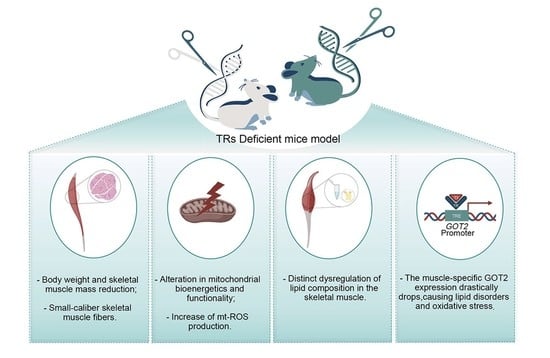
Thyroid Hormone Receptor Isoforms Alpha and Beta Play Convergent Roles in Muscle Physiology and Metabolic Regulation
Abstract
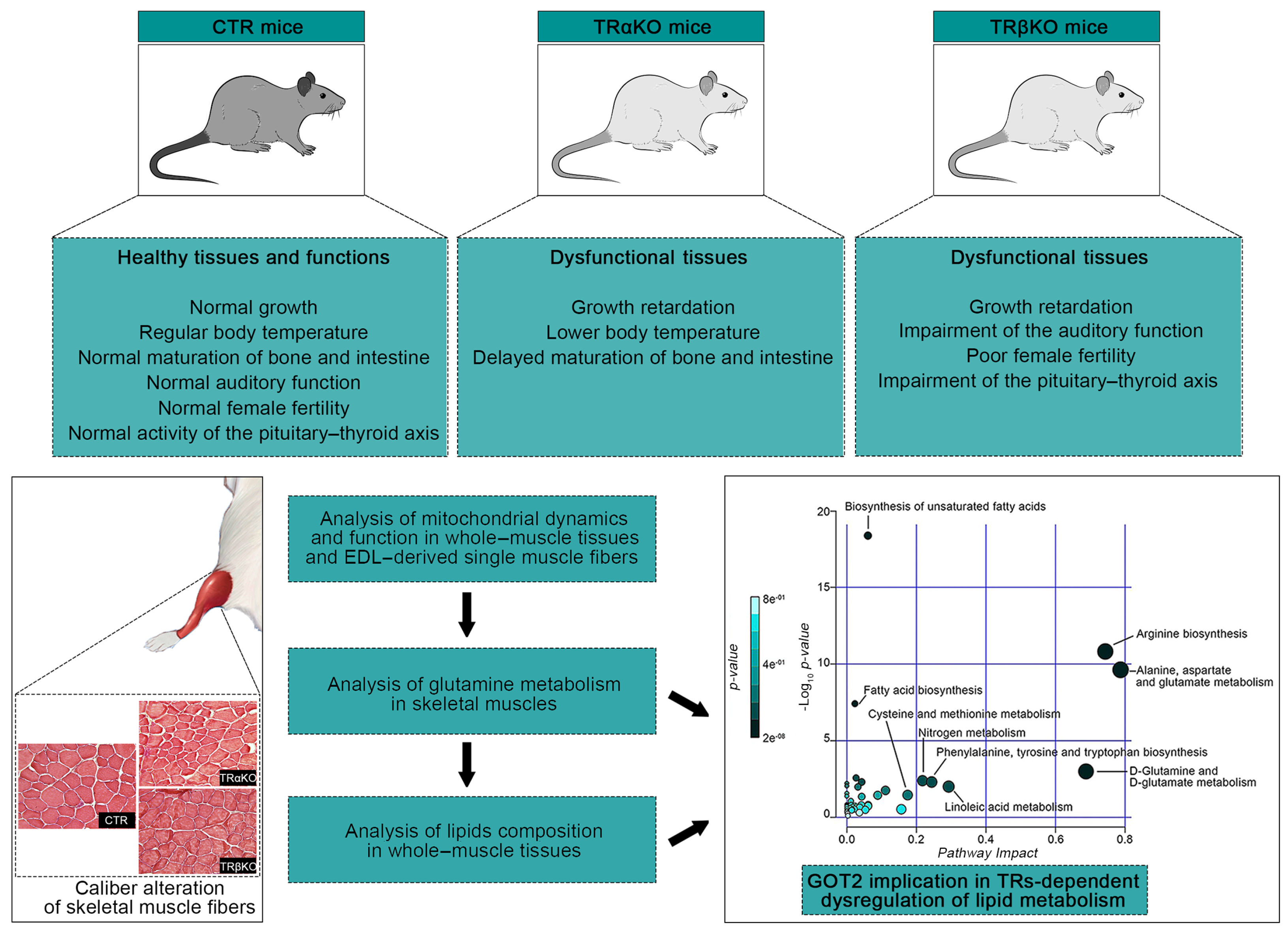
1. Introduction
2. Results
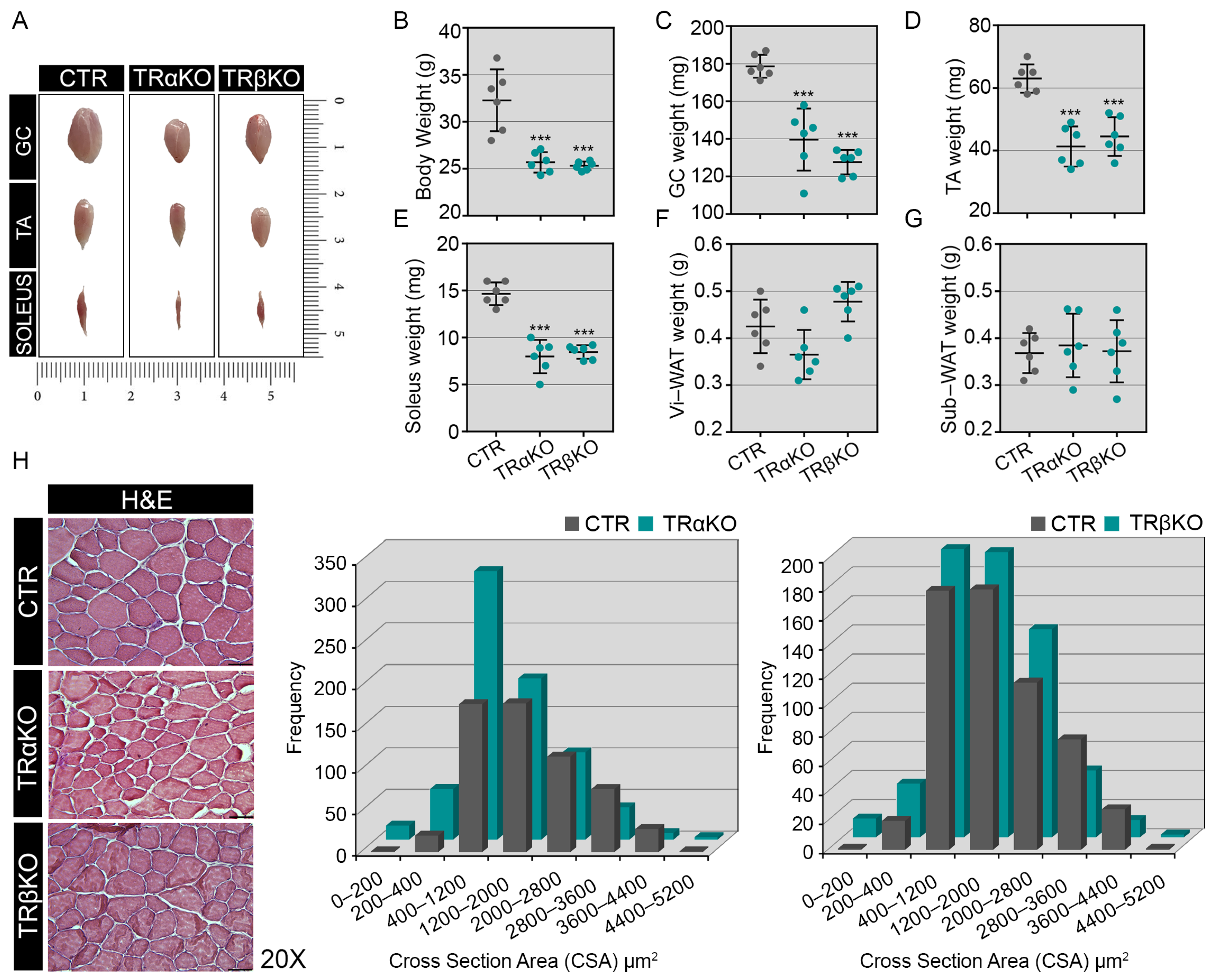
2.1. Phenotypical Analysis of TRα- and TRβKO Muscles
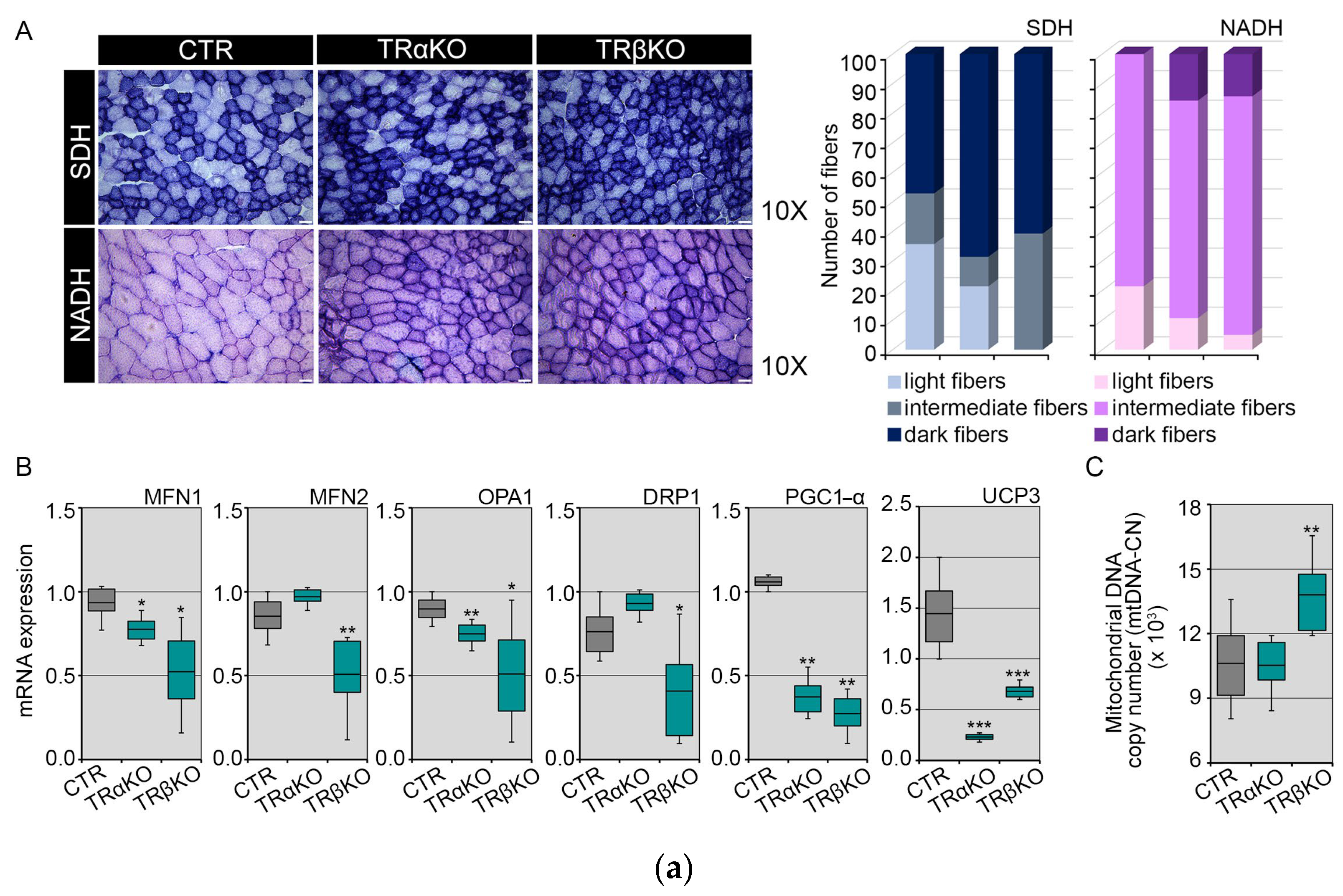
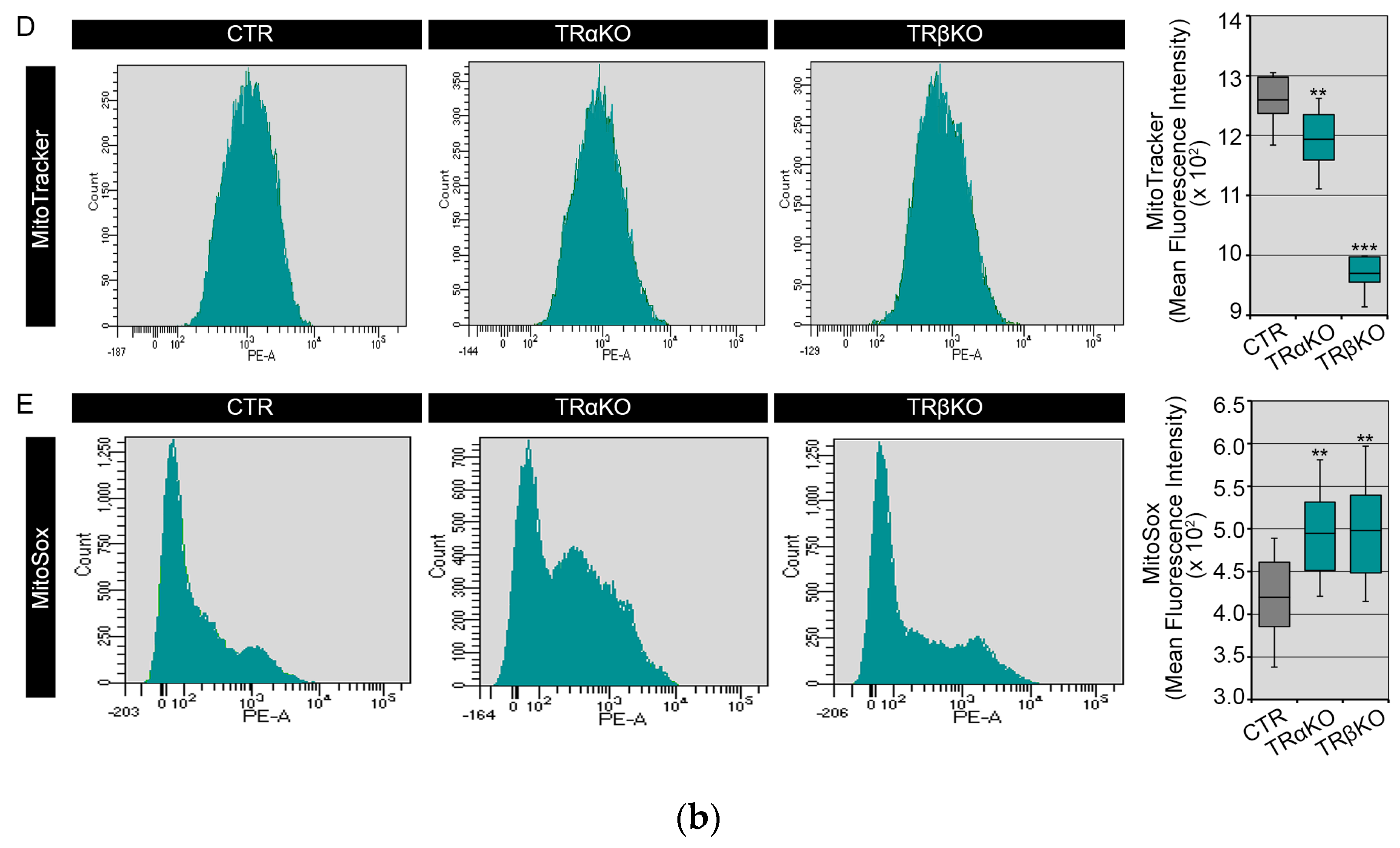
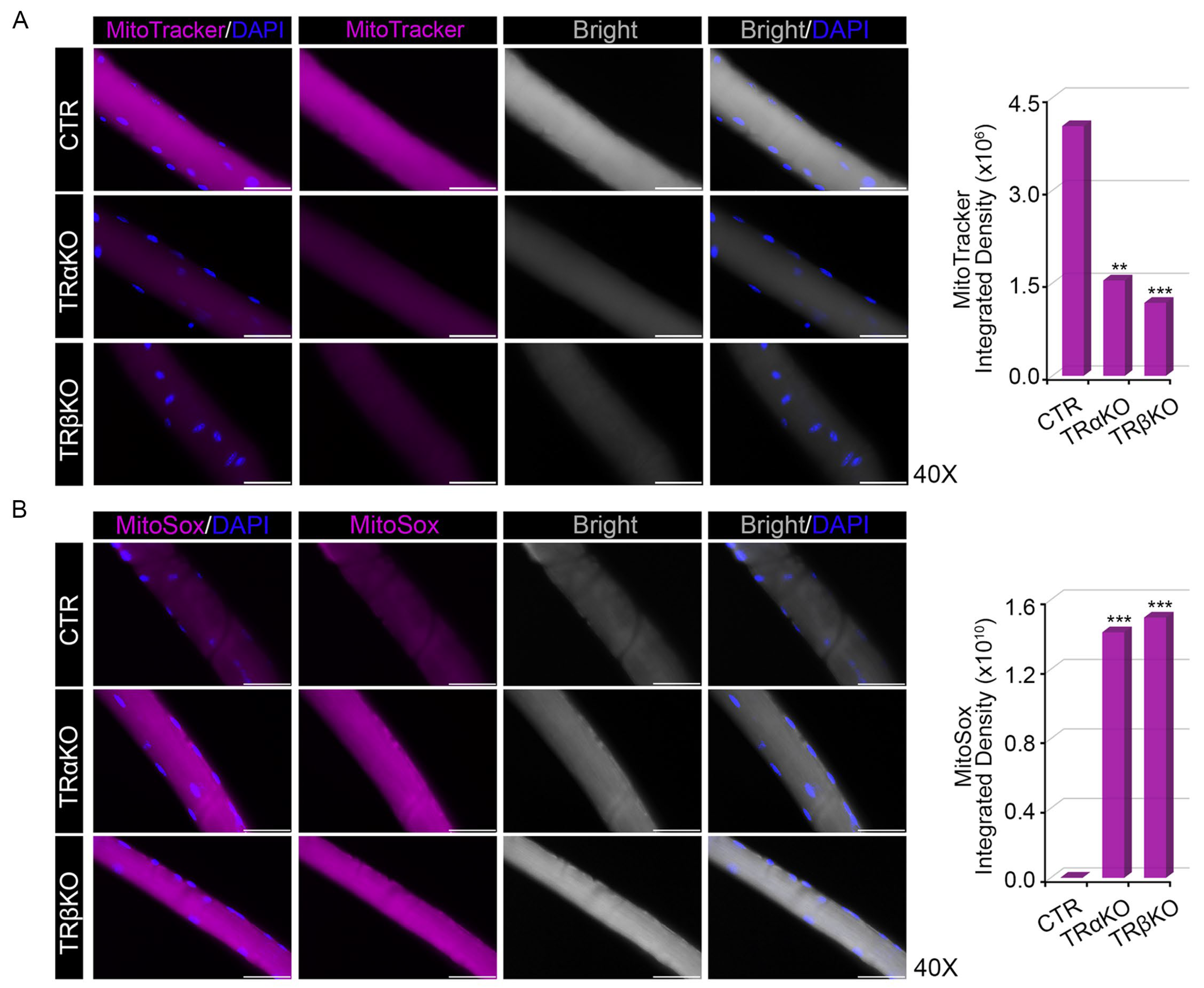
2.2. THRα and THRβ Deficiency Affects Mitochondrial Dynamics and Function
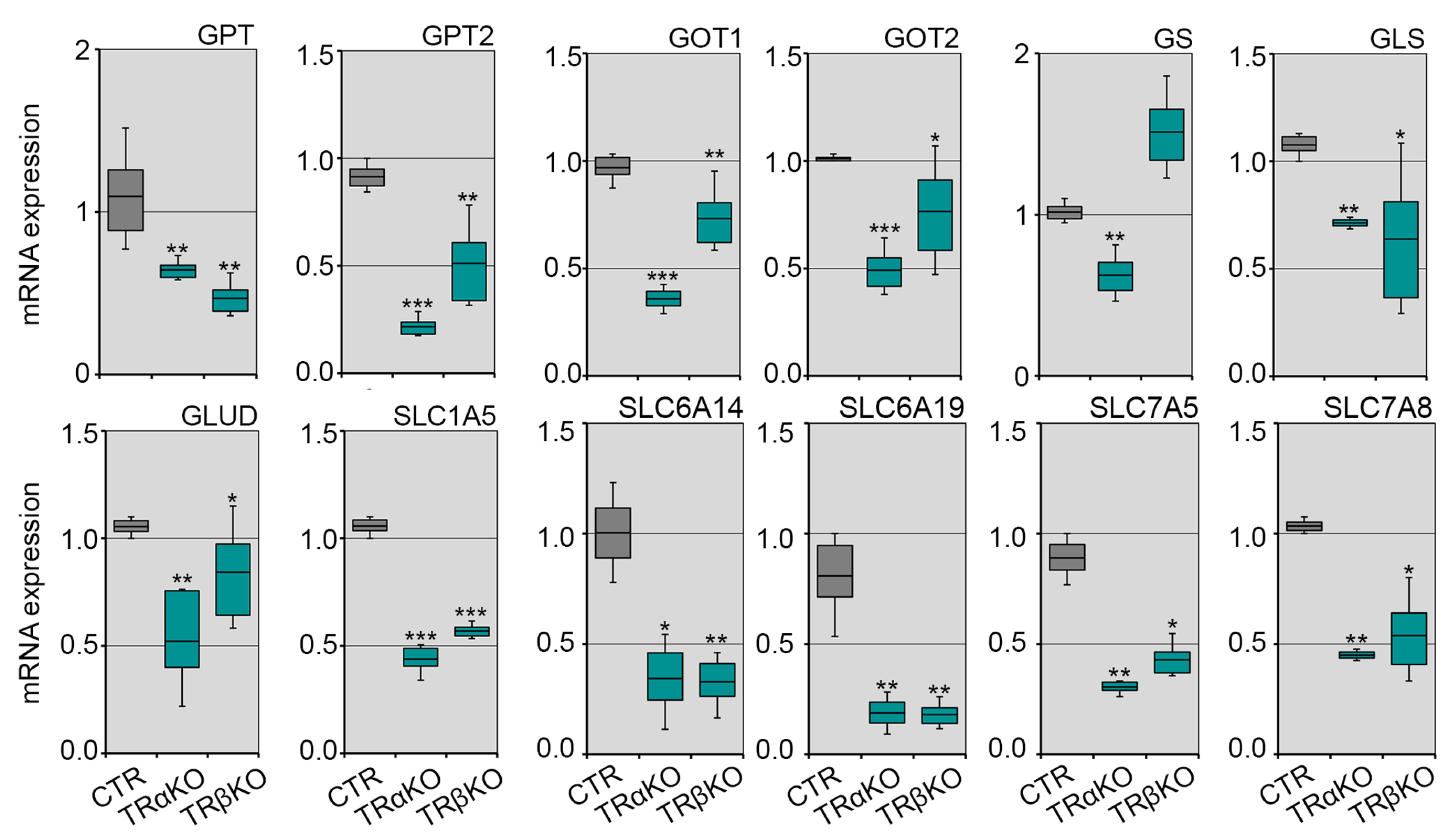
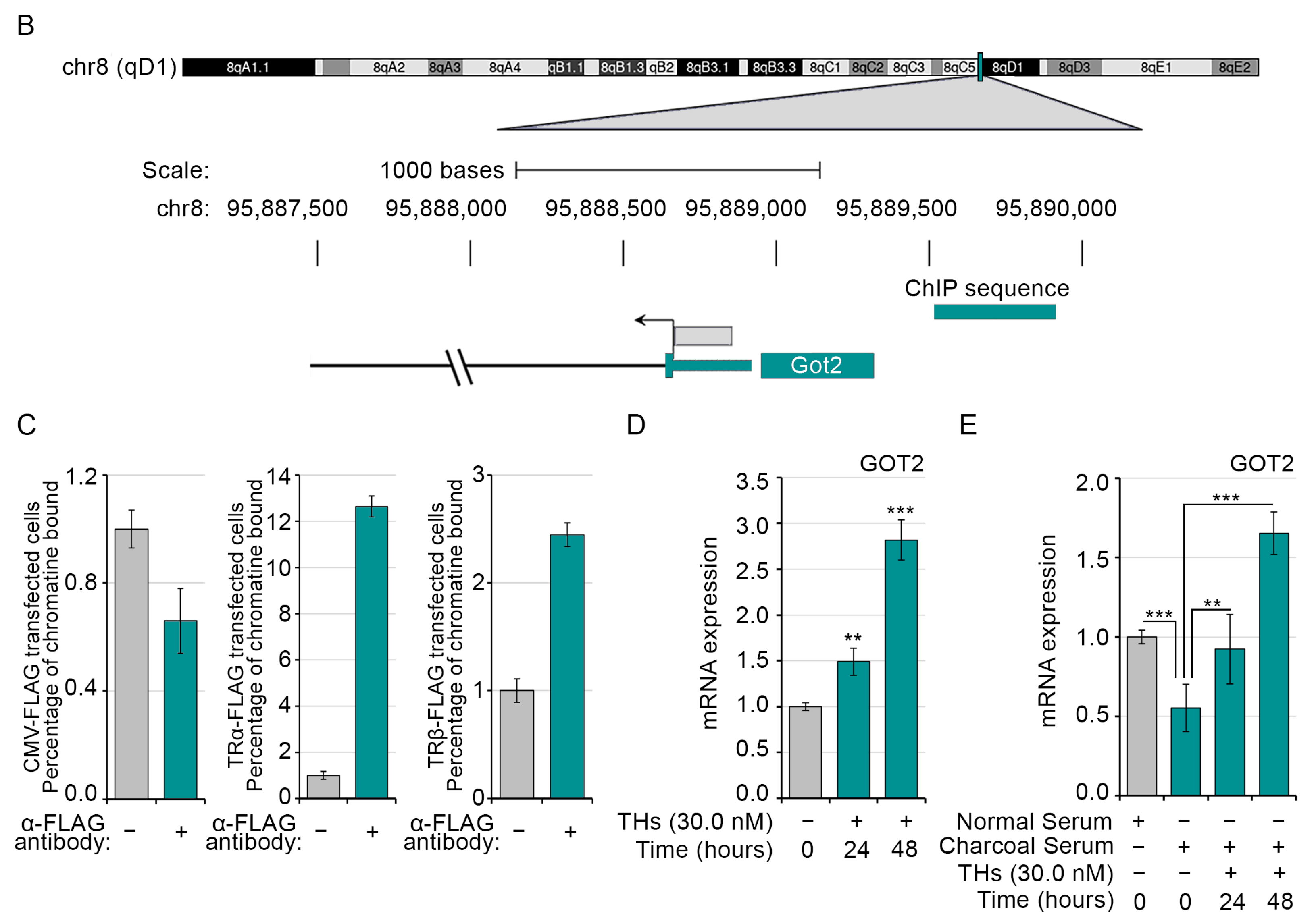
2.3. Loss of Both TRα and TRβ Isoforms Affects the Glutamine Metabolism in Skeletal Muscle
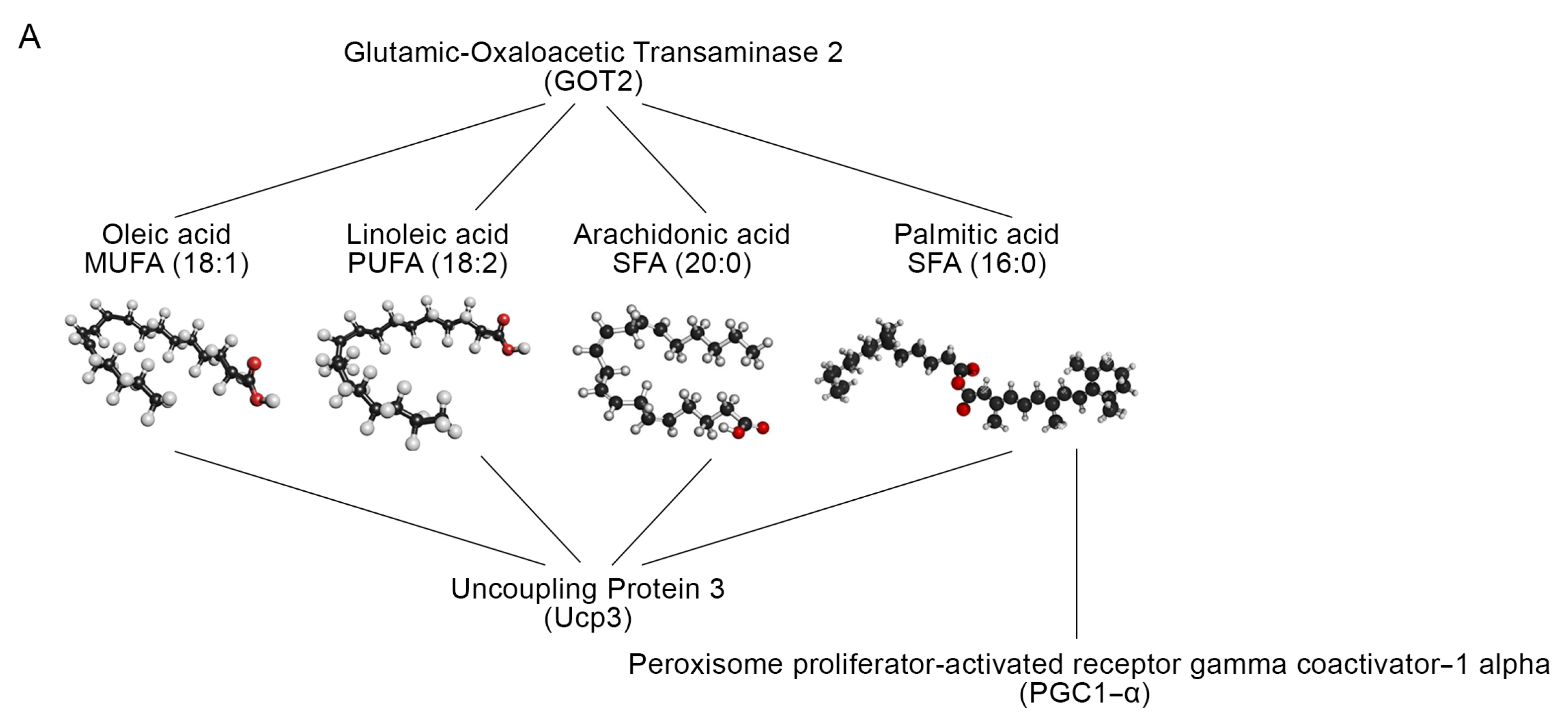
2.4. Skeletal Muscle TRs Deficiency Impacts on Lipids Composition
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Reagents and Transfection
4.2. Mouse Strains
4.3. Animals and Histology
4.4. Histochemical SDH Staining
4.5. Histochemical NADH Staining
4.6. Isolation and Loading of Single Skeletal Muscle Fibers with Mito-Sox and Mito-Tracker
4.7. Real-Time PCR
4.8. LC-MS/MS Analysis of Muscle Tissue
4.9. Transcriptomic and Metabolomic Analysis
4.10. In Silico Promoter Analysis for Searching Transition Factor Binding Sites
4.11. Chromatin Immuno-Precipitation (ChIP) Assay
4.12. Quantification and Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sagliocchi, S.; Cicatiello, A.G.; Di Cicco, E.; Ambrosio, R.; Miro, C.; Di Girolamo, D.; Nappi, A.; Mancino, G.; De Stefano, M.A.; Luongo, C.; et al. The thyroid hormone activating enzyme, type 2 deiodinase, induces myogenic differentiation by regulating mitochondrial metabolism and reducing oxidative stress. Redox Biol. 2019, 24, 101228. [Google Scholar] [CrossRef] [PubMed]
- Mancino, G.; Miro, C.; Di Cicco, E.; Dentice, M. Thyroid hormone action in epidermal development and homeostasis and its implications in the pathophysiology of the skin. J. Endocrinol. Investig. 2021, 44, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Zhu, X.G.; Ge, K.; Cheng, S.Y. Adipogenesis is differentially impaired by thyroid hormone receptor mutant isoforms. J. Mol. Endocrinol. 2010, 44, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Ma, X.L.; Wang, X.; Chen, H.; Huang, B.R. Insulin-like growth factor binding protein-6 interacts with the thyroid hormone receptor alpha1 and modulates the thyroid hormone-response in osteoblastic differentiation. Mol. Cell. Biochem. 2012, 361, 197–208. [Google Scholar] [CrossRef]
- Chatonnet, F.; Picou, F.; Fauquier, T.; Flamant, F. Thyroid hormone action in cerebellum and cerebral cortex development. J. Thyroid Res. 2011, 2011, 145762. [Google Scholar] [CrossRef]
- Gereben, B.; Zeöld, A.; Dentice, M.; Salvatore, D.; Bianco, A.C. Activation and inactivation of thyroid hormone by deiodinases: Local action with general consequences. Cell. Mol. Life Sci. 2008, 65, 570–590. [Google Scholar] [CrossRef]
