIR Identified Species- and Variety-Specific Variation of Ocimum Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bungãu, S.G.; Popa, V.-C. Between religion and science some aspects concerning illness and healing in antiquity. Transylv. Rev. 2015, 24, 3–18. [Google Scholar]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- General Guidelines for Methodologies on Research and Evaluation of Traditional Medicine; World Health Organization: Geneva, Switzerland, 2000.
- Huck, C. Infrared Spectroscopic Technologies for the Quality Control of Herbal Medicines. In Evidence-Based Validation of Herbal Medicine; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 477–493. [Google Scholar] [CrossRef]
- Glevitzky, I.; Dumitrel, G.A.; Glevitzky, M.; Pasca, B.; Otrisal, P.; Bungau, S.; Cioca, G.; Pantis, C.; Popa, M. Statistical Analysis of the Relationship between Antioxidant Activity and the Structure of Flavonoid Compounds. Rev. Chim. 2019, 70, 3103–3107. [Google Scholar] [CrossRef]
- Ekor, M. The Growing Use of Herbal Medicines: Issues Relating to Adverse Reactions and Challenges in Monitoring Safety. Front. Neurol. 2014, 4, 177. [Google Scholar] [CrossRef]
- Singh, D.; Chaudhuri, P.K. A Review on Phytochemical and Pharmacological Properties of Holy Basil (Ocimum sanctum L.). Ind. Crops Prod. 2018, 118, 367–382. [Google Scholar] [CrossRef]
- Nahak, G.; Mishra, R.C.; Sahu, R.K. Taxonomic Distribution, Medicinal Properties and Drug Development Potentiality of Ocimum (Tulsi). Drug Invent. Today 2011, 3, 95–113. [Google Scholar]
- Sestili, P.; Ismail, T.; Calcabrini, C.; Guescini, M.; Catanzaro, E.; Turrini, E.; Layla, A.; Akhtar, S.; Fimognari, C. The Potential Effects of Ocimum Basilicum on Health: A Review of Pharmacological and Toxicological Studies. Expert Opin. Drug Metab. Toxicol. 2018, 14, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Chaiyana, W.; Anuchapreeda, S.; Punyoyai, C.; Neimkhum, W.; Lee, K.H.; Lin, W.C.; Lue, S.C.; Viernstein, H.; Mueller, M. Ocimum Sanctum Linn. as a Natural Source of Skin Anti-Ageing Compounds. Ind. Crops Prod. 2019, 127, 217–224. [Google Scholar] [CrossRef]
- Abidoyea, A.O.; Ojedokunb, F.O.; Fasogbonc, B.M.; Bamidele, O.P. Effects of Sweet Basil Leaves (Ocimum basilicum L.) Addition on the Chemical, Antioxidant, and Storage Stability of Roselle Calyces (Hibiscus sabdariffa) Drink. Food Chem. 2022, 371, 131170. [Google Scholar] [CrossRef]
- Maria Simona, C.; Muste, S.; Viman, I.; Georgiana, F. Basil (Ocimum basilicum L.) Chemical Compositions and Applications in Food Processing. Hop and Medicinal Plants. January 2020. Available online: https://www.researchgate.net/publication/351370972 (accessed on 1 December 2022).
