A Combination of Metabolomics and Machine Learning Results in the Identification of a New Cyst Nematode Hatching Factor
Abstract
:1. Introduction
2. Results
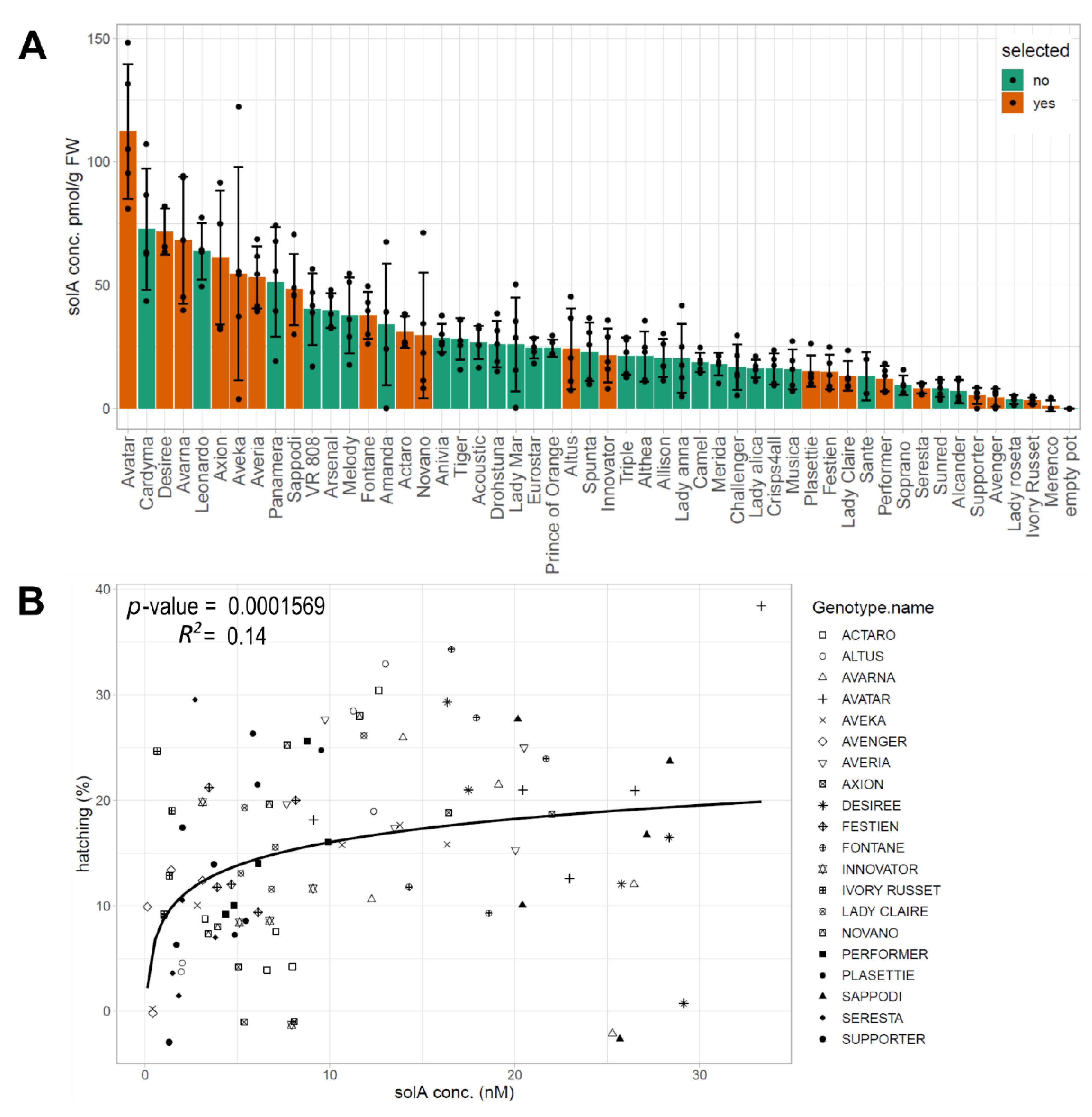
2.1. Natural Variation in the SolA Content of Root Exudates Obtained from Potato Cultivars Is Associated with Differences in Hatching Activity
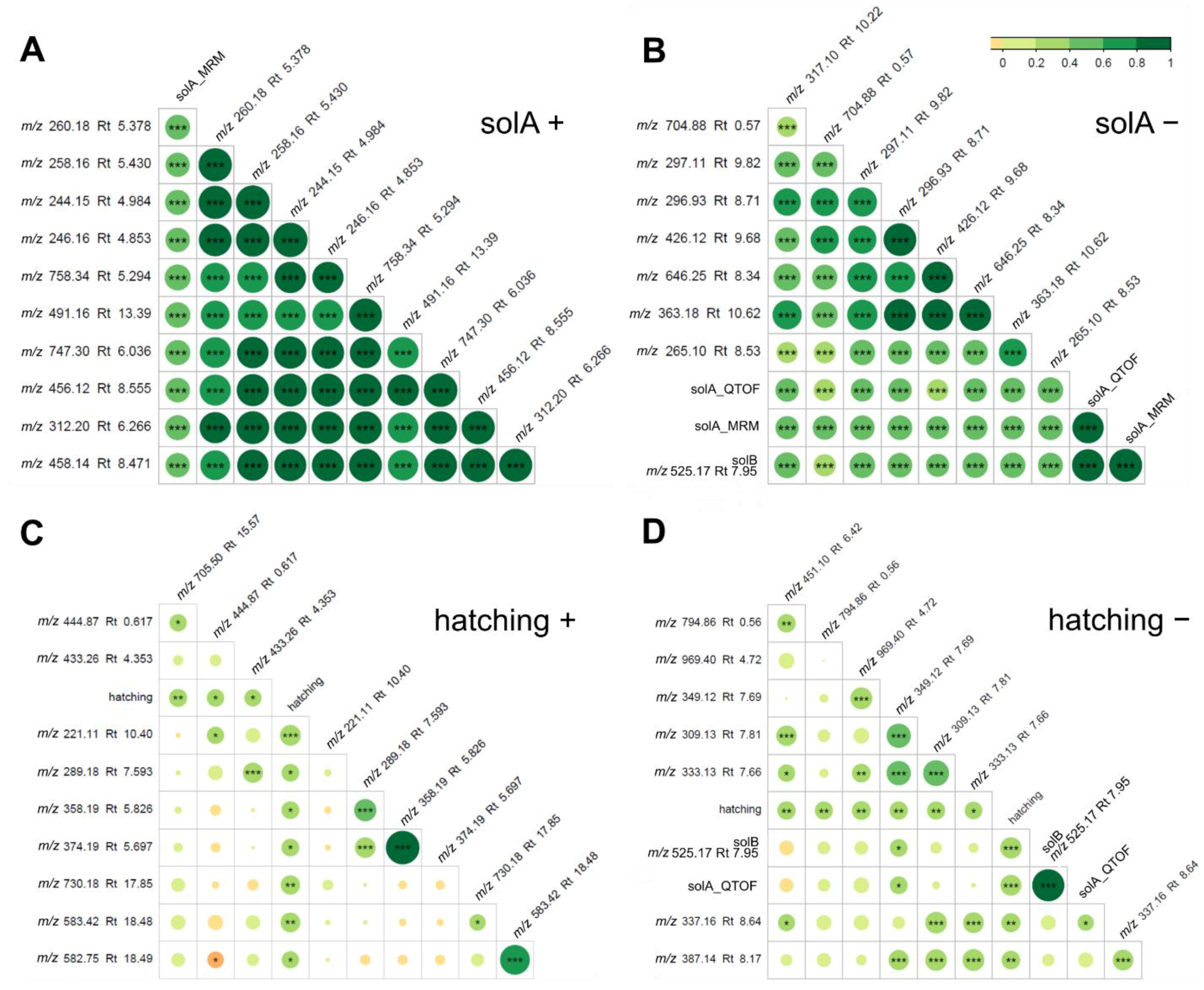
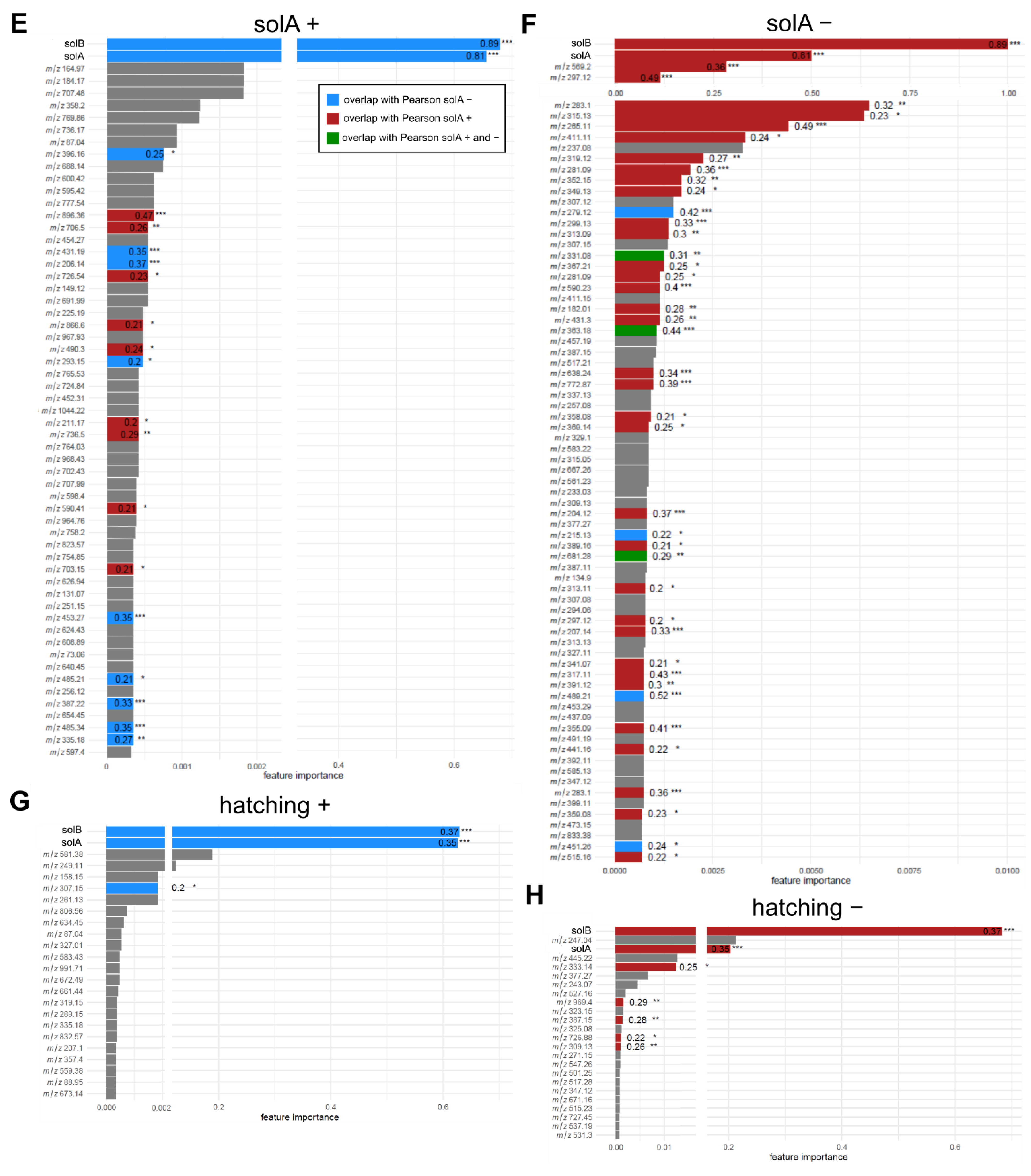
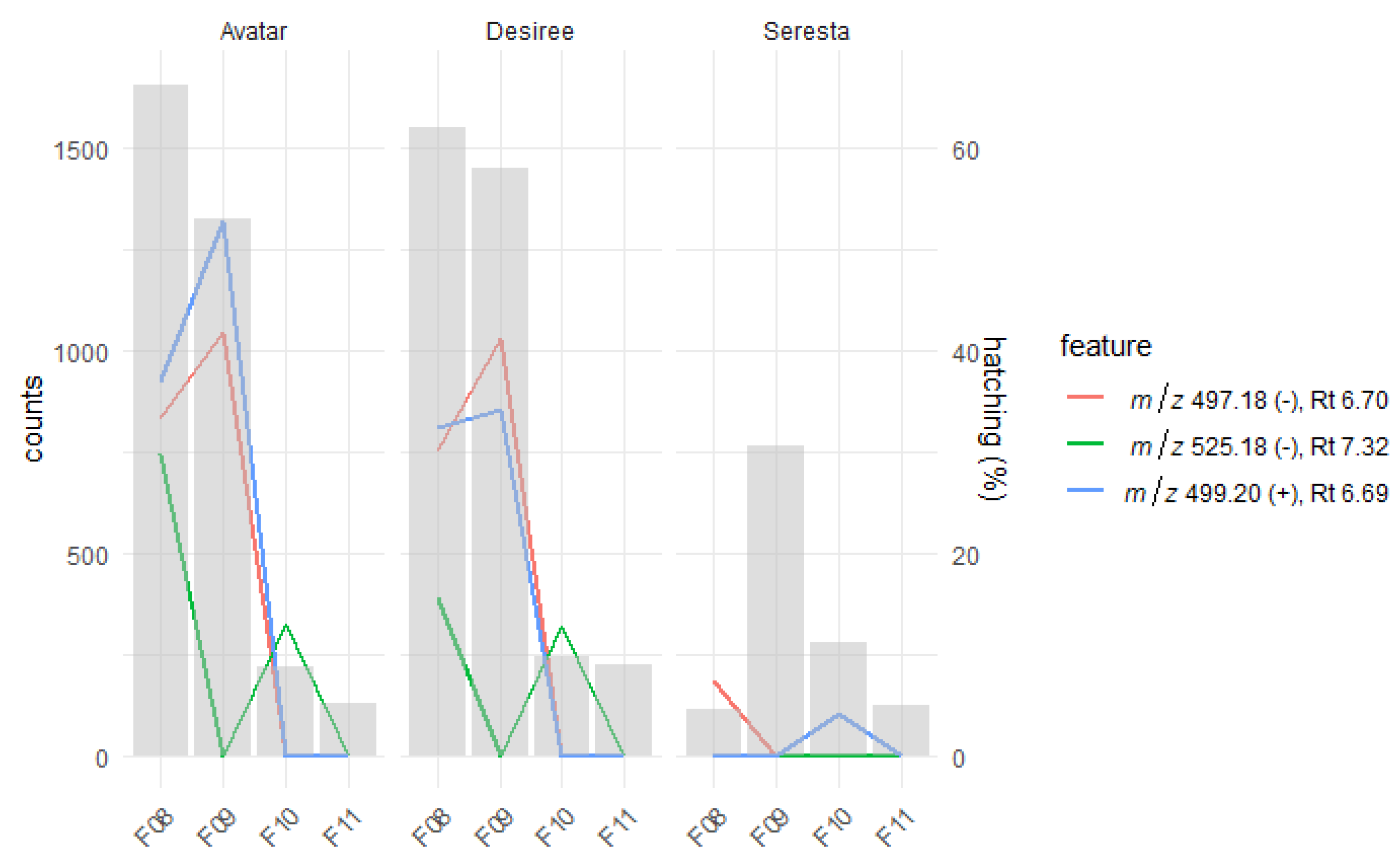