- Boelen, A.; van der Spek, A.H.; Bloise, F.; de Vries, E.M.; Surovtseva, O.V.; van Beeren, M.; Ackermans, M.T.; Kwakkel, J.; Fliers, E. Tissue thyroid hormone metabolism is differentially regulated during illness in mice. J. Endocrinol. 2017, 233, 25–36. [Google Scholar] [CrossRef]
- Gereben, B.; Zavacki, A.M.; Ribich, S.; Kim, B.W.; Huang, S.A.; Simonides, W.S.; Zeöld, A.; Bianco, A.C. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr. Rev. 2008, 29, 898–938. [Google Scholar] [CrossRef]
- Nappi, A.; Murolo, M.; Sagliocchi, S.; Miro, C.; Cicatiello, A.G.; Di Cicco, E.; Di Paola, R.; Raia, M.; D’Esposito, L.; Stornaiuolo, M.; et al. Selective Inhibition of Genomic and Non-Genomic Effects of Thyroid Hormone Regulates Muscle Cell Differentiation and Metabolic Behavior. Int. J. Mol. Sci. 2021, 22, 7175. [Google Scholar] [CrossRef]
- Davis, P.J.; Ashur-Fabian, O.; Incerpi, S.; Mousa, S.A. Editorial: Non Genomic Actions of Thyroid Hormones in Cancer. Front. Endocrinol. 2019, 10, 847. [Google Scholar] [CrossRef]
- Davis, P.J.; Goglia, F.; Leonard, J.L. Nongenomic actions of thyroid hormone. Nat. Rev. Endocrinol. 2016, 12, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.J.; Leonard, J.L.; Davis, F.B. Mechanisms of nongenomic actions of thyroid hormone. Front. Neuroendocrinol. 2008, 29, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Hiroi, Y.; Kim, H.H.; Ying, H.; Furuya, F.; Huang, Z.; Simoncini, T.; Noma, K.; Ueki, K.; Nguyen, N.H.; Scanlan, T.S.; et al. Rapid nongenomic actions of thyroid hormone. Proc. Natl. Acad. Sci. USA 2006, 103, 14104–14109. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev. Endocr. Metab. Disord 2000, 1, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Umesono, K.; Giguere, V.; Glass, C.K.; Rosenfeld, M.G.; Evans, R.M. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature 1988, 336, 262–265. [Google Scholar] [CrossRef]
- Glass, C.K.; Rosenfeld, M.G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000, 14, 121–141. [Google Scholar] [CrossRef]
- Forrest, D.; Sjoberg, M.; Vennstrom, B. Contrasting developmental and tissue-specific expression of alpha and beta thyroid hormone receptor genes. EMBO J. 1990, 9, 1519–1528. [Google Scholar] [CrossRef]
- Minakhina, S.; Bansal, S.; Zhang, A.; Brotherton, M.; Janodia, R.; De Oliveira, V.; Tadepalli, S.; Wondisford, F.E. A Direct Comparison of Thyroid Hormone Receptor Protein Levels in Mice Provides Unexpected Insights into Thyroid Hormone Action. Thyroid 2020, 30, 1193–1204. [Google Scholar] [CrossRef]
- Milanesi, A.; Lee, J.W.; Kim, N.H.; Liu, Y.Y.; Yang, A.; Sedrakyan, S.; Kahng, A.; Cervantes, V.; Tripuraneni, N.; Cheng, S.Y.; et al. Thyroid Hormone Receptor alpha Plays an Essential Role in Male Skeletal Muscle Myoblast Proliferation, Differentiation, and Response to Injury. Endocrinology 2016, 157, 4–15. [Google Scholar] [CrossRef]