- Collin, H. Herbs, Spices and Cardiovascular Disease; Woodhead Publishing: Sawston, UK, 2006. [Google Scholar] [CrossRef]
- Dharsono, H.D.A.; Putri, S.A.; Kurnia, D.; Dudi, D.; Satari, M.H. Ocimum Species: A Review on Chemical Constituents and Antibacterial Activity. Molecules 2022, 27, 6350. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Moura, E.; Faroni, L.R.D.A.; Heleno, F.F.; Rodrigues, A.A.Z.; Prates, L.H.F.; De Queiroz, M.E.L.R. Optimal Extraction of Ocimum basilicum Essential Oil by Association of Ultrasound and Hydrodistillation and Its Potential as a Biopesticide against a Major Stored Grains Pest. Molecules 2020, 25, 2781. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, T.; Mandal, A.; Roy, S.C.; De Sarker, D. Diversity of the Genus Ocimum (Lamiaceae) through Morpho-Molecular (RAPD) and Chemical (GC–MS) Analysis. J. Genet. Eng. Biotechnol. 2017, 15, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Balekundri, A.; Mannur, V. Quality Control of the Traditional Herbs and Herbal Products: A Review. Futur. J. Pharm. Sci. 2020, 6, 67. [Google Scholar] [CrossRef]
- Yeshi, K.; Crayn, D.; Ritmejerytė, E.; Wangchuk, P. Plant Secondary Metabolites Produced in Response to Abiotic Stresses Has Potential Application in Pharmaceutical Product Development. Molecules 2022, 27, 313. [Google Scholar] [CrossRef]
- Tangpao, T.; Charoimek, N.; Teerakitchotikan, P.; Leksawasdi, N.; Jantanasakulwong, K.; Rachtanapun, P.; Seesuriyachan, P.; Phimolsiripol, Y.; Chaiyaso, T.; Ruksiriwanich, W.; et al. Volatile Organic Compounds from Basil Essential Oils: Plant Taxonomy, Biological Activities, and Their Applications in Tropical Fruit Productions. Horticulturae 2022, 8, 144. [Google Scholar] [CrossRef]
- Avetisyan, A.; Markosian, A.; Petrosyan, M.; Sahakyan, N.; Babayan, A.; Aloyan, S.; Trchounian, A. Chemical Composition and Some Biological Activities of the Essential Oils from Basil Ocimum Different Cultivars. BMC Complement. Altern. Med. 2017, 17, 60. [Google Scholar] [CrossRef]
- Gurav, T.P.; Dholakia, B.B.; Giri, A.P. A Glance at the Chemodiversity of Ocimum Species: Trends, Implications, and Strategies for the Quality and Yield Improvement of Essential Oil. Phytochem. Rev. 2022, 21, 879–913. [Google Scholar] [CrossRef]
- Patel, M.K.; Pandey, S.; Kumar, M.; Haque, M.I.; Pal, S.; Yadav, N.S. Plants Metabolome Study: Emerging Tools and Techniques. Plants 2021, 10, 2409. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.G.; Singh, D.; Lee, J.S.; Lee, S.; Lee, C.H. Exploring the Metabolomic Diversity of Plant Species across Spatial (Leaf and Stem) Components and Phylogenic Groups. BMC Plant Biol. 2020, 20, 39. [Google Scholar] [CrossRef]
- Ch, R.; Chevallier, O.; McCarron, P.; McGrath, T.F.; Wu, D.; Nguyen Doan Duy, L.; Kapil, A.P.; McBride, M.; Elliott, C.T. Metabolomic Fingerprinting of Volatile Organic Compounds for the Geographical Discrimination of Rice Samples from China, Vietnam and India. Food Chem. 2021, 334, 127553. [Google Scholar] [CrossRef]
- Villate, A.; San Nicolas, M.; Gallastegi, M.; Aulas, P.A.; Olivares, M.; Usobiaga, A.; Etxebarria, N.; Aizpurua-Olaizola, O. Review: Metabolomics as a Prediction Tool for Plants Performance under Environmental Stress. Plant Sci. 2021, 303, 110789. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Nagana Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. Nmr Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Salem, M.A.; De Souza, L.P.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Beć, K.B.; Grabska, J.; Huck, C.W. NIR Spectroscopy of Natural Medicines Supported by Novel Instrumentation and Methods for Data Analysis and Interpretation. J. Pharm. Biomed. Anal. 2021, 193, 113686. [Google Scholar] [CrossRef] [PubMed]
