Analysis of Serum Proteome after Treatment of Osteoporosis with Anabolic or Antiresorptive Drugs
Abstract
:1. Introduction
2. Results
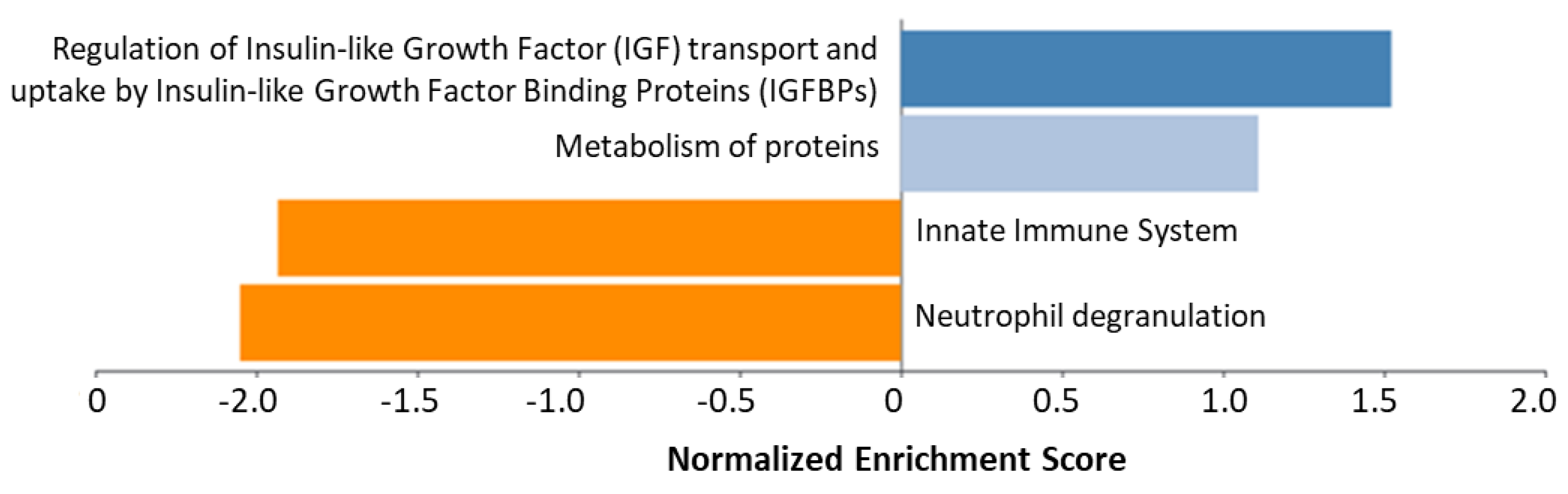
2.1. Teriparatide
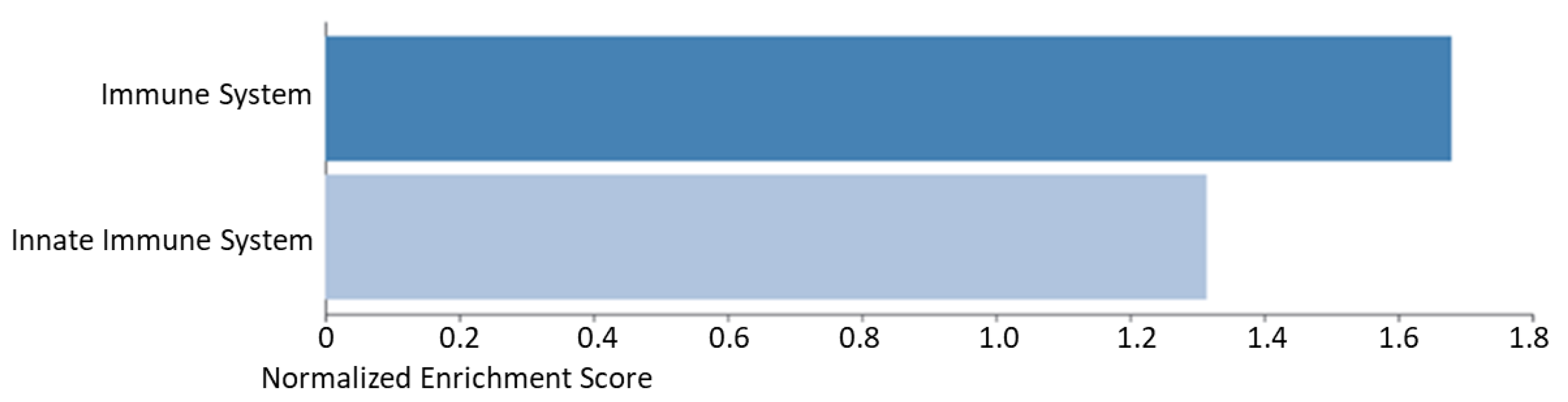
2.2. Denosumab
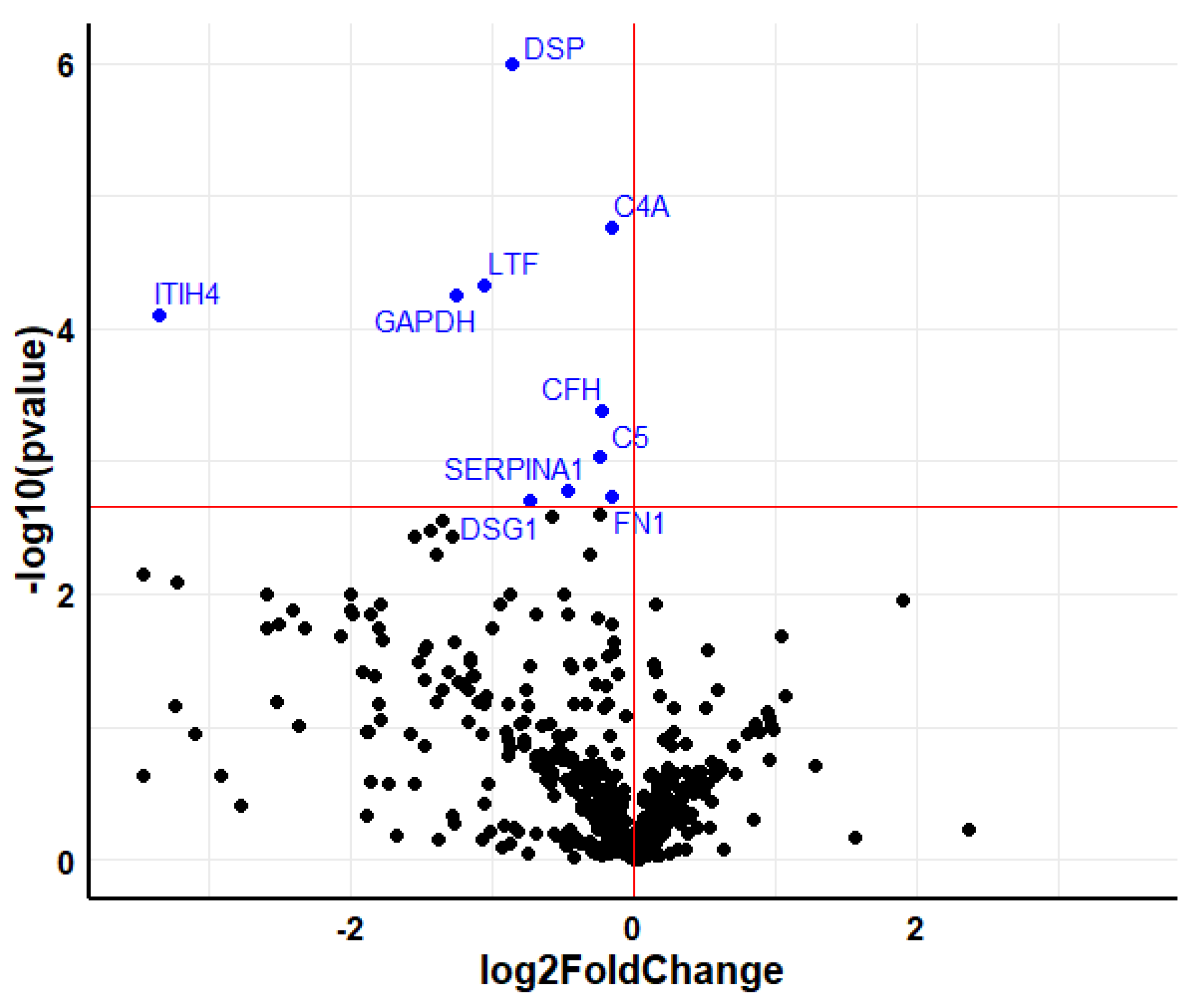
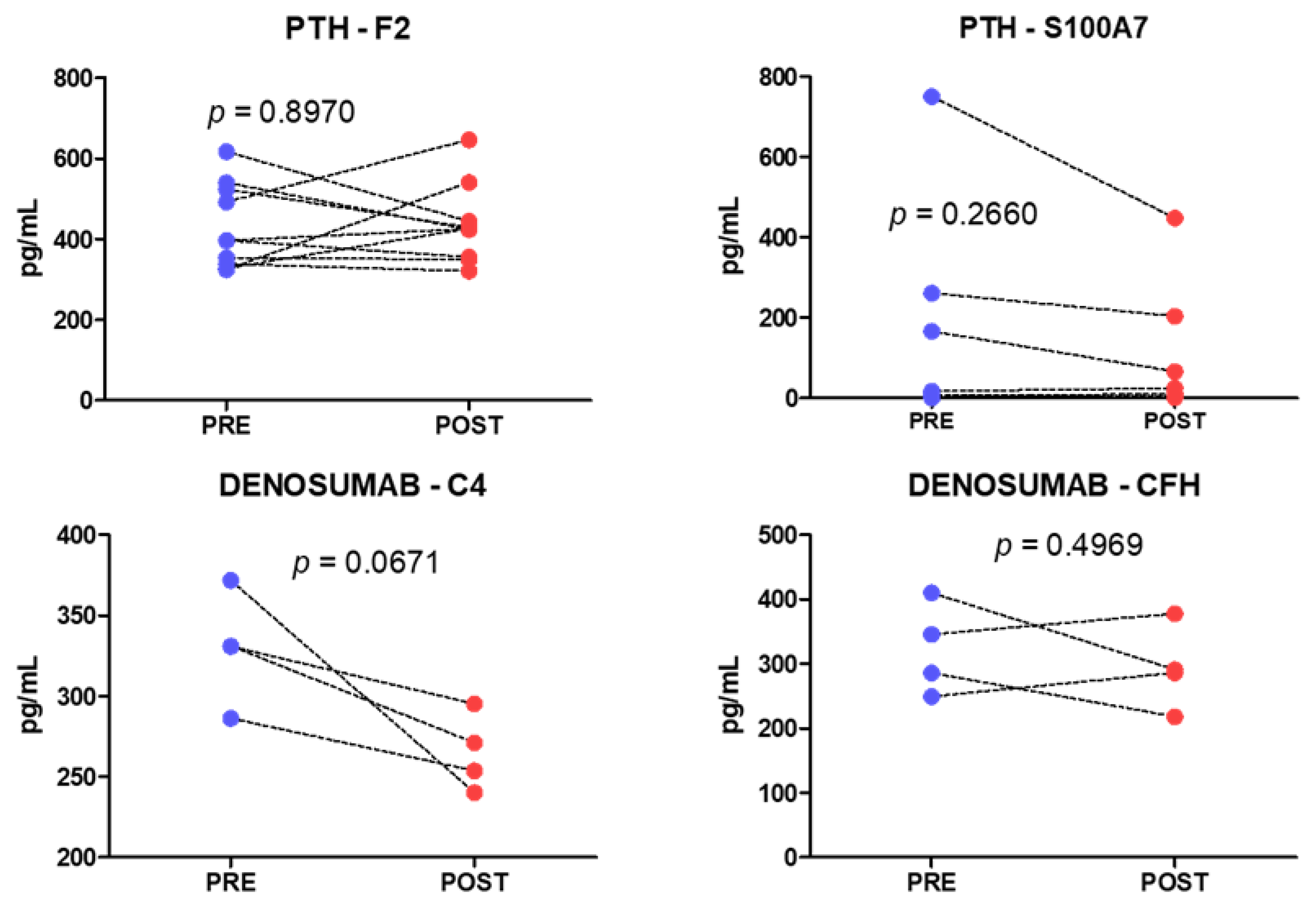
2.3. Analysis of Individual Proteins
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouxsein, M.L.; Eastell, R.; Lui, L.Y.; Wu, L.A.; de Papp, A.E.; Grauer, A.; Marin, F.; Cauley, J.A.; Bauer, D.C.; Black, D.M. Change in Bone Density and Reduction in Fracture Risk: A Meta-Regression of Published Trials. J. Bone Miner. Res. 2019, 34, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, P.; Kapoor, E.; Asi, N.; Alahdab, F.; Mohammed, K.; Benkhadra, K.; Almasri, J.; Farah, W.; Sarigianni, M.; Muthusamy, K.; et al. Efficacy of Pharmacological Therapies for the Prevention of Fractures in Postmenopausal Women: A Network Meta-Analysis. J. Clin. Endocrinol. Metab. 2019, 104, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Shabestari, M.; Shabestari, Y.R.; Landin, M.A.; Pepaj, M.; Cleland, T.P.; Reseland, J.E.; Eriksen, E.F. Altered protein levels in bone marrow lesions of hip osteoarthritis: Analysis by proteomics and multiplex immunoassays. Int. J. Rheum. Dis. 2020, 23, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Garnero, P. The Utility of Biomarkers in Osteoporosis Management. Mol. Diagn. Ther. 2017, 21, 401–418. [Google Scholar] [CrossRef] [PubMed]
- Nielson, C.M.; Wiedrick, J.; Shen, J.; Jacobs, J.; Baker, E.S.; Baraff, A.; Piehowski, P.; Lee, C.G.; Baratt, A.; Petyuk, V.; et al. Identification of Hip BMD Loss and Fracture Risk Markers Through Population-Based Serum Proteomics. J. Bone Miner. Res. 2017, 32, 1559–1567. [Google Scholar] [CrossRef]