- Ambrosio, R.; De Stefano, M.A.; Di Girolamo, D.; Salvatore, D. Thyroid hormone signaling and deiodinase actions in muscle stem/progenitor cells. Mol. Cell. Endocrinol. 2017, 459, 79–83. [Google Scholar] [CrossRef]
- Dentice, M.; Marsili, A.; Ambrosio, R.; Guardiola, O.; Sibilio, A.; Paik, J.H.; Minchiotti, G.; DePinho, R.A.; Fenzi, G.; Larsen, P.R.; et al. The FoxO3/type 2 deiodinase pathway is required for normal mouse myogenesis and muscle regeneration. J. Clin. Investig. 2010, 120, 4021–4030. [Google Scholar] [CrossRef] [PubMed]
- Dentice, M.; Ambrosio, R.; Damiano, V.; Sibilio, A.; Luongo, C.; Guardiola, O.; Yennek, S.; Zordan, P.; Minchiotti, G.; Colao, A.; et al. Intracellular inactivation of thyroid hormone is a survival mechanism for muscle stem cell proliferation and lineage progression. Cell Metab. 2014, 20, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Udayakumar, N.; Rameshkumar, A.C.; Srinivasan, A.V. Hoffmann syndrome: Presentation in hypothyroidism. J. Postgrad. Med. 2005, 51, 332–333. [Google Scholar] [PubMed]
- Ramsay, I.D. Muscle dysfunction in hyperthyroidism. Lancet 1966, 2, 931–934. [Google Scholar] [CrossRef]
- Fraichard, A.; Chassande, O.; Plateroti, M.; Roux, J.P.; Trouillas, J.; Dehay, C.; Legrand, C.; Gauthier, K.; Kedinger, M.; Malaval, L.; et al. The T3R alpha gene encoding a thyroid hormone receptor is essential for post-natal development and thyroid hormone production. EMBO J. 1997, 16, 4412–4420. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, K.; Chassande, O.; Plateroti, M.; Roux, J.P.; Legrand, C.; Pain, B.; Rousset, B.; Weiss, R.; Trouillas, J.; Samarut, J. Different functions for the thyroid hormone receptors TRalpha and TRbeta in the control of thyroid hormone production and post-natal development. EMBO J. 1999, 18, 623–631. [Google Scholar] [CrossRef]
- Wikström, L.; Johansson, C.; Saltó, C.; Barlow, C.; Campos Barros, A.; Baas, F.; Forrest, D.; Thorén, P.; Vennström, B. Abnormal heart rate and body temperature in mice lacking thyroid hormone receptor alpha 1. EMBO J. 1998, 17, 455–461. [Google Scholar] [CrossRef]
- Bochukova, E.; Schoenmakers, N.; Agostini, M.; Schoenmakers, E.; Rajanayagam, O.; Keogh, J.M.; Henning, E.; Reinemund, J.; Gevers, E.; Sarri, M.; et al. A mutation in the thyroid hormone receptor alpha gene. N. Engl. J. Med. 2012, 366, 243–249. [Google Scholar] [CrossRef]
- Moran, C.; Agostini, M.; Visser, W.E.; Schoenmakers, E.; Schoenmakers, N.; Offiah, A.C.; Poole, K.; Rajanayagam, O.; Lyons, G.; Halsall, D.; et al. Resistance to thyroid hormone caused by a mutation in thyroid hormone receptor (TR)alpha1 and TRalpha2: Clinical, biochemical, and genetic analyses of three related patients. Lancet Diabetes Endocrinol. 2014, 2, 619–626. [Google Scholar] [CrossRef]