2.2. Root Exudates Contain SolA and SolA-Like Compounds That Predict Hatching
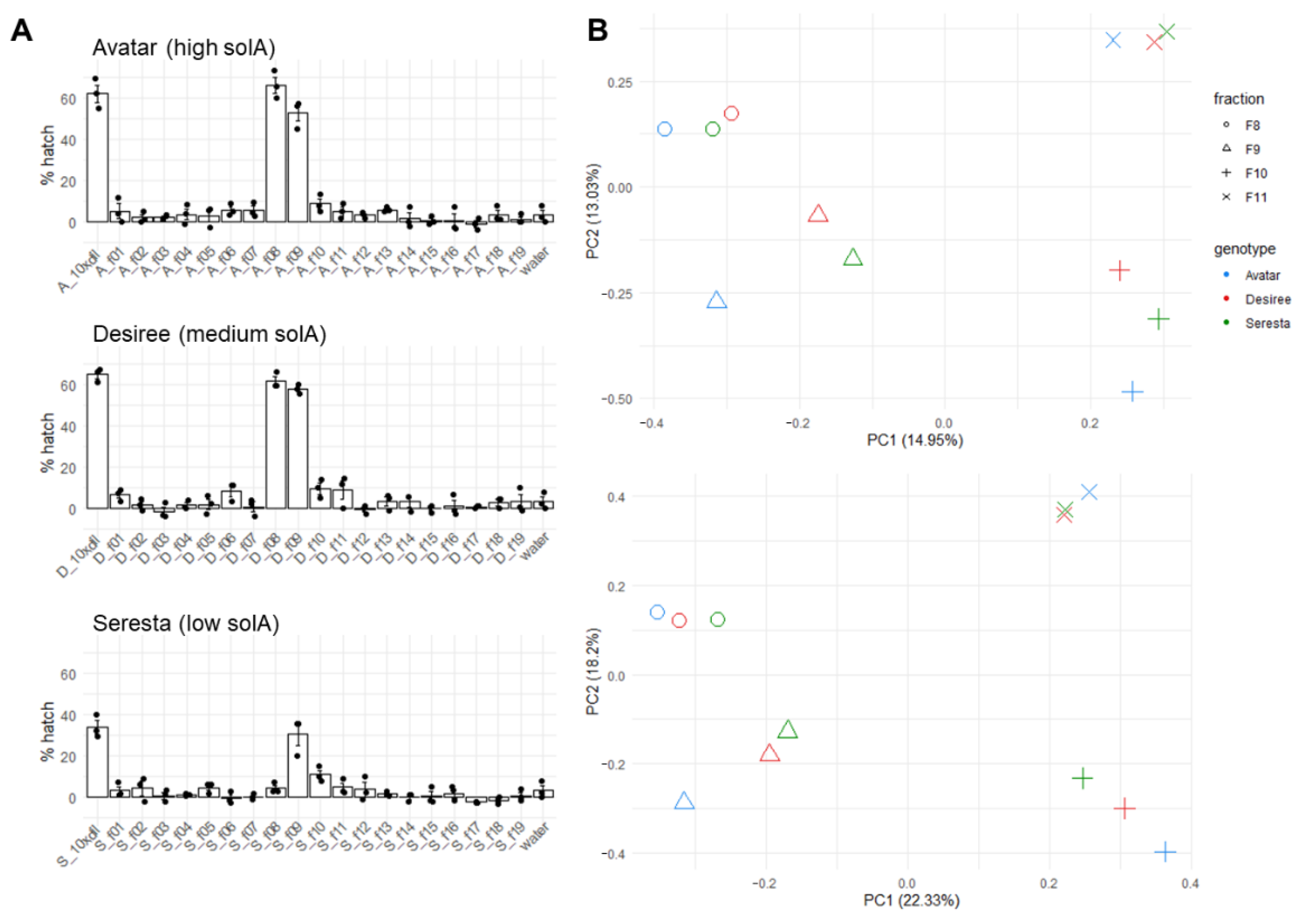
2.3. Fractionation of Root Exudates Shows Specific Hatching Activity
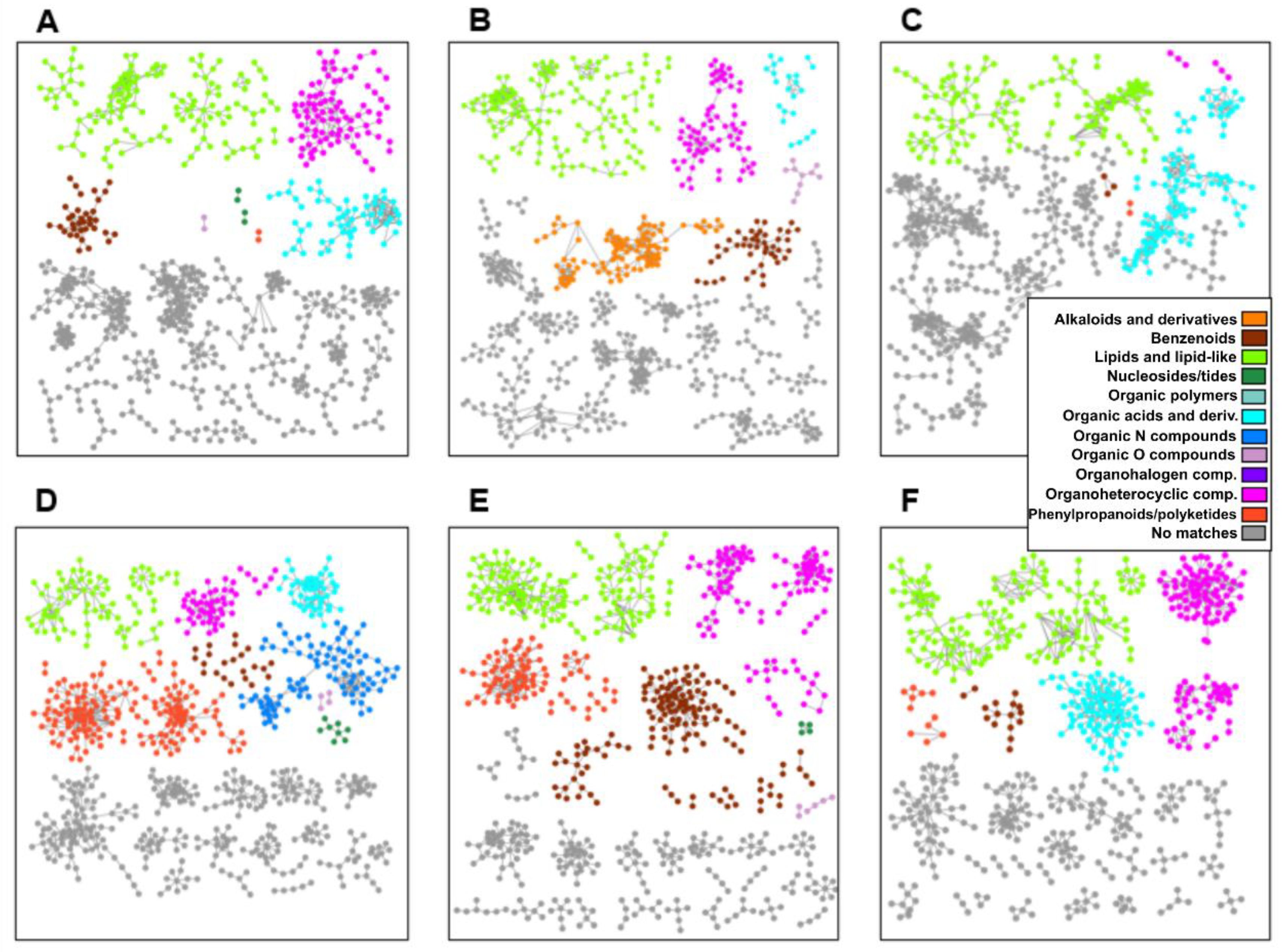
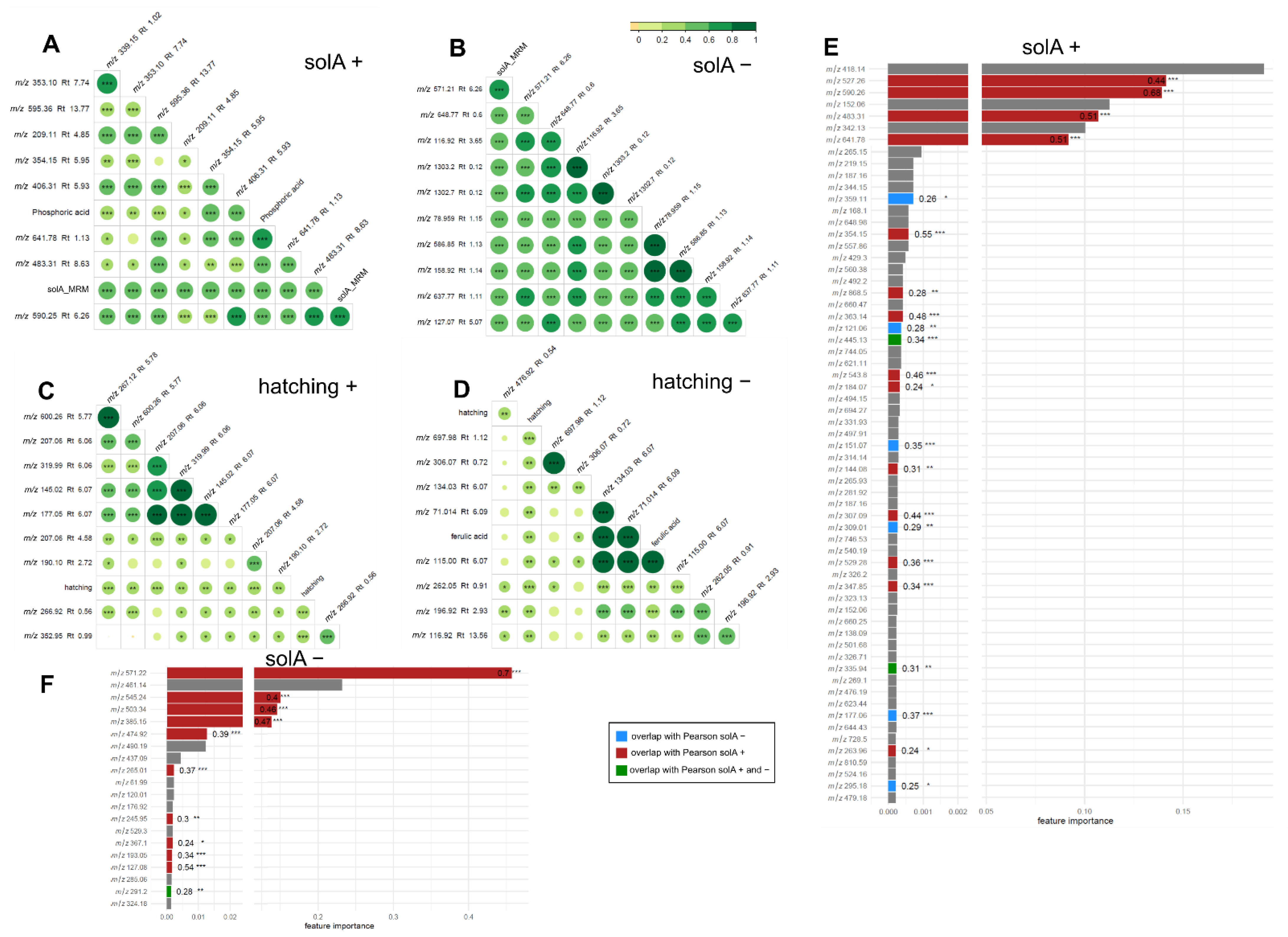
2.4. Metabolomics of Fractions with High Hatching Activity Confirm Discovery of a New Hatching Factor
2.5. Root Extracts Do Not Contain SolA, but Some Metabolites That Correlate with Hatching May Be Its Precursors