- Takayanagi, H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef]
- Omata, Y.; Frech, M.; Saito, T.; Schett, G.; Zaiss, M.M.; Tanaka, S. Inflammatory Arthritis and Bone Metabolism Regulated by Type 2 Innate and Adaptive Immunity. Int. J. Mol. Sci. 2022, 23, 1104. [Google Scholar] [CrossRef]
- Saxena, Y.; Routh, S.; Mukhopadhaya, A. Immunoporosis: Role of Innate Immune Cells in Osteoporosis. Front. Immunol. 2021, 12, 3168. [Google Scholar] [CrossRef]
- Dahl, J.; Gulseth, H.L.; Forsén, L.; Hoff, M.; Forsmo, S.; Åsvold, B.O.; Schei, B.; Midthjell, K.; Meyer, H.E. Risk of hip and forearm fracture in subjects with type 2 diabetes mellitus and latent autoimmune diabetes of adults—The HUNT Study, Norway. Bone 2021, 153, 116110. [Google Scholar] [CrossRef]
- Toscani, D.; Bolzoni, M.; Accardi, F.; Aversa, F.; Giuliani, N. The osteoblastic niche in the context of multiple myeloma. Ann. N. Y. Acad. Sci. 2015, 1335, 45–62. [Google Scholar] [CrossRef]
- Donham, C.; Manilay, J.O. The Effects of Sclerostin on the Immune System. Curr. Osteoporos. Rep. 2020, 18, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Cronin, O.; Lanham-New, S.A.; Corfe, B.M.; Gregson, C.L.; Darling, A.L.; Ahmadi, K.R.; Gibson, P.S.; Tobias, J.H.; Ward, K.A.; Traka, M.H.; et al. Role of the Microbiome in Regulating Bone Metabolism and Susceptibility to Osteoporosis. Calcif. Tissue Int. 2022, 110, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Hsu, E.; Pacifici, R. From Osteoimmunology to Osteomicrobiology: How the Microbiota and the Immune System Regulate Bone. Calcif. Tissue Int. 2018, 102, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Malik Tyagi, A.; Li, J.Y.; Adams, J.; Denning, T.L.; Weitzmann, M.N.; Jones, R.M.; Pacifici, R. PTH induces bone loss via microbial-dependent expansion of intestinal TNF + T cells and Th17 cells. Nat. Commun. 