- Di Cicco, E.; Moran, C.; Visser, W.E.; Nappi, A.; Schoenmakers, E.; Todd, P.; Lyons, G.; Dattani, M.; Ambrosio, R.; Parisi, S.; et al. Germ Line Mutations in the Thyroid Hormone Receptor Alpha Gene Predispose to Cutaneous Tags and Melanocytic Nevi. Thyroid 2021, 31, 1114–1126. [Google Scholar] [CrossRef]
- Forrest, D.; Hanebuth, E.; Smeyne, R.J.; Everds, N.; Stewart, C.L.; Wehner, J.M.; Curran, T. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor beta: Evidence for tissue-specific modulation of receptor function. EMBO J. 1996, 15, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Göthe, S.; Wikström, L.; Forrest, D.; Vennström, B.; Larsson, L. Effects of thyroid hormone receptor gene disruption on myosin isoform expression in mouse skeletal muscles. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 278, R1545–R1554. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef] [PubMed]
- Iwen, K.A.; Schroder, E.; Brabant, G. Thyroid hormones and the metabolic syndrome. Eur. Thyroid J. 2013, 2, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Cicatiello, A.G.; Sagliocchi, S.; Nappi, A.; Di Cicco, E.; Miro, C.; Murolo, M.; Stornaiuolo, M.; Dentice, M. Thyroid hormone regulates glutamine metabolism and anaplerotic fluxes by inducing mitochondrial glutamate aminotransferase GPT2. Cell Rep. 2022, 38, 110562. [Google Scholar] [CrossRef]
- Wulf, A.; Harneit, A.; Kröger, M.; Kebenko, M.; Wetzel, M.G.; Weitzel, J.M. T3-mediated expression of PGC-1alpha via a far upstream located thyroid hormone response element. Mol. Cell. Endocrinol. 2008, 287, 90–95. [Google Scholar] [CrossRef]
- de Lange, P.; Feola, A.; Ragni, M.; Senese, R.; Moreno, M.; Lombardi, A.; Silvestri, E.; Amat, R.; Villarroya, F.; Goglia, F.; et al. Differential 3,5,3′-triiodothyronine-mediated regulation of uncoupling protein 3 transcription: Role of Fatty acids. Endocrinology 2007, 148, 4064–4072. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Silva, J.E. Thermogenic mechanisms and their hormonal regulation. Physiol. Rev. 2006, 86, 435–464. [Google Scholar] [CrossRef]
- Warner, A.; Mittag, J. Thyroid hormone and the central control of homeostasis. J. Mol. Endocrinol. 2012, 49, R29–R35. [Google Scholar] [CrossRef]
- Yen, P.M. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 2001, 81, 1097–1142. [Google Scholar] [CrossRef] [PubMed]
- Cicatiello, A.G.; Di Girolamo, D.; Dentice, M. Metabolic Effects of the Intracellular Regulation of Thyroid Hormone: Old Players, New Concepts. Front. Endocrinol. 2018, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Zurlo, F.; Larson, K.; Bogardus, C.; Ravussin, E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J. Clin. Investig. 1990, 86, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Argov, Z.; Renshaw, P.F.; Boden, B.; Winokur, A.; Bank, W.J. Effects of thyroid hormones on skeletal muscle bioenergetics. In vivo phosphorus-31 magnetic resonance spectroscopy study of humans and rats. J. Clin. Investig. 1988, 81, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Bloise, F.F.; Cordeiro, A.; Ortiga-Carvalho, T.M. Role of thyroid hormone in skeletal muscle physiology. J. Endocrinol. 2018, 236, R57–R68. [Google Scholar] [CrossRef]
