Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics of the Patients Enrolled in This Study
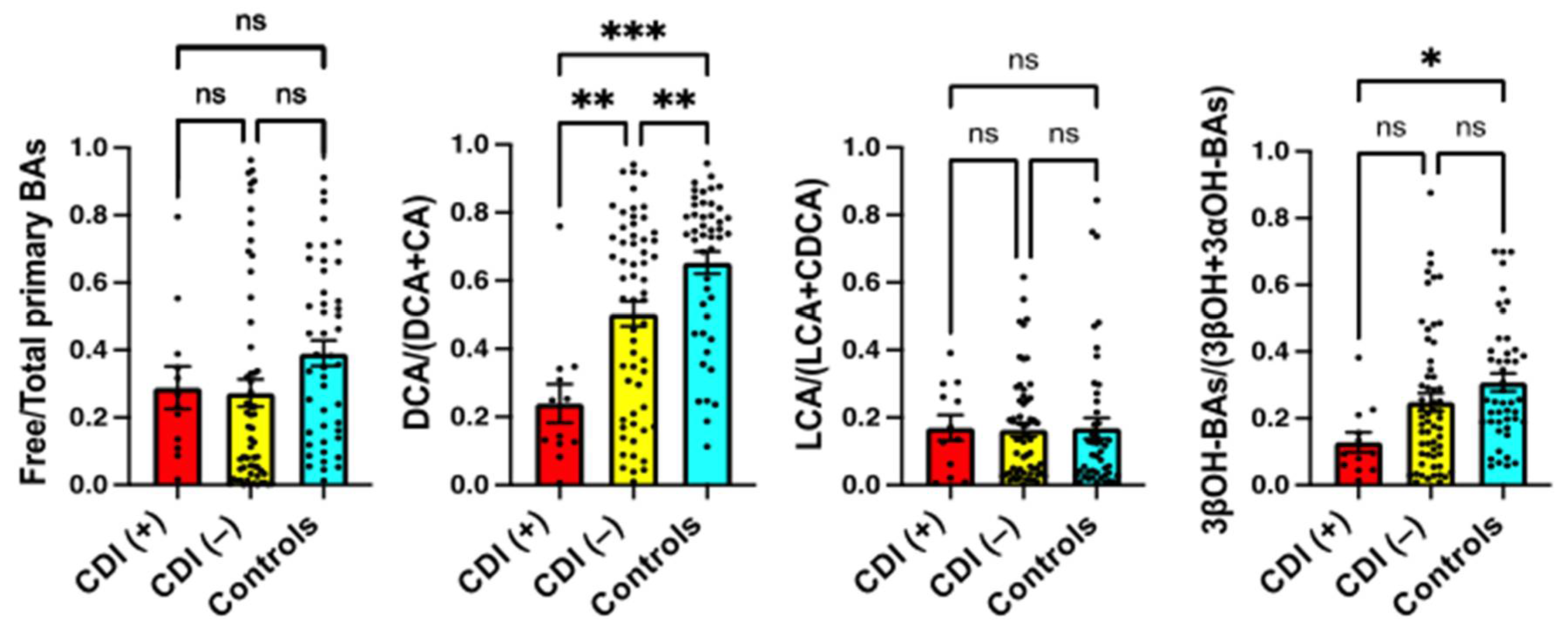
2.2. Serum BA Composition in CDI Patients
2.3. Serum BA Transformation Markers in CDI Patients
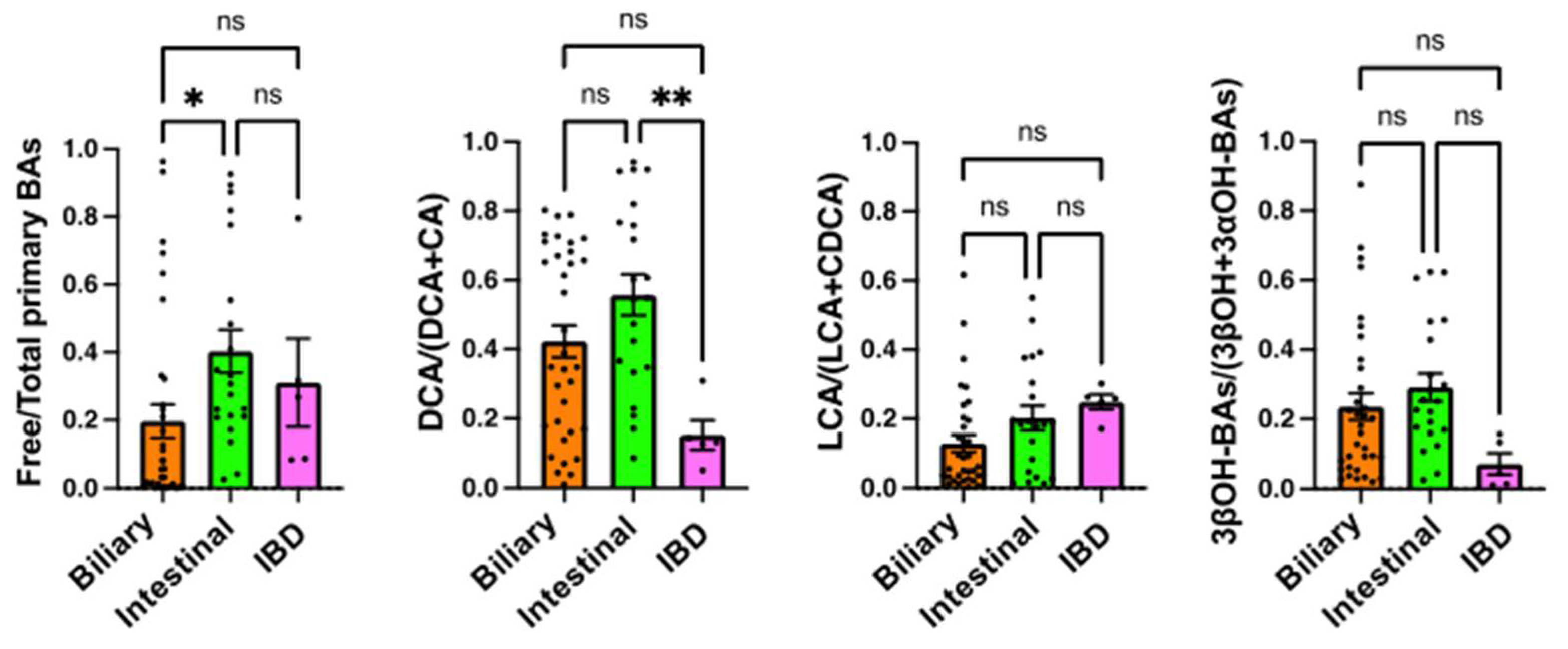
2.4. Comparison of Serum BA Markers among the Underlying Diseases
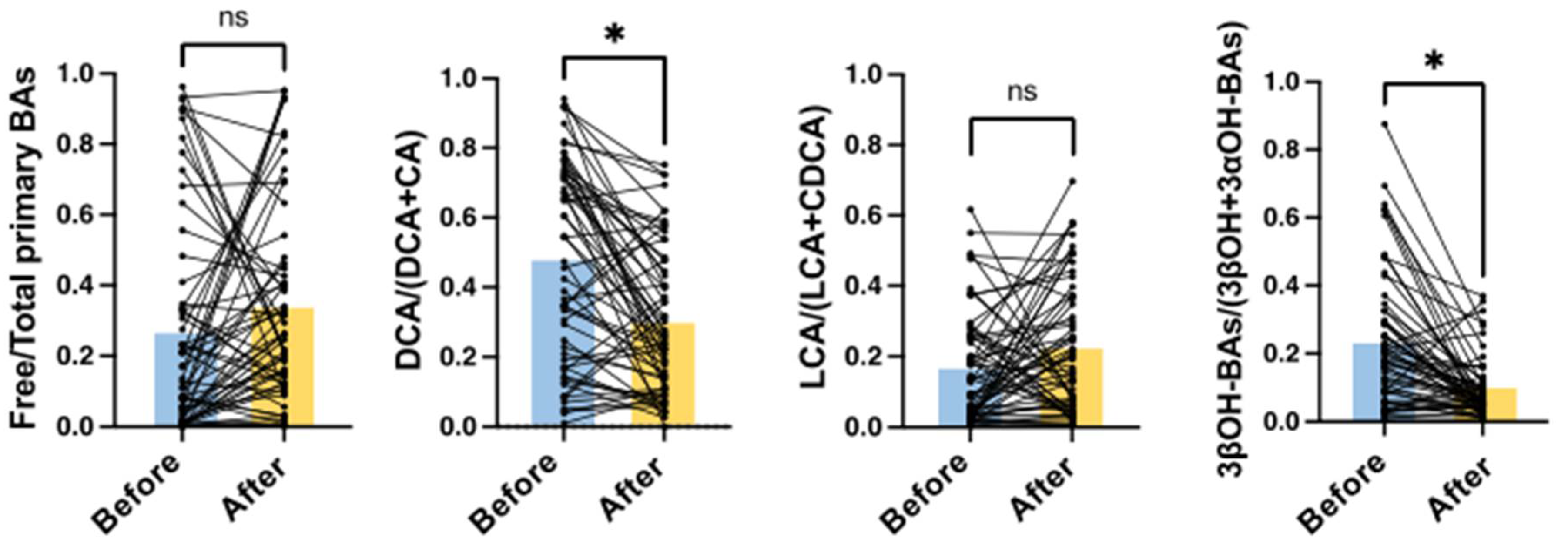
2.5. Effects of Treatment with Antibiotics on Serum BA Markers
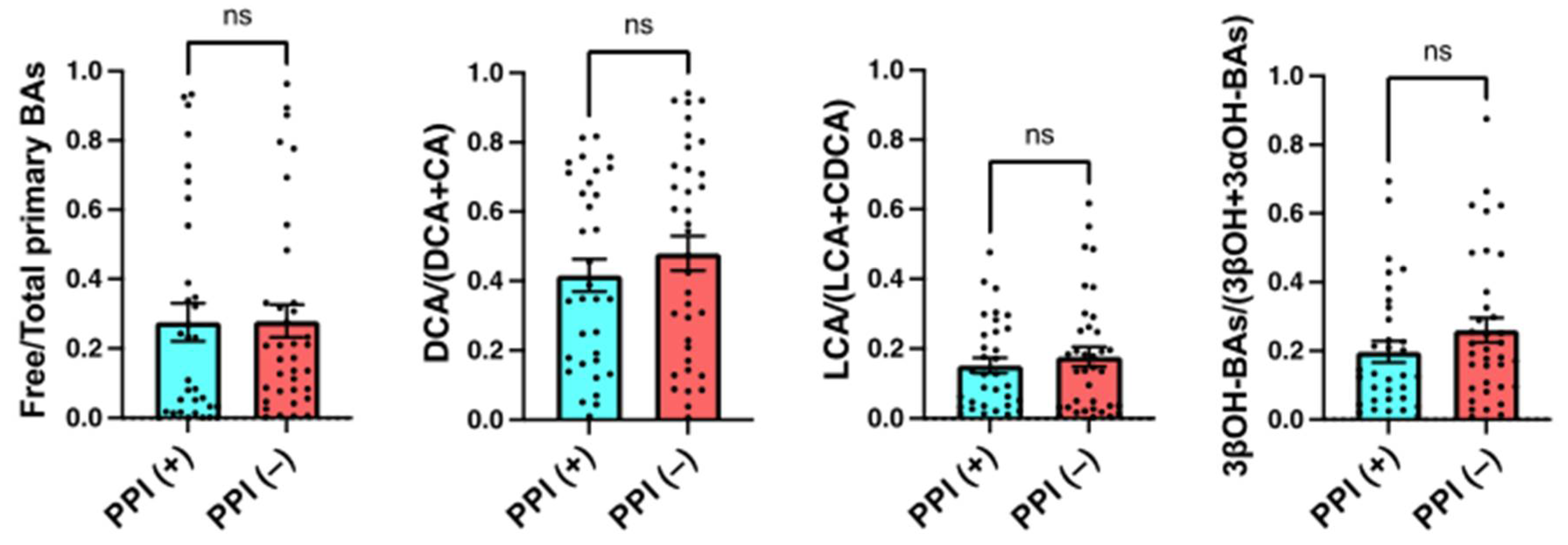
2.6. Effects of the Use of PPIs on Serum BA Markers
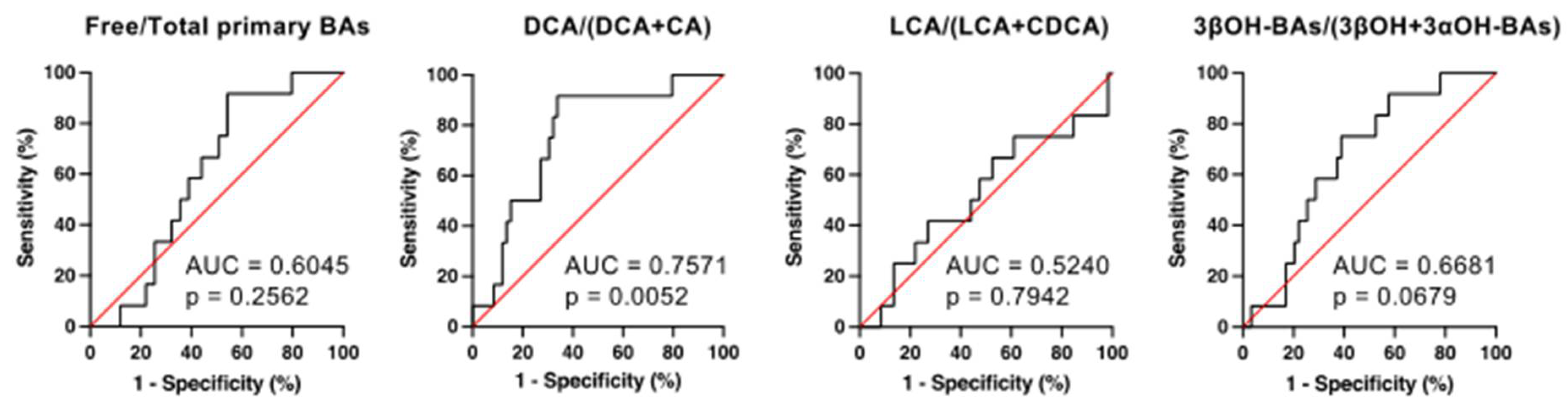
2.7. The Receiver Operating Characteristic (ROC) Analyses for the Prediction of CDI Development by Serum BA Markers