2020, 11, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabacco, G.; Bilezikian, J.P. Osteoanabolic and dual action drugs. Br. J. Clin. Pharmacol. 2019, 85, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; D’Amelio, P.; Tyagi, A.M.; Vaccaro, C.; Li, J.; Hsu, E.; Buondonno, I.; Sassi, F.; Adams, J.; Weitzmann, M.N.; et al. Regulatory T cells are expanded by Teriparatide treatment in humans and mediate intermittent PTH-induced bone anabolism in mice. EMBO Rep. 2018, 19, 156–171. [Google Scholar] [CrossRef]
- Pacifici, R. Role of Gut Microbiota in the Skeletal Response to PTH. J. Clin. Endocrinol. Metab. 2021, 106, 636–645. [Google Scholar] [CrossRef]
- Li, J.Y.; Yu, M.; Pal, S.; Tyagi, A.M.; Dar, H.; Adams, J.; Neale Weitzmann, M.; Jones, R.M.; Pacifici, R. Parathyroid hormone-dependent bone formation requires butyrate production by intestinal microbiota. J. Clin. Investig. 2020, 130, 1767–1781. [Google Scholar] [CrossRef]
- Takegahara, N.; Kim, H.; Choi, Y. RANKL biology. Bone 2022, 159, 116353. [Google Scholar] [CrossRef]
- McClung, M.R.; Lewiecki, E.M.; Cohen, S.B.; Bolognese, M.A.; Woodson, G.C.; Moffett, A.H.; Peacock, M.; Miller, P.D.; Lederman, S.N.; Chesnut, C.H.; et al. Denosumab in postmenopausal women with low bone mineral density. N. Engl. J. Med. 2006, 354, 821–831. [Google Scholar] [CrossRef]
- Cummings, S.R.; Martin, J.S.; McClung, M.R.; Siris, E.S.; Eastell, R.; Reid, I.R.; Delmas, P.; Zoog, H.B.; Austin, M.; Wang, A.; et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N. Engl. J. Med. 2009, 361, 756–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diker-Cohen, T.; Rosenberg, D.; Avni, T.; Shepshelovich, D.; Tsvetov, G.; Gafter-Gvili, A. Risk for Infections During Treatment with Denosumab for Osteoporosis: A Systematic Review and Meta-analysis. J. Clin. Endocrinol. Metab. 