- Simonides, W.S.; van Hardeveld, C. Thyroid hormone as a determinant of metabolic and contractile phenotype of skeletal muscle. Thyroid 2008, 18, 205–216. [Google Scholar] [CrossRef]
- Caiozzo, V.J.; Haddad, F.; Baker, M.; McCue, S.; Baldwin, K.M. MHC polymorphism in rodent plantaris muscle: Effects of mechanical overload and hypothyroidism. Am. J. Physiol. Cell Physiol. 2000, 278, C709–C717. [Google Scholar] [CrossRef]
- Miyabara, E.H.; Aoki, M.S.; Soares, A.G.; Saltao, R.M.; Vilicev, C.M.; Passarelli, M.; Scanlan, T.S.; Gouveia, C.H.; Moriscot, A.S. Thyroid hormone receptor-beta-selective agonist GC-24 spares skeletal muscle type I to II fiber shift. Cell Tissue Res. 2005, 321, 233–241. [Google Scholar] [CrossRef]
- Miyabara, E.H.; Aoki, M.S.; Soares, A.G.; Moriscot, A.S. Expression of tropism-related genes in regenerating skeletal muscle of rats treated with cyclosporin-A. Cell Tissue Res. 2005, 319, 479–489. [Google Scholar] [CrossRef]
- Salvatore, D.; Simonides, W.S.; Dentice, M.; Zavacki, A.M.; Larsen, P.R. Thyroid hormones and skeletal muscle—New insights and potential implications. Nat. Rev. Endocrinol. 2014, 10, 206–214. [Google Scholar] [CrossRef]
- Johansson, C.; Göthe, S.; Forrest, D.; Vennström, B.; Thorén, P. Cardiovascular phenotype and temperature control in mice lacking thyroid hormone receptor-beta or both alpha1 and beta. Am. J. Physiol. 1999, 276, H2006–H2012. [Google Scholar] [PubMed]
- Ribeiro, M.O.; Carvalho, S.D.; Schultz, J.J.; Chiellini, G.; Scanlan, T.S.; Bianco, A.C.; Brent, G.A. Thyroid hormone--sympathetic interaction and adaptive thermogenesis are thyroid hormone receptor isoform—Specific. J. Clin. Investig. 2001, 108, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, H.; Rudling, M.; Saltó, C.; Forrest, D.; Angelin, B.; Vennström, B. Requirement for thyroid hormone receptor beta in T3 regulation of cholesterol metabolism in mice. Mol. Endocrinol. 2002, 16, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Marrif, H.; Schifman, A.; Stepanyan, Z.; Gillis, M.A.; Calderone, A.; Weiss, R.E.; Samarut, J.; Silva, J.E. Temperature homeostasis in transgenic mice lacking thyroid hormone receptor-alpha gene products. Endocrinology 2005, 146, 2872–2884. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, J.; Gauthier, K.; Ho, J.P.; Lim, A.; Zhu, X.G.; Han, C.R.; Sinha, R.A.; Cheng, S.Y.; Yen, P.M. Thyroid Hormone Receptor alpha Regulates Autophagy, Mitochondrial Biogenesis, and Fatty Acid Use in Skeletal Muscle. Endocrinology 2021, 162, bqab112. [Google Scholar] [CrossRef]