- Muráriková, A.; Ťažký, A.; Neugebauerová, J.; Planková, A.; Jampílek, J.; Mučaji, P.; Mikuš, P. Characterization of Essential Oil Composition in Different Basil Species and Pot Cultures by a GC-MS Method. Molecules 2017, 22, 1221. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Shah, S.; Kumar, R.; Kumar, A.; Shasany, A.K. Comparative Temporal Metabolomics Studies to Investigate Interspecies Variation in Three Ocimum Species. Sci. Rep. 2020, 10, 5234. [Google Scholar] [CrossRef]
- Pandey, R.; Kumar, B. HPLC-QTOF-MS/MS-Based Rapid Screening of Phenolics and Triterpenic Acids in Leaf Extracts of Ocimum Species and Their Interspecies Variation. J. Liq. Chromatogr. Relat. Technol. 2016, 39, 225–238. [Google Scholar] [CrossRef]
- Bhuvaneshwari, K.; Gokulanathan, A.; Jayanthi, M.; Govindasamy, V.; Milella, L.; Lee, S.; Yang, D.C.; Girija, S. Can Ocimum basilicum L. and Ocimum tenuiflorum L. in Vitro Culture Be a Potential Source of Secondary Metabolites? Food Chem. 2016, 194, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Ratti, C. Hot Air and Freeze-Drying of High-Value Foods: A Review. J. Food Eng. 2001, 49, 311–319. [Google Scholar] [CrossRef]
- Ch, R.; Rey, G.; Ray, S.; Jha, P.K.; Driscoll, P.C.; Dos Santos, M.S.; Malik, D.M.; Lach, R.; Weljie, A.M.; MacRae, J.I.; et al. Rhythmic Glucose Metabolism Regulates the Redox Circadian Clockwork in Human Red Blood Cells. Nat. Commun. 2021, 12, 377. [Google Scholar] [CrossRef]
- Pomerantsev, A.L.; Rodionova, O.Y. Concept and Role of Extreme Objects in PCA/SIMCA. J. Chemom. 2014, 28, 429–438. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA Cross Validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. Springer Series in Statistics the Elements of Statistical Learning Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Koo, I.; Kim, S.; Zhang, X. Comparative Analysis of Mass Spectral Matching-Based Compound Identification in Gas Chromatography-Mass Spectrometry. J. Chromatogr. A 2013, 1298, 132–138. [Google Scholar] [CrossRef]
- Granato, D.; Putnik, P.; Kovačević, D.B.; Santos, J.S.; Calado, V.; Rocha, R.S.; Da Cruz, A.G.; Jarvis, B.; Rodionova, O.Y.; Pomerantsev, A. Trends in Chemometrics: Food Authentication, Microbiology, and Effects of Processing. Compr. Rev. Food Sci. Food Saf. 2018, 17, 663–677. [Google Scholar] [CrossRef]
- Shannon, M.; Ratnasekhar, C.H.; McGrath, T.F.; Kapil, A.P.; Elliott, C.T. A Two-Tiered System of Analysis to Tackle Rice Fraud: The Indian Basmati Study. Talanta 2021, 225, 122038. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, Biological Functions, and Biotechnological Applications. Frontiers in Plant Science. Front. Res. Found. 2012, 3, 222. [Google Scholar] [CrossRef]
- Khalid, M.; Saeed-ur-Rahman; Bilal, M.; Huang, D. Role of Flavonoids in Plant Interactions with the Environment and against Human Pathogens—A Review. Journal of Integrative Agriculture. Chin. Acad. Agric. Sci. 2019, 18, 211–230. [Google Scholar] [CrossRef]
- Prinsi, B.; Morgutti, S.; Negrini, N.; Faoro, F.; Espen, L. Insight into Composition of Bioactive Phenolic Compounds in Leaves and Flowers of Green and Purple Basil. Plants 2020, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Majdi, M.; Malekzadeh-Mashhady, A.; Maroufi, A.; Crocoll, C. Tissue-Specific Gene-Expression Patterns of Genes Associated with Thymol/Carvacrol Biosynthesis in Thyme (Thymus vulgaris L.) and Their Differential Changes upon Treatment with Abiotic Elicitors. Plant Physiol. Biochem. 2017, 115, 152–162. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, P.; Deng, Z.; Jin, G.; Zhang, Y. Chicoric Acid Provides Better Ultraviolet Protection than the Sum of Its Substrates in Purple Coneflower Plants. Ind. Crops Prod. 2021, 170, 113778. [Google Scholar] [CrossRef]
- Daloso, D.M.; Medeiros, D.B.; dos Anjos, L.; Yoshida, T.; Araújo, W.L.; Fernie, A.R. Metabolism within the Specialized Guard Cells of Plants. New Phytol. 2017, 216, 1018–1033. [Google Scholar] [CrossRef]