2020, 105, 1641–1658. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Fernández, A.; Paradela, A.; Navajas, R.; Albar, J.P. Generalized method for probability-based peptide and protein identification from tandem mass spectrometry data and sequence database searching. Mol. Cell Proteom. 2008, 7, 1748–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Serra, P.; Marcilla, M.; Villanueva, A.; Ramos-Fernandez, A.; Palau, A.; Leal, L.; Wahi, J.E.; Setien-Baranda, F.; Szczesna, K.; Moutinho, C.; et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014, 5, 3608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]



| Gene Symbol | Gene Full Name | Log2 Fold Change | q-Value |
|---|---|---|---|
| FN1 | Fibronectin | −0.241 | 1.3 × 10−5 |
| F2 | Prothrombin F2 | −0.173 | 3.9 × 10−5 |
| SERPINF2 | Alpha-2-antiplasmin | −0.219 | 1.0 × 10−3 |
| C3 | Complement C3 | −0.049 | 1.0 × 10−3 |
| ANXA2 | Annexin A2 | −1.024 | 4.0 × 10−3 |
| HRNR | Hornerin | −0.973 | 4.0 × 10−3 |
| JUP | Junction plakoglobin | -0.81 | 4.0 × 10−3 |
| DSP | Desmoplakin | −0.756 | 4.0 × 10−3 |
| S100A7 | Protein S100-A7 | −1.262 | 5.0 × 10−3 |
| GC | Vitamin D-binding protein | −0.099 | 6.0 × 10−3 |
| A2M | Alpha-2-macroglobulin | 0.093 | 6.0 × 10−3 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | −1.353 | 1.0 × 10−2 |
| FABP5 | Fatty acid-binding protein 5 | −2.106 | 1.2 × 10−2 |
| CALML5 | Calmodulin-like protein 5 | −1.447 | 1.2 × 10−2 |
| FLG2 | Filaggrin-2 | −0.928 | 1.3 × 10−2 |
| DSG1 | Desmoglein-1 | −1.067 | 1.6 × 10−2 |
| DSC1 | Desmocollin-1 | −0.919 | 2.0 × 10−2 |
| CTSD | Cathepsin D | −0.901 | 2.1 × 10−2 |
| SBSN | Suprabasin | −0.62 | 2.2 × 10−2 |
| S100A8 | Protein S100-A8 | −0.852 | 2.6 × 10−2 |
| SAA1 | Serum amyloid A-1 protein | 0.764 | 3.1 × 10−2 |
| SERPINC1 | Antithrombin-III | −0.109 | 3.2 × 10−2 |
| SAA2 | Serum amyloid A-2 protein | 2.684 | 3.3 × 10−2 |
| HBD | Hemoglobin subunit delta | −0.517 | 3.3 × 10−2 |
| ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 | −0.103 | 3.5 × 10−2 |
| CFB | Complement factor B | 0.09 | 4.0 × 10−2 |
| DCD | Dermcidin | −0.63 | 4.3 × 10−2 |
| HBA1 | Hemoglobin subunit alpha | −0.472 | 4.3 × 10−2 |
| Biological Process | |||
| Gene Set | Description | FDR | Gene ID |
| GO:0002446 | Neutrophil mediated immunity | 1.06 × 10−11 | F2; C3; ANXA2; HRNR; JUP; DSP; S100A7; FABP5; CALML5; FLG2; DSG1; DSC1; CTSD; S100A8 |
| GO:0002526 | Acute inflammatory response | 2.16 × 10−11 | FN1; F2; SERPINF2; C3; A2M; S100A8; SAA1; SERPINC1; SAA2; CFB |
| GO:0006959 | Humoral immune response | 1.17 × 10−6 | F2; C3; S100A7; A2M; GAPDH; S100A8; CFB; DCD |
| GO:0052547 | Regulation of peptidase activity | 6.40 × 10−5 | FN1; SERPINF2; C3; A2M; GAPDH; S100A8; SERPINC1; ITIH2 |
| Molecular Function | |||
| Gene Set | Description | FDR | Gene ID |
| GO:0061134 | Peptidase regulator activity | 4.81 × 10−6 | FN1; SERPINF2; C3; A2M; GAPDH; SERPINC1; ITIH2 |
| GO:0098631 | Cell adhesion mediator activity | 6.74 × 10−3 | ANXA2; JUP; DSP |
| GO:0017171 | Serine hydrolase activity | 1.41 × 10−2 | F2; C3; CTSD; CFB |
| GO:0005539 | Glycosaminoglycan binding | 1.43 × 10−2 | FN1; F2; SAA1; SERPINC1 |
| Gene Symbol | Gene Full Name | Log2 Fold Change | q-Value |
|---|---|---|---|
| DSP | Desmoplakin | −0.856 | 3.2 × 10−5 |
| C4A | Complement C4-A | −0.157 | 1.0 × 10−3 |
| LTF | Lactotransferrin | −1.057 | 2.0 × 10−3 |
| ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | −3.358 | 3.0 × 10−3 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | −1.253 | 3.0 × 10−3 |
| CFH | Complement factor H | −0.224 | 1.3 × 10−2 |
| C5 | Complement C5 | −0.238 | 2.4 × 10−2 |
| SERPINA1 | Alpha-1-antitrypsin | −0.47 | 4.2 × 10−2 |
| FN1 | Fibronectin | −0.159 | 4.3 × 10−2 |
| DSG1 | Desmoglein-1 | −0.734 | 4.5 × 10−2 |
| DSP | Desmoplakin | −0.856 | 3.2 × 10−5 |
| Biological Process | |||
| Gene Set | Description | FDR | Gene ID |
| GO:0002526 | Acute inflammatory response | 1.98 × 10−7 | C4A; ITIH4; CFH; C5; SERPINA1; FN1 |
| GO:0052547 | Regulation of peptidase activity | 6.44 × 10−7 | C4A; LTF; ITIH4; GAPDH; C5; SERPINA1; FN1 |
| GO:0002446 | Neutrophil mediated immunity | 1.95 × 10−2 | DSP; LTF; SERPINA1; DSG1 |
| Molecular Function | |||
| Gene Set | Description | FDR | Gene ID |
| GO:0061134 | Peptidase regulator activity | 3.98 × 10−9 | C4A; LTF; ITIH4; GAPDH; C5; SERPINA1; FN1 |
| GO:0043394 | Proteoglycan binding | 1.96 × 10−2 | CFH; FN1 |
| Lab Code | Treatment | Sex | Age | T-Score Lumbar Spine | T-Score Femoral Neck | Fractures | Treatment in Previous 12 Months | BMI |
|---|---|---|---|---|---|---|---|---|
| P1 | Teriparatide | Female | 89 | −4.9 | −3.1 | Vertebral | None | 27.5 |
| P2 | Teriparatide | Female | 63 | −0.8 | −0.7 | Vertebral | None | 30.5 |
| P3 | Teriparatide | Female | 65 | −2.8 | −1.6 | Vertebral | None | 28.8 |
| P4 | Teriparatide | Female | 79 | −0.9 | −2.2 | Vertebral, peripheral | Denosumab | 27.4 |
| P5 | Teriparatide | Female | 81 | - | - | Vertebral | None | 33.3 |
| P6 | Teriparatide | Female | 84 | −2.6 | −2.3 | Vertebral, peripheral | None | 28.1 |
| P7 | Teriparatide | Female | 91 | −1.4 | −3.7 | Vertebral | None | 24.5 |