- Zaidi, M.; New, M.I.; Blair, H.C.; Zallone, A.; Baliram, R.; Davies, T.F.; Cardozo, C.; Iqbal, J.; Sun, L.; Rosen, C.J.; et al. Actions of pituitary hormones beyond traditional targets. J. Endocrinol. 2018, 237, R83–R98. [Google Scholar] [CrossRef]
- Pellegrini, G.; Moggio, M. Hypothyroid myopathy: Histochemical and ultrastructural features with physiopatological correlations (author’s transl). Riv. Patol. Nerv. Ment. 1979, 99, 275–288. [Google Scholar]
- Malik, A.N.; Czajka, A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef]
- van Karnebeek, C.D.M.; Ramos, R.J.; Wen, X.Y.; Tarailo-Graovac, M.; Gleeson, J.G.; Skrypnyk, C.; Brand-Arzamendi, K.; Karbassi, F.; Issa, M.Y.; van der Lee, R.; et al. Bi-allelic GOT2 Mutations Cause a Treatable Malate-Aspartate Shuttle-Related Encephalopathy. Am. J. Hum. Genet. 2019, 105, 534–548. [Google Scholar] [CrossRef]
- Yang, S.; Hwang, S.; Kim, M.; Seo, S.B.; Lee, J.H.; Jeong, S.M. Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 2018, 9, 55. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, L.; Shi, Q.; Zhao, Y.; Lin, H.; Zhang, M.; Zhao, S.; Yang, Y.; Ling, Z.Q.; Guan, K.L.; et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015, 34, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Dubowitz, V. Muscle biopsy—Technical and diagnostic aspects. Ann. Clin. Res. 1974, 6, 69–79. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Gulley, M.L. DNA extraction from fresh or frozen tissues. Methods Mol. Med. 2001, 49, 5–10. [Google Scholar] [PubMed]
- Sommella, E.; Badolati, N.; Riccio, G.; Salviati, E.; Bottone, S.; Dentice, M.; Campiglia, P.; Tenore, G.C.; Stornaiuolo, M.; Novellino, E. A Boost in Mitochondrial Activity Underpins the Cholesterol-Lowering Effect of Annurca Apple Polyphenols on Hepatic Cells. Nutrients 2019, 11, 163. [Google Scholar] [CrossRef]
- Lapi, D.; Stornaiuolo, M.; Sabatino, L.; Sommella, E.; Tenore, G.; Daglia, M.; Scuri, R.; Di Maro, M.; Colantuoni, A.; Novellino, E. The Pomace Extract Taurisolo Protects Rat Brain From Ischemia-Reperfusion Injury. Front. Cell. Neurosci. 2020, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]

| CTR | TRαKO | TRβKO | ||
|---|---|---|---|---|
| Oleic acid | MUFA (18:1) | 63.9 ± 1.15 | 12.6 ± 0.91 | 14.1 ± 3.10 |
| Palmitoleic acid | MUFA (16:1) | 8.1 ± 0.23 | 1.5 ± 0.31 | 2.3 ± 0.24 |
| Palmitic acid | SFA (16:0) | 71.0 ± 1.35 | 25.2 ± 1.79 | 12.9 ± 1.97 |
| Arachidic acid | SFA (20:0) | 1.2 ± 0.13 | 0.8 ± 0.12 | 0.1 ± 0.00 |
| Linoleic acid | PUFA (18:2) | 31.9 ± 0.37 | 15.4 ± 1.32 | 6.4 ± 0.56 |
| Cholesterol | Cholesterol | 379.1 ± 0.28 | 366.5 ± 2.64 | 336.6 ± 2.91 |
| Docosahexaenoic acid | PUFA (22:6) | 0.2 ± 0.02 | 0.2 ± 0.16 | 0.2 ± 0.25 |
| Behenic acid | SFA (22:0) | 0.2 ± 0.02 | 0.4 ± 0.37 | 0.2 ± 0.15 |
| Docosadienoic acid | PUFA (22:2) | 0.4 ± 0.02 | 0.4 ± 0.10 | 1.3 ± 0.32 |
| Myristic acid | SFA (14:0) | 2.2 ± 0.18 | 1.2 ± 0.11 | 11.4 ± 0.81 |
| Eicosadienoic acid | PUFA (20:2) | 0.2 ± 0.02 | 0.5 ± 0.09 | 2.1 ± 0.38 |
| Hexadecenoic acid | PUFA (16:2) | 0.2 ± 0.02 | 1.0 ± 0.40 | 2.7 ± 0.66 |
| Docosatetraenoic acid | PUFA (22:4) | 0.3 ± 0.02 | 0.2 ± 0.25 | 1.5 ± 1.47 |
| Eicosapentaenoic acid | PUFA (20:5) | 0.1 ± 0.01 | 1.0 ± 0.35 | 1.3 ± 0.22 |
| Docosenoic acid | MUFA (22:1) | 0.3 ± 0.13 | 0.8 ± 0.71 | 1.6 ± 0.12 |
| Eicosatetraenoic acid | PUFA (20:4) | 0.3 ± 0.12 | 0.2 ± 0.00 | 0.5 ± 0.08 |
| Octadecatrienoic acid | PUFA (18:3) | 1.1 ± 0.08 | 1.2 ± 0.79 | 2.4 ± 0.71 |
| Eicosatrienoic acid | PUFA (20:3) | 0.2 ± 0.02 | 0.5 ± 0.34 | 0.4 ± 0.30 |
| Docosapentaenoic acid | PUFA (22:5) | 0.2 ± 0.02 | 0.5 ± 0.37 | 0.3 ± 0.21 |
| Lauric acid | SFA (12:0) | 0.2 ± 0.02 | 0.3 ± 0.12 | 0.2 ± 0.01 |
| Stearic acid | SFA (18:0) | 13.7 ± 0.39 | 45.0 ± 0.82 | 18.6 ± 1.08 |
| Oligos | Sequences | Amplicon Length |
|---|---|---|
| CyA | Forward CGCCACTGTCGCTTTTCG Reverse AACTTTGTCTGCAAACAGCTC | 120 bp |
| DRP1 | Forward CTAGAAGAGCCCAGCCTACG Reverse AGCAAAGTCGGGGTGTTTTG | 237 bp |
| GOT1 | Forward CTCCTCCGGTTCTGGTCTTT Reverse CCCCAAGAACTAGGCGAGAA | 234 bp |
| GOT2 | Forward CAGCCGAGATGTCTTTCTGC Reverse GGACACTCTGCTCTGGGATT | 167 bp |
| GOT2-ChIP | Forward CCCAGAGAGTCTATACAGTTC Reverse GGGGGCCACAGCATGTGCTGC | 390 bp |
| GLS | Forward GCTGTGCTCTATTGAAGTGA Reverse GCAAACTGCCCTGAGAAGTC | 175 bp |
| GLUD | Forward GAGATGTCCTGGATCGCTGA Reverse GGCCCACATTACCAAATCCC | 274 bp |
| GPT | Forward TTCAAGAAGGTGCTCACGGA Reverse CATCTGTTTCTGCACCTCGG | 278 bp |
| GPT2 | Forward GCTTTGAATGTGGACGAGCT Reverse TCGTGCCCCATCTGGTAAAG | 254 bp |
| GS | Forward ACAGCGACATGTACCTCCAT Reverse CTGCTCCATTCCAAACCAGG | 185 bp |
| MFN1 | Forward GATGACCTGGTTTTAGTAGAC Reverse TGAAGATGTTGGGCTTGGAGA | 187 bp |
| MFN2 | Forward TCGAGAGGCAGTTTGAGGAG Reverse GCAGCTTGTAGTCTTGAGCC | 187 bp |
| mtND-1 | Forward GTGAGTGATAGGGTAGGTGCA Reverse AACACTCCTCGTCCCCATTC | 219 bp |
| OPA1 | Forward GAAGGACGACAAAGGCATCC Reverse AGTCACCTTCACTGGAGAGC | 244 bp |
| PGC1-α | Forward TGATGTGAATGACTTGGATACAGACA Reverse GCTCATTGTTGTACTGGTTGGATATG | 95 bp |
| RNAaseP | Forward CTGACCACACGAGCTGGTAGAA Reverse GCCTACACTGGAGTCGTGCTACT | 61 bp |
| SLC1A5 | Forward CGGGACCTCTTCTAGCTCTG Reverse GGACACCCCGTTTAGTTGTG | 181 bp |
| SLC6A14 | Forward ATTCCCTTTCTGTGGCTTGG Reverse AGAGCTTCAGGATAGGCAATGA | 235 bp |
| SLC6A19 | Forward GGCCTCATCTCCTTCTCCAG Reverse AGTCATCAAAGCGCTCAGTG | 154 bp |
| SLC7A5 | Forward CCCTGGCCCTCATCATTTTG Reverse GAAGAGGCCGCTGTACAAAG | 149 bp |
| SLC7A8 | Forward CAGCGCCTGTGGTATCATTG Reverse AGACTTAGGGATGGTGACGC | 181 bp |
| UCP3 | Forward GCCTACAGAACCATCGCCAG Reverse GCCACCATCTTCAGCATACA | 302 bp |
| AC | Score | Loc. | Strand | Consensus | Sequence |
|---|---|---|---|---|---|
| M00239 | 0.77 | 10 | (−) | SNNTRAGGTCACGSNN | ACTGCTGACCTAACTC |
| M00239 | 0.73 | 33 | (+) | SNNTRAGGTCACGSNN | CCTTATGGTCAATTAT |