- Riggs, J.W.; Rockwell, N.C.; Cavales, P.C.; Callis, J. Identification of the Plant Ribokinase and Discovery of a Role for Arabidopsis Ribokinase in Nucleoside Metabolism. J. Biol. Chem. 2016, 291, 22572–22582. [Google Scholar] [CrossRef] [PubMed]
- Seifert, J.G.; Shecterle, L.M. Use of Ribose to Enhance Plant Growth. U.S. Patent US 8486859b2, 16 July 2013. [Google Scholar]
- Cho, Y.H.; Yoo, S.D. Signaling Role of Fructose Mediated by FINS1/FBP in Arabidopsis Thaliana. PLoS Genet. 2011, 7, e1001263. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.A.; Rahman, U.U.; Khan, M.R.; Sahar, A.; Mehmood, T.; Khan, M. Essential Oil Eugenol: Sources, Extraction Techniques and Nutraceutical Perspectives. RSC Advances. R. Soc. Chem. 2017, 7, 32669–32681. [Google Scholar] [CrossRef]
- Ma, L.; Peng, Y.; Pei, Y.; Zeng, J.; Shen, H.; Cao, J.; Qiao, Y.; Wu, Z. Systematic Discovery about NIR Spectral Assignment from Chemical Structural Property to Natural Chemical Compounds. Sci. Rep. 2019, 9, 9503. [Google Scholar] [CrossRef] [PubMed]

| Complete Model | ||||||
|---|---|---|---|---|---|---|
| Training | ||||||
| Variety | Sen (%) | TFN (%) | Spe (%) | TFP (%) | Acc (%) | Rel (%) |
| HT-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| HT-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OA | 100 | 0 | 100 | 0 | 100 | 100 |
| OB-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OB-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| OK | 100 | 0 | 100 | 0 | 100 | 100 |
| OS-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OS-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| TEST | ||||||
| HT-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| HT-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OA | 100 | 0 | 100 | 0 | 100 | 100 |
| OB-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OB-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| OK | 100 | 0 | 100 | 0 | 100 | 100 |
| OS-V2 | 100 | 0 | 100 | 0 | 100 | 100 |
| OS-V1 | 100 | 0 | 100 | 0 | 100 | 100 |
| Types | PC | α | γ | DOF (SD) | DOF (OD) | SEN | SPE |
|---|---|---|---|---|---|---|---|
| HT-v1 | 2 | 0.010 | 0.01 | 1 | 1 | 100 | 100 |
| HT-v2 | 2 | 0.010 | 0.01 | 1 | 2 | 100 | 100 |
| OA | 2 | 0.010 | 0.01 | 2 | 2 | 100 | 100 |
| OB-v1 | 2 | 0.010 | 0.01 | 2 | 3 | 100 | 100 |
| OB-v2 | 2 | 0.010 | 0.01 | 1 | 1 | 100 | 100 |
| OK | 2 | 0.010 | 0.01 | 1 | 2 | 100 | 100 |
| OS-v2 | 2 | 0.010 | 0.01 | 1 | 1 | 100 | 100 |
| OS-v1 | 2 | 0.010 | 0.01 | 3 | 3 | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, A.K.; Khan, S.; Kumar, A.; Dubey, B.K.; Lal, R.K.; Tiwari, A.; Trivedi, P.K.; Elliott, C.T.; Ch, R. Comprehensive Metabolomic Fingerprinting Combined with Chemometrics Identifies Species- and Variety-Specific Variation of Medicinal Herbs: An Ocimum Study. Metabolites 2023, 13, 122. https://doi.org/10.3390/metabo13010122
Rai AK, Khan S, Kumar A, Dubey BK, Lal RK, Tiwari A, Trivedi PK, Elliott CT, Ch R. Comprehensive Metabolomic Fingerprinting Combined with Chemometrics Identifies Species- and Variety-Specific Variation of Medicinal Herbs: An Ocimum Study. Metabolites. 2023; 13(1):122. https://doi.org/10.3390/metabo13010122
Chicago/Turabian StyleRai, Abhishek Kumar, Samreen Khan, Akhilesh Kumar, Basant Kumar Dubey, R. K. Lal, Ashutosh Tiwari, Prabodh Kumar Trivedi, Christopher T. Elliott, and Ratnasekhar Ch. 2023. "Comprehensive Metabolomic Fingerprinting Combined with Chemometrics Identifies Species- and Variety-Specific Variation of Medicinal Herbs: An Ocimum Study" Metabolites 13, no. 1: 122. https://doi.org/10.3390/metabo13010122
APA StyleRai, A. K., Khan, S., Kumar, A., Dubey, B. K., Lal, R. K., Tiwari, A., Trivedi, P. K., Elliott, C. T., & Ch, R. (2023). Comprehensive Metabolomic Fingerprinting Combined with Chemometrics Identifies Species- and Variety-Specific Variation of Medicinal Herbs: An Ocimum Study. Metabolites, 13(1), 122. https://doi.org/10.3390/metabo13010122