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant and Nematode Materials and Growth Conditions
4.3. Sample Extraction and Purification
4.4. UPLC-Multiple Reaction Monitoring-MS/MS Analysis
4.5. LC-ESI-QTOF-MS Analysis
4.6. Fractionation of Root Exudates
4.7. Nematode Hatching Assay
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birch, P.R.J.; Bryan, G.; Fenton, B.; Gilroy, E.M.; Hein, I.; Jones, J.T.; Prashar, A.; Taylor, M.A.; Torrance, L.; Toth, I.K. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 2012, 4, 477–508, ISBN 1257101202201. [Google Scholar] [CrossRef]
- Nicol, J.M.; Turner, S.J.; Coyne, D.L.; den Nijs, L.; Hockland, S.; Tahna Maafi, Z. Current nematode threats to world agriculture. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Springer: Dordrecht, The Netherlands, 2011; pp. 369–393. ISBN 978-94-007-0433-6. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef]
- Perry, R.N.; Wharton, D.A. Molecular and Physiological Basis of Nematode Survival, 1st ed.; Perry, R.N., Wharton, D.A., Eds.; CAB International: Cambridge, MA, USA, 2011; ISBN 9781845936877. [Google Scholar]
- Perry, R.N.; Clarke, A.J. Hatching mechanisms of nematodes. Parasitology 1981, 83, 435–449. [Google Scholar] [CrossRef]
- Byrne, J.; Twomey, U.; Maher, N.; Devine, K.J.; Jones, P.W. Detection of hatching inhibitors and hatching factor stimulants for golden potato cyst nematode, Globodera rostochiensis, in potato root leachate. Ann. Appl. Biol. 1998, 132, 463–472. [Google Scholar] [CrossRef]
- Mulder, J.G.; Diepenhorst, P.; Plieger, P.; Brüggemann-Rotgans, I.E.M. Hatching Agent for the potato cyst nematode 1996 (patent). Available online: https://patents.google.com/patent/US5585505A/en (accessed on 2 May 2022).