| M00239 | 0.82 | 352 | (+) | SNNTRAGGTCACGSNN | GGATAAGGTCATATTA |
| M00239 | 0.73 | 364 | (−) | SNNTRAGGTCACGSNN | ATTAGTGACTCCATCT |
| M00239 | 0.75 | 409 | (+) | SNNTRAGGTCACGSNN | ACTTAAGATCTTGGCC |
| M00239 | 0.76 | 543 | (−) | SNNTRAGGTCACGSNN | CTCAGTCGCCTCAGTC |
| M00239 | 0.74 | 552 | (−) | SNNTRAGGTCACGSNN | CTCAGTCGCCTCAGTT |
| M00239 | 0.74 | 618 | (+) | SNNTRAGGTCACGSNN | CAACAAGCTCAAAGCA |
| M00239 | 0.81 | 642 | (+) | SNNTRAGGTCACGSNN | CTGTAAGCTCATTATA |
| M00239 | 0.75 | 653 | (+) | SNNTRAGGTCACGSNN | TTATAAGTTCAAAGCA |
| M00239 | 0.73 | 705 | (−) | SNNTRAGGTCACGSNN | ACCTGTGGCTTCATCC |
| M00239 | 0.75 | 783 | (+) | SNNTRAGGTCACGSNN | ATGAAAGCTCACTGTA |
| M00239 | 0.73 | 935 | (+) | SNNTRAGGTCACGSNN | CTGTGAAATCACTAAT |
| M00239 | 0.77 | 1467 | (+) | SNNTRAGGTCACGSNN | GTTTGAGGCCAGTCTG |
| M00239 | 0.75 | 1490 | (+) | SNNTRAGGTCACGSNN | AAGTGAGTTCCAGGAC |
| M00239 | 0.75 | 1611 | (+) | SNNTRAGGTCACGSNN | GGTTAAGAGCACTGTC |
| M00239 | 0.74 | 1633 | (+) | SNNTRAGGTCACGSNN | TCCAGAGGTCATGAGT |
| M00239 | 0.75 | 1639 | (−) | SNNTRAGGTCACGSNN | GGTCATGAGTTCAATT |
| M00239 | 0.77 | 1773 | (+) | SNNTRAGGTCACGSNN | GCCCAAGGTCAGGGAG |
| M00239 | 0.73 | 1845 | (−) | SNNTRAGGTCACGSNN | GATCTGGACTTCATGC |
| M00239 | 0.83 | 2045 | (−) | SNNTRAGGTCACGSNN | ATCCATCACCTTATAG |
| M00239 | 0.78 | 2166 | (−) | SNNTRAGGTCACGSNN | TAGTGTGGCCACATTG |
| M00239 | 0.77 | 2283 | (+) | SNNTRAGGTCACGSNN | GATTAAAGGCATGCAC |
| M00239 | 0.78 | 2357 | (−) | SNNTRAGGTCACGSNN | ACCAGTGACCTGGGCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nappi, A.; Murolo, M.; Cicatiello, A.G.; Sagliocchi, S.; Di Cicco, E.; Raia, M.; Stornaiuolo, M.; Dentice, M.; Miro, C. Thyroid Hormone Receptor Isoforms Alpha and Beta Play Convergent Roles in Muscle Physiology and Metabolic Regulation. Metabolites 2022, 12, 405. https://doi.org/10.3390/metabo12050405
Nappi A, Murolo M, Cicatiello AG, Sagliocchi S, Di Cicco E, Raia M, Stornaiuolo M, Dentice M, Miro C. Thyroid Hormone Receptor Isoforms Alpha and Beta Play Convergent Roles in Muscle Physiology and Metabolic Regulation. Metabolites. 2022; 12(5):405. https://doi.org/10.3390/metabo12050405
Chicago/Turabian StyleNappi, Annarita, Melania Murolo, Annunziata Gaetana Cicatiello, Serena Sagliocchi, Emery Di Cicco, Maddalena Raia, Mariano Stornaiuolo, Monica Dentice, and Caterina Miro. 2022. "Thyroid Hormone Receptor Isoforms Alpha and Beta Play Convergent Roles in Muscle Physiology and Metabolic Regulation" Metabolites 12, no. 5: 405. https://doi.org/10.3390/metabo12050405
APA StyleNappi, A., Murolo, M., Cicatiello, A. G., Sagliocchi, S., Di Cicco, E., Raia, M., Stornaiuolo, M., Dentice, M., & Miro, C. (2022). Thyroid Hormone Receptor Isoforms Alpha and Beta Play Convergent Roles in Muscle Physiology and Metabolic Regulation. Metabolites, 12(5), 405. https://doi.org/10.3390/metabo12050405