| P8 | Teriparatide | Female | 77 | −1.4 | −2.1 | Peripheral | None | 28.8 |
| P9 | Teriparatide | Female | 79 | −4.2 | −2.7 | Vertebral | None | 20.4 |
| P10 | Teriparatide | Female | 78 | −3.3 | −2.7 | Vertebral | None | 21.5 |
| D1 | Denosumab | Female | 80 | −1.4 | −0.8 | Vertebral | None | 21.9 |
| D2 | Denosumab | Female | 80 | - | - | Vertebral | None | 28.2 |
| D3 | Denosumab | Female | 72 | −2.7 | −2.5 | Vertebral | None | 22.7 |
| D4 | Denosumab | Female | 79 | −2.9 | −3.6 | Vertebral | None | 23.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Real, A.; Ciordia, S.; Sañudo, C.; Garcia-Ibarbia, C.; Roa-Bautista, A.; Ocejo-Viñals, J.G.; Corrales, F.; Riancho, J.A. Analysis of Serum Proteome after Treatment of Osteoporosis with Anabolic or Antiresorptive Drugs. Metabolites 2022, 12, 399. https://doi.org/10.3390/metabo12050399
del Real A, Ciordia S, Sañudo C, Garcia-Ibarbia C, Roa-Bautista A, Ocejo-Viñals JG, Corrales F, Riancho JA. Analysis of Serum Proteome after Treatment of Osteoporosis with Anabolic or Antiresorptive Drugs. Metabolites. 2022; 12(5):399. https://doi.org/10.3390/metabo12050399
Chicago/Turabian Styledel Real, Alvaro, Sergio Ciordia, Carolina Sañudo, Carmen Garcia-Ibarbia, Adriel Roa-Bautista, Javier G. Ocejo-Viñals, Fernando Corrales, and Jose A. Riancho. 2022. "Analysis of Serum Proteome after Treatment of Osteoporosis with Anabolic or Antiresorptive Drugs" Metabolites 12, no. 5: 399. https://doi.org/10.3390/metabo12050399
APA Styledel Real, A., Ciordia, S., Sañudo, C., Garcia-Ibarbia, C., Roa-Bautista, A., Ocejo-Viñals, J. G., Corrales, F., & Riancho, J. A. (2022). Analysis of Serum Proteome after Treatment of Osteoporosis with Anabolic or Antiresorptive Drugs. Metabolites, 12(5), 399. https://doi.org/10.3390/metabo12050399