- Sakata, I.; Kushida, A.; Tanino, K. The hatching-stimulation activity of solanoeclepin a toward the eggs of Globodera (Tylenchida: Heteroderidae) species. Appl. Entomol. Zool. 2020, 56, 51–57. [Google Scholar] [CrossRef]
- Guerrieri, A.; Floková, K.; Vlaar, L.E.; Chojnacka, A.; van Dijk, Y.R.; Kramer, G.; Bouwmeester, H.J.; Dong, L. UPLC-MS/MS analysis and biological activity of the potato cyst nematode hatching stimulant, solanoeclepin A, in the root exudate of Solanum spp. Planta 2021, 254, 1–13. [Google Scholar] [CrossRef]
- Masamune, T.; Anetai, M.; Takasugi, M.; Katsui, N. Isolation of a natural hatching stimulus, glycinoeclepin A, for the soybean cyst nematode. Nature 1982, 297, 495–496. [Google Scholar] [CrossRef]
- Fukuzawa, A.; Matsue, H.; Ikura, M.; Masamune, T. Glycinoeclepins B and C, Nortriterpenes related to Glycinoeclepin A. Tetrahedron Lett. 1985, 26, 5539–5542. [Google Scholar] [CrossRef]
- Ochola, J.; Cortada, L.; Ng’ang’a, M.; Hassanali, A.; Coyne, D.; Torto, B. Mediation of Potato—Potato Cyst Nematode, G. rostochiensis Interaction by Specific Root Exudate Compounds. Front. Plant Sci. 2020, 11, 649. [Google Scholar] [CrossRef]
- Shimizu, K.; Kushida, A.; Akiyama, R.; Lee, H.J.; Okamura, Y.; Masuda, Y.; Sakata, I.; Tanino, K.; Matsukida, S.; Inoue, T.; et al. Hatching stimulation activity of steroidal glycoalkaloids toward the potato cyst nematode, Globodera rostochiensis. Plant Biotechnol. 2020, 37, 319–325. [Google Scholar] [CrossRef]
- Sun, M.; Qiu, F.; Li, W.-D. Recent progresses in the synthesis of solanoeclepin A. Chin. J. Org. Chem. 2019, 39, 2759–2770. [Google Scholar] [CrossRef] [Green Version]
- Byrne, J.T.; Maher, N.J.; Jones, P.W. Comparative responses of Globodera rostochiensis and G. pallida to hatching chemicals. J. Nematol. 2001, 33, 195–202. [Google Scholar]
- Perry, R.N. Hatching. In The Biology of Nematodes; Lee, D.L., Ed.; Taylor & Francis: Abingdon-on-Thames, UK, 2002; pp. 297–337. [Google Scholar]
- Devine, K.J.; Jones, P.W. Purification and partial characterisation of hatching factors for the potato cyst nematode Globodera rostochiensis from potato root leachate. Nematology 2000, 2, 231–236. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Fowler, M.; Isaac, R.E. Partial purification of hatching activity for Globodera rostochiensis from potato root diffusate. Ann. Appl. Biol. 1987, 110, 115–125. [Google Scholar] [CrossRef]
- Gobena, D.; Shimels, M.; Rich, P.J.; Ruyter-Spira, C.; Bouwmeester, H.; Kanuganti, S.; Mengiste, T.; Ejeta, G. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. USA 2017, 114, 4471–4476. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.; Enge, S.; Jensen, K.R.; Müller, C.; Kiær, L.P.; Agerbirk, N.; Heimes, C.; Hauser, T.P. Different herbivore responses to two co-occurring chemotypes of the wild crucifer Barbarea vulgaris. Arthropod-Plant Interact. 2019, 13, 19–30. [Google Scholar] [CrossRef]
- Deliopoulos, T.; Devine, K.J.; Haydock, P.P.J.; Jones, P.W. Studies on the effect of mycorrhization of potato roots on the hatching activity of potato root leachate towards the potato cyst nematodes, Globodera pallida and G. rostochiensis. Nematology 2007, 9, 719–729. [Google Scholar] [CrossRef]
- Devine, K.J.; Byrne, J.; Maher, N.; Jones, P.W. Resolution of natural hatching factors for golden potato cyst nematode, Globodera rostochiensis. Ann. Appl. Biol. 1996, 129, 323–334. [Google Scholar] [CrossRef]
- Dandurand, L.M.; Knudsen, G.R. Effect of the trap crop Solanum sisymbriifolium and two biocontrol fungi on reproduction of the potato cyst nematode, Globodera pallida. Ann. Appl. Biol. 2016, 169, 180–189. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18 (Suppl. S1), S96–S104. [Google Scholar] [CrossRef]
- Kucheryavskiy, S. mdatools—R package for chemometrics. Chemom. Intell. Lab. Syst. 2020, 198, 103937. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix 2021. Available online: https://cran.r-project.org/web/packages/corrplot/index.html (accessed on 29 December 2021).
- Shi, L.; Westerhuis, J.A.; Rose, J.; Landberg, R.; Brunius, C. Gene expression Variable selection and validation in multivariate modelling. Bioinformatics 2019, 35, 972–980. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.J.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with GNPS. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Ernst, M.; Kang, K.B.; Nothias, L.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; Hooft, J.J.J. Van Der MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
| Molecular m/z | Rt | Measured m/z | Calculated Mass | Δ m/z (ppm) | Ion | Predicted Formula | Detected in | Putative Name | Correlation |
|---|---|---|---|---|---|---|---|---|---|
| 248.0508 | 9.86 | 247.0435 | 247.0448 | 5.4 | [M-H]− | C9H12O8 | Exudate | hatch | |
| 248.11 | 6.38 | 249.1108 | 249.1121 | 5.4 | [M-H]+ | C14H17O4 | Exudate | prenyl caffeate | hatch |
| 263.0641 | 0.91 | 262.0568 | 262.0557 | −4.0 | [M-H]− | C9H13NO8 | Extract | ascorbalamic acid | hatch |
| 265.9186 | 0.56 | 266.9259 | [M-H]+ | Extract | hatch | ||||
| 498.1887 | 7.66 | 497.1814 | 497.18061 | −1.6 | [M-H]− | C27H30O9 | Exudate, fractions | solA | solA, hatch |
| 498.1887 | 7.66 | 499.1944 | 499.1963 | 3.7 | [M-H]+ | C27H30O9 | Exudate, fractions | solA | solA, hatch |
| 526.1843 | 7.95 | 525.1771 | 525.1755 | −3.0 | [M-H]− | C28H30O10 | Exudate, fractions | solB | solA, hatch |
| 526.1843 | 7.94 | 527.1893 | 527.1912 | 3.6 | [M-H]+ | C28H30O10 | Exudate, fractions | solB | solA, hatch |
| 572.2254 | 6.26 | 571.2182 | 571.2174 | −1.4 | [M-H]− | C30H36O11 | Extract | solA precursor? | solA |
| 580.38 | 13.95 | 581.3835 | 581.3837 | 0.3 | [M-H]+ | C36H52O6 | Exudate | hugonone A | hatch |
| 589.2527 | 6.26 | 590.2600 | 590.2596 | −0.7 | [M-H]+ | C30H39NO11 | Extract | platencin A13 | solA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlaar, L.E.; Thiombiano, B.; Abedini, D.; Schilder, M.; Yang, Y.; Dong, L. A Combination of Metabolomics and Machine Learning Results in the Identification of a New Cyst Nematode Hatching Factor. Metabolites 2022, 12, 551. https://doi.org/10.3390/metabo12060551
Vlaar LE, Thiombiano B, Abedini D, Schilder M, Yang Y, Dong L. A Combination of Metabolomics and Machine Learning Results in the Identification of a New Cyst Nematode Hatching Factor. Metabolites. 2022; 12(6):551. https://doi.org/10.3390/metabo12060551
Chicago/Turabian StyleVlaar, Lieke E., Benjamin Thiombiano, Davar Abedini, Mario Schilder, Yuting Yang, and Lemeng Dong. 2022. "A Combination of Metabolomics and Machine Learning Results in the Identification of a New Cyst Nematode Hatching Factor" Metabolites 12, no. 6: 551. https://doi.org/10.3390/metabo12060551
APA StyleVlaar, L. E., Thiombiano, B., Abedini, D., Schilder, M., Yang, Y., & Dong, L. (2022). A Combination of Metabolomics and Machine Learning Results in the Identification of a New Cyst Nematode Hatching Factor. Metabolites, 12(6), 551. https://doi.org/10.3390/metabo12060551