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Diagnosis of CDI
4.3. Serum BA Analyses
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bai | bile acid-inducible |
| BAs | bile acids |
| CA | cholic acid |
| CDCA | chenodeoxycholic acid |
| CDI | Clostridium difficile infection |
| CD toxins | Clostridium difficile-toxins |
| DCA | deoxycholic acid |
| GDH | glutamate dehydrogenase |
| GUDCA | glycoursodeoxycholic acid |
| IBD | inflammatory bowel diseases |
| LCA | lithocholic acid |
| 3αOH | 3α-hydroxy |
| 3βOH | 3β-hydroxy |
| PPIs | proton pump inhibitors |
| ROC | receiver operating characteristic |
| SEM | standard error of the mean |
| SIBO | small intestinal bacterial overgrowth |
References
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czepiel, J.; Drozdz, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultanska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksi, J.; Anttila, V.J.; Mattila, E. Treatment of Clostridioides (Clostridium) difficile infection. Ann. Med. 2020, 52, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorg, J.A.; Sonenshein, A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010, 192, 4983–4990. [Google Scholar] [CrossRef] [Green Version]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef] [Green Version]
- Winston, J.A.; Theriot, C.M. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016, 41, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Zhu, L.; Faden, H.S. Interactions of bile acids and the gut microbiota: Learning from the differences in Clostridium difficile infection between children and adults. Physiol. Genom. 2019, 51, 218–223. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Kearney, S.; Li, N.; Bogart, E.; Bullock, K.; Gerber, G.K.; Bry, L.; Clish, C.B.; Alm, E.; Korzenik, J.R. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016, 43, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Joyce, S.A.; Gahan, C.G. Disease-Associated Changes in Bile Acid Profiles and Links to Altered Gut Microbiota. Dig. Dis. 2017, 35, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studer, N.; Desharnais, L.; Beutler, M.; Brugiroux, