Metabolomic Analysis of Plasma from Breast Cancer Patients Using Ultra-High-Performance Liquid Chromatography Coupled with Mass Spectrometry: An Untargeted Study
Abstract
:1. Introduction
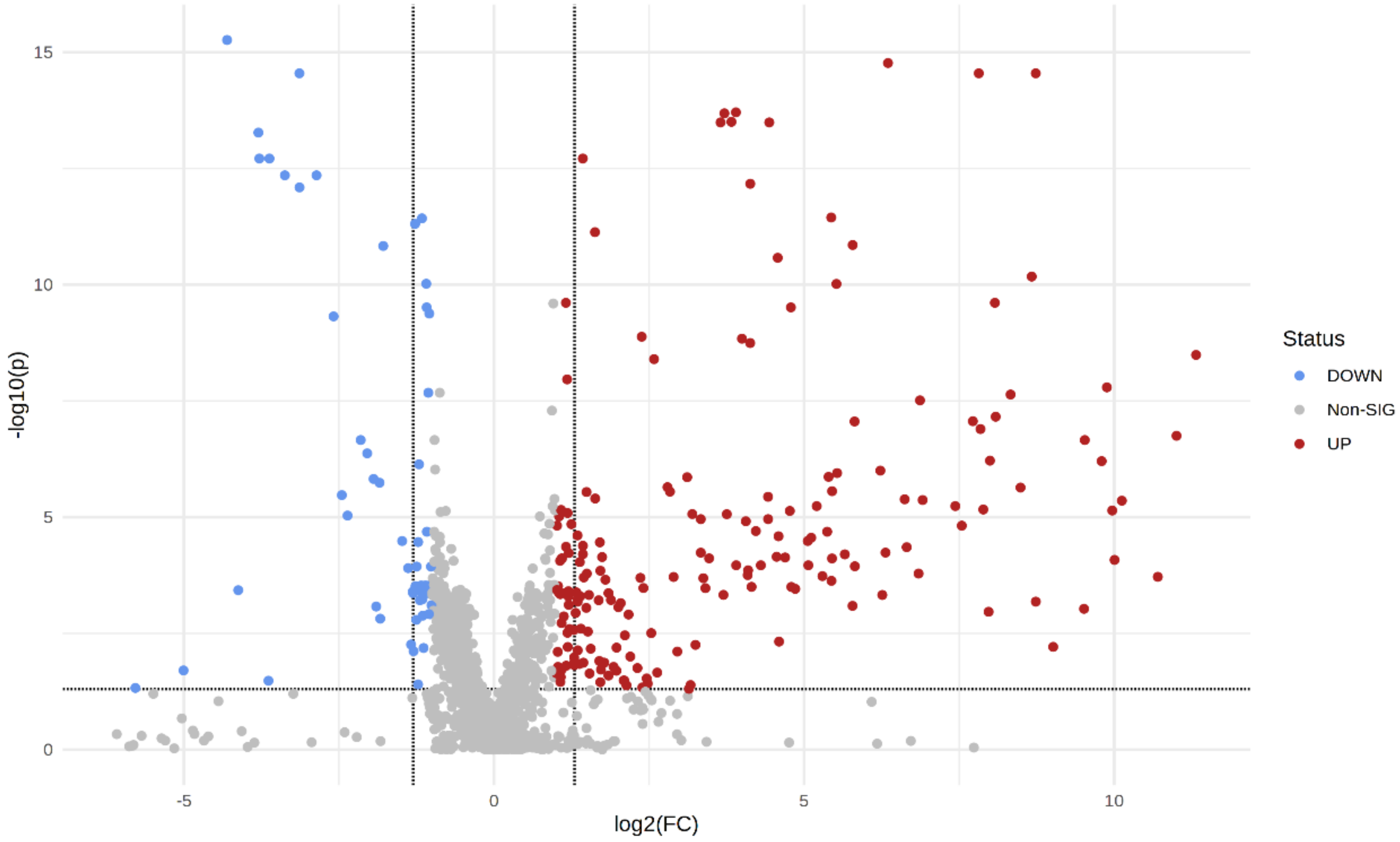
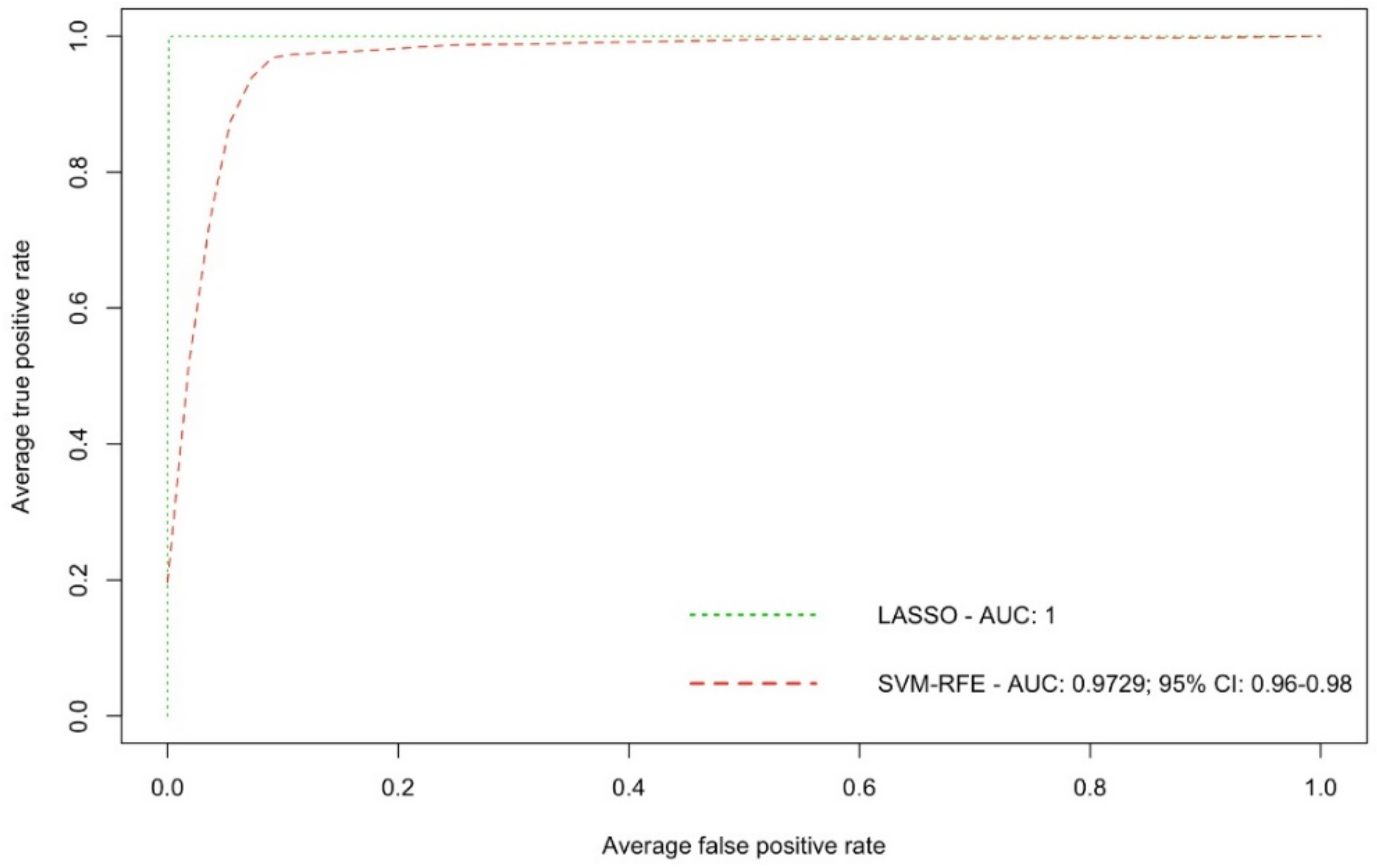
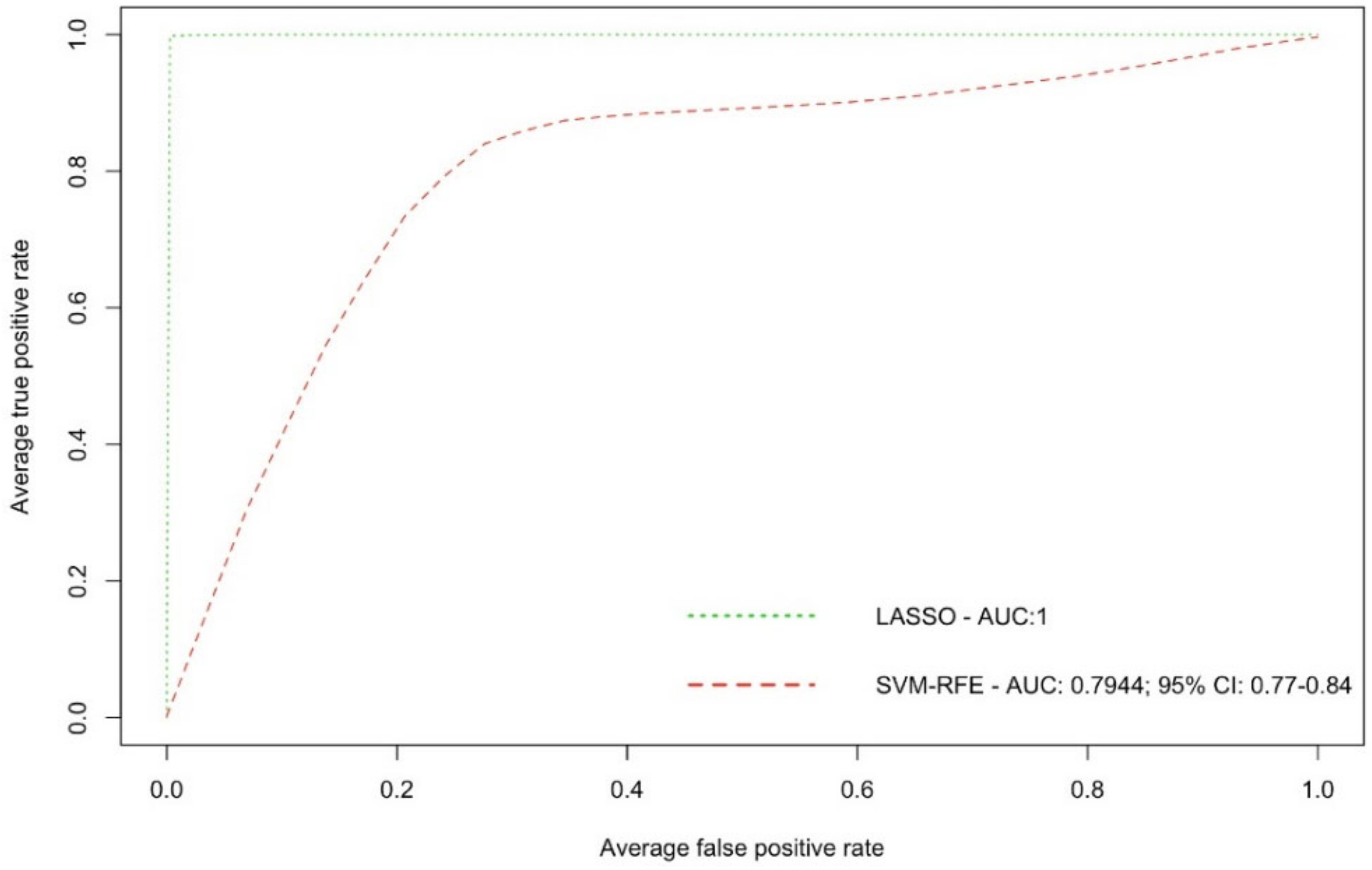
2. Results
3. Discussion
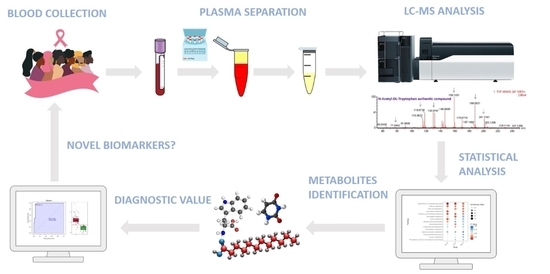
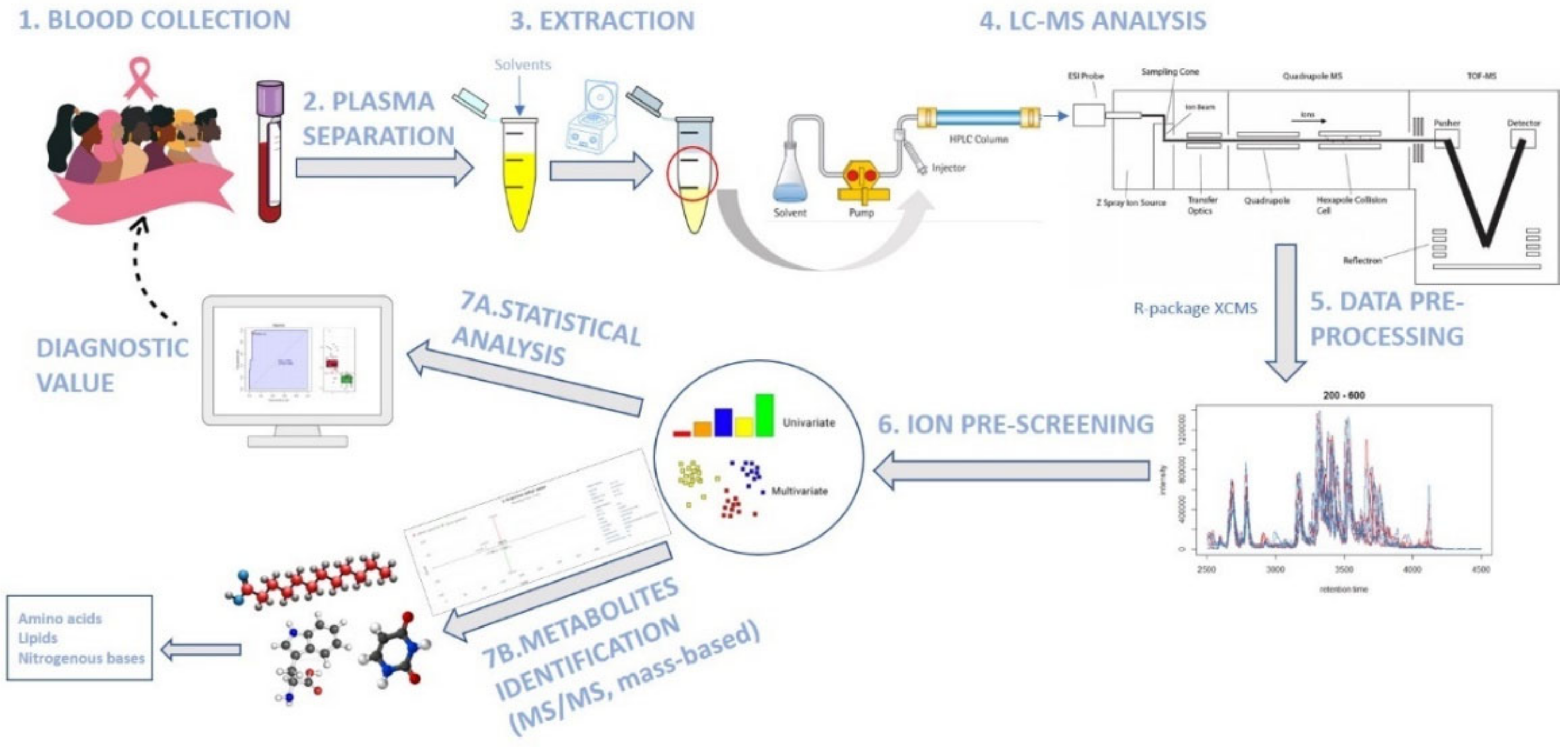
4. Materials and Methods
4.1. Patient Population and Plasma Samples
4.2. Metabolite Extraction from Plasma and LC-MS Analysis
4.3. Data Pre-Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2020; American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Berry, D.A.; Cronin, K.A.; Plevritis, S.K.; Fryback, D.G.; Clarke, L.; Zelen, M.; Mandelblatt, J.S.; Yakovlev, A.Y.; Habbema, J.D.F.; Feuer, E.J. Effect of screening and adjuvant therapy on mortality from breast cancer. N. Engl. J. Med. 2005, 353, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Rosmawati, N.H. Knowledge, attitudes and practice of breast self-examination among women in a suburban area in Terengganu, Malaysia. Asian Pac. J. Cancer Prev. 2010, 11, 1503–1508. [Google Scholar]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef]
- Hirschey, M.D.; DeBerardinis, R.J.; Diehl, A.M.E.; Drew, J.E.; Frezza, C.; Green, M.F.; Jones, L.W.; Ko, Y.H.; Le, A.; Lea, M.A.; et al. Dysregulated metabolism contributes to oncogenesis. Semin. Cancer Biol. 2015, 35, S129–S150. [Google Scholar] [CrossRef] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- McCartney, A.; Vignoli, A.; Biganzoli, L.; Love, R.; Tenori, L.; Luchinat, C.; Di Leo, A. Metabolomics in breast cancer: A decade in review. Cancer Treat. Rev. 2018, 67, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhou, X.; Xia, T.S.; Chen, Z.; Li, J.; Liu, Q.; Alolga, R.N.; Chen, Y.; Lai, M.D.; Li, P.; et al. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget 2016, 7, 9925–9938. [Google Scholar] [CrossRef] [Green Version]
- Tsoukalas, D.; Sarandi, E.; Thanasoula, M.; Docea, A.O.; Tsilimidos, G.; Calina, D.; Tsatsakis, A. Metabolic Fingerprint of Chronic Obstructive Lung Diseases: A New Diagnostic Perspective. Metabolites 2019, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Tsoukalas, D.; Fragoulakis, V.; Sarandi, E.; Docea, A.O.; Papakonstaninou, E.; Tsilimidos, G.; Anamaterou, C.; Fragkiadaki, P.; Aschner, M.; Tsatsakis, A.; et al. Targeted Metabolomic Analysis of Serum Fatty Acids for the Prediction of Autoimmune Diseases. Front. Mol. Biosci. 2019, 6, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Jové, M.; Collado, R.; Quiles, J.L.; Ramírez-Tortosa, M.C.; Sol, J.; Ruiz-Sanjuan, M.; Fernandez, M.; de la Torre Cabrera, C.; Ramírez-Tortosa, C.; Granados-Principal, S.; et al. A plasma metabolomic signature discloses human breast cancer. Oncotarget 2017, 8, 19522–19533. [Google Scholar] [CrossRef] [Green Version]
- Jasbi, P.; Wang, D.; Cheng, S.L.; Fei, Q.; Cui, J.Y.; Liu, L.; Wei, Y.; Raftery, D.; Gu, H. Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1105, 26–37. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, X.; Chou, J.; Yu, J.; Yang, T.; Liu, L.; Zhang, F. Taurine, glutamic acid and ethylmalonic acid as important metabolites for detecting human breast cancer based on the targeted metabolomics. Cancer Biomark. 2018, 23, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yan, L.; Liu, S.; Ambrosone, C.B.; Zhao, H. Plasma metabolomic profiles in breast cancer patients and healthy controls: By race and tumor receptor subtypes. Transl. Oncol. 2013, 6, 757–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougan, M.M.; Li, Y.; Chu, L.W.; Haile, R.W.; Whittemore, A.S.; Han, S.S.; Moore, S.C.; Sampson, J.N.; Andrulis, I.L.; John, E.M.; et al. Metabolomic profiles in breast cancer:a pilot case-control study in the breast cancer family registry. BMC Cancer 2018, 18, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budhu, A.; Terunuma, A.; Zhang, G.; Hussain, S.P.; Ambs, S.; Wang, X.W. Metabolic profiles are principally different between cancers of the liver, pancreas and breast. Int. J. Biol. Sci. 2014, 10, 966–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobard, E.; Pontoizeau, C.; Blaise, B.J.; Bachelot, T.; Elena-Herrmann, B.; Trédan, O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014, 343, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Ambs, S. Metabolic Signatures of Human Breast Cancer. Mol. Cell. Oncol. 2015, 2, e992217. [Google Scholar] [CrossRef] [Green Version]
- Do Canto, L.M.; Marian, C.; Willey, S.; Sidawy, M.; Da Cunha, P.A.; Rone, J.D.; Li, X.; Gusev, Y.; Haddad, B.R. MicroRNA analysis of breast ductal fluid in breast cancer patients. Int. J. Oncol. 2016, 48, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Do Canto, L.M.; Marian, C.; Varghese, R.S.; Ahn, J.; Da Cunha, P.A.; Willey, S.; Sidawy, M.; Rone, J.D.; Cheema, A.K.; Luta, G.; et al. Metabolomic profiling of breast tumors using ductal fluid. Int. J. Oncol. 2016, 49, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, C.; Cavalli, L.R.; Cohen, Y.; Pennanen, M.; Shankar, L.K.; Freedman, M.; Singh, B.; Liu, M.; Gallagher, A.; Rone, J.D.; et al. Detection of LOH and mitochondrial DNA alterations in ductal lavage and nipple aspirate fluids from hngh-risk patients. Breast Cancer Res. Treat. 2004, 84, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.G.; Qin, X.B.; Liu, F.F.; Zhou, L.L. Tryptophan-induced pathogenesis of breast cancer. Afr. Health Sci. 2015, 15, 982–985. [Google Scholar] [CrossRef] [Green Version]
- Juhász, C.; Nahleh, Z.; Zitron, I.; Chugani, D.C.; Janabi, M.Z.; Bandyopadhyay, S.; Ali-Fehmi, R.; Mangner, T.J.; Chakraborty, P.K.; Mittal, S.; et al. Tryptophan metabolism in breast cancers: Molecular imaging and immunohistochemistry studies. Nucl. Med. Biol. 2012, 39, 926–932. [Google Scholar] [CrossRef] [Green Version]
- Travers, M.T.; Gow, I.F.; Barber, M.C.; Thomson, J.; Shennan, D.B. Indoleamine 2,3-dioxygenase activity and L-tryptophan transport in human breast cancer cells. Biochim. Biophys. Acta 2004, 1661, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, K.; Amano, S.; Enomoto, K.; Kashio, M.; Saito, Y.; Sakamoto, A.; Matsuo, S.; Suzuki, M.; Kitajima, A.; Hirano, T.; et al. Study of indoleamine 2,3-dioxygenase expression in patients with breast cancer. Gan To Kagaku Ryoho 2005, 32, 1546–1549. [Google Scholar]
- Fuchs, B.C.; Bode, B.P. Amino acid transporters ASCT2 and LAT1 in cancer: Partners in crime? Semin. Cancer Biol. 2005, 15, 254–266. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef]
- Louie, S.M.; Roberts, L.S.; Mulvihill, M.M.; Luo, K.; Nomura, D.K. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim. Biophys. Acta 2013, 1831, 1566–1572. [Google Scholar] [CrossRef] [Green Version]
- Kuemmerle, N.B.; Rysman, E.; Lombardo, P.S.; Flanagan, A.J.; Lipe, B.C.; Wells, W.A.; Pettus, J.R.; Froehlich, H.M.; Memoli, V.A.; Morganelli, P.M.; et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol. Cancer Ther. 2011, 10, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.W.; Cravatt, B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, J.C.A.; Collins, J.W.; Nakayama, J.; Horak, C.E.; Liewehr, D.J.; Steinberg, S.M.; Albaugh, M.; Vidal-Vanaclocha, F.; Palmieri, D.; Barbier, M.; et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. J. Natl. Cancer Inst. 2012, 104, 1306–1319. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, M.; Ramachandran, S.; Kim, E.Y.; Allegood, J.C.; Rashid, O.M.; Yamada, A.; Zhao, R.; Milstien, S.; Zhou, H.; Spiegel, S.; et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012, 72, 726–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suman, S.; Sharma, R.K.; Kumar, V.; Sinha, N.; Shukla, Y. Metabolic fingerprinting in breast cancer stages through 1H NMR spectroscopy-based metabolomic analysis of plasma. J. Pharm. Biomed. Anal. 2018, 160, 38–45. [Google Scholar] [CrossRef]
- Available online: https://www.ebi.ac.uk/training/online/courses/metabolomics-introduction/designing-a-metabolomics-study/comparison-of-nmr-and-ms/ (accessed on 27 April 2022).
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [Green Version]


| Characteristics | N = 55 | % * |
|---|---|---|
| Age (mean ± SD) | 53.35 (±12.26) | |
| Menopausal status | ||
| Pre-menopause | 18 | 32.73% |
| Peri-menopause | 2 | 3.64% |
| Post-menopause | 35 | 63.64% |
| Race | ||
| Asian | 5 | 9.09% |
| African American | 15 | 27.27% |
| Caucasian | 33 | 60.00% |
| Hispanic | 2 | 3.64% |
| Smoking history | ||
| Current smoker | 7 | 12.73% |
| Former smoker | 5 | 9.09% |
| Never smoked | 43 | 78.18% |
| Histology | ||
| Ductal carcinoma in situ (DCIS) | 10 | 18.18% |
| Invasive ductal carcinoma (IDC) | 35 | 63.64% |
| Invasive lobular carcinoma (ILC) | 5 | 9.09% |
| Mixed | 2 | 3.63% |
| BC stage | ||
| 0 | 9 | 16.36% |
| I | 21 | 38.18% |
| II | 15 | 27.27% |
| III | 4 | 7.27% |
| BC grade | ||
| Low | 7 | 12.72% |
| Intermediate | 21 | 38.18% |
| High | 25 | 45.45% |
| Lymph node involvement | ||
| Yes | 14 | 25.45% |
| No | 40 | 72.72% |
| Estrogen receptor (ER) | ||
| Positive | 45 | 81.82% |
| Negative | 9 | 16.36% |
| Progesterone receptor (PR) | ||
| Positive | 33 | 60.00% |
| Negative | 21 | 38.18% |
| HER2 status | ||
| Positive | 11 | 20.00% |
| Negative | 39 | 70.90% |
| Operative procedure | ||
| Bilateral mastectomy (BM) | 18 | 32.73% |
| Preventive mastectomy (PM) | 14 | 25.45% |
| Mastectomy | 22 | 40.00% |
| Endoscopy-assisted breast surgery (EABS) | 1 | 1.82% |
| Palpable tumor | ||
| Yes | 25 | 45.45% |
| No | 30 | 54.55% |
| Ion Mode | Number of Ions Detected | Number of Ions with Adjusted p Value < 0.05 |
|---|---|---|
| Positive (POS) | 1930 | 603 |
| Negative (NEG) | 564 | 189 |
| Metabolite ID | Formula | m/z | Exact Mass | Ion Mode | Retention Time (RT) | BC vs. HC | Fold Change (FC) | Adjusted p Value |
|---|---|---|---|---|---|---|---|---|
| Caproleic acid | C10H18O2 | 171.139 | 170.131 (M + H) | + | 314.469 | ↓ | −8.62 | 1.00 × 10−24 |
| L-Arginine (ester) * | C8H18N4O2 | 235.176 | 202.143 (M + CH3OH + H) | + | 150.534 | ↑ | 98.43 | 1.21 × 10−22 |
| N-stearoyl tryptophan | C29H46N2O3 | 236.184 | 470.354 (M + 2H) | + | 135.323 | ↑ | 50.15 | 5.23 × 10−29 |
| Ile-Ser * | C9H18N2O4 | 236.184 | 426.386 (M + 2Na) | + | 135.323 | ↑ | 50.15 | 5.23 × 10−29 |
| Uracil (derivative) | C15H20ClN3O2 | 310.129 | 309.124 (M + H) | + | 55.740 | ↓ | −17.82 | 6.85 × 10−24 |
| Met-His-OH | C16H18N4O6S | 395.103 | 394.095 (M + H) | + | 361.000 | ↑ | 2.06 | 1.64 × 10−11 |
| 5-[(4-Nitrobenzoyl)amino]isophthalic acid * | C15H10N2O7 | 329.046 | 330.049 (M − H) | − | 323.114 | ↑ | 5.84 | 2.72 × 10−13 |
| Metabolite | Train Samples (40 vs. 40) | Test Samples (15 vs. 15) | All Samples (55 vs. 55) | ||||
|---|---|---|---|---|---|---|---|
| m/z | Ion Mode | AUC | 95% CI AUC | AUC | 95% CI AUC | AUC | 95% CI AUC |
| 171.139 | + | 0.971 | 0.922–1 | 0.916 | 0.787–1 | 0.969 | 0.931–1 |
| 203.107 | + | 0.925 | 0.864–0.985 | 0.809 | 0.620–0.997 | 0.904 | 0.843–0.964 |
| 221.118 | + | 0.970 | 0.939–1 | 0.844 | 0.666–1 | 0.940 | 0.886–0.992 |
| 223.064 | + | 0.884 | 0.801–0.967 | 0.920 | 0.788–1 | 0.911 | 0.850–0.971 |
| 235.176 | + | 0.968 | 0.925–1 | 1.000 | 1 | 0.976 | 0.944–1 |
| 236.184 | + | 0.974 | 0.941–1 | 1.000 | 1 | 0.980 | 0.954–1 |
| 256.942 | + | 0.746 | 0.638–0.853 | 0.987 | 0.957–1 | 0.681 | 0.581–0.780 |
| 302.122 | + | 0.940 | 0.881–0.998 | 0.893 | 0.774–1 | 0.929 | 0.877–0.981 |
| 306.977 | + | 0.913 | 0.842–0.983 | 0.711 | 0.501–0.920 | 0.875 | 0.803–0.947 |
| 310.129 | + | 0.951 | 0.883–1 | 0.858 | 0.720–0.995 | 0.937 | 0.882–0.990 |
| 395.103 | + | 0.855 | 0.764–0.945 | 0.769 | 0.572–0.965 | 0.842 | 0.761–0.922 |
| 451.165 | + | 0.898 | 0.831–0.963 | 0.764 | 0.580–0.948 | 0.876 | 0.810–0.940 |
| 223.028 | − | 0.897 | 0.817–0.975 | 0.907 | 0.772–1 | 0.912 | 0.851–0.972 |
| 317.948 | − | 0.949 | 0.893–1 | 0.667 | 0.451–0.881 | 0.889 | 0.819–0.959 |
| 329.046 | − | 0.988 | 0.971–1 | 0.836 | 0.657–1 | 0.953 | 0.907–0.998 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Cunha, P.A.; Nitusca, D.; Canto, L.M.D.; Varghese, R.S.; Ressom, H.W.; Willey, S.; Marian, C.; Haddad, B.R. Metabolomic Analysis of Plasma from Breast Cancer Patients Using Ultra-High-Performance Liquid Chromatography Coupled with Mass Spectrometry: An Untargeted Study. Metabolites 2022, 12, 447. https://doi.org/10.3390/metabo12050447
Da Cunha PA, Nitusca D, Canto LMD, Varghese RS, Ressom HW, Willey S, Marian C, Haddad BR. Metabolomic Analysis of Plasma from Breast Cancer Patients Using Ultra-High-Performance Liquid Chromatography Coupled with Mass Spectrometry: An Untargeted Study. Metabolites. 2022; 12(5):447. https://doi.org/10.3390/metabo12050447
Chicago/Turabian StyleDa Cunha, Patricia A., Diana Nitusca, Luisa Matos Do Canto, Rency S. Varghese, Habtom W. Ressom, Shawna Willey, Catalin Marian, and Bassem R. Haddad. 2022. "Metabolomic Analysis of Plasma from Breast Cancer Patients Using Ultra-High-Performance Liquid Chromatography Coupled with Mass Spectrometry: An Untargeted Study" Metabolites 12, no. 5: 447. https://doi.org/10.3390/metabo12050447
APA StyleDa Cunha, P. A., Nitusca, D., Canto, L. M. D., Varghese, R. S., Ressom, H. W., Willey, S., Marian, C., & Haddad, B. R. (2022). Metabolomic Analysis of Plasma from Breast Cancer Patients Using Ultra-High-Performance Liquid Chromatography Coupled with Mass Spectrometry: An Untargeted Study. Metabolites, 12(5), 447. https://doi.org/10.3390/metabo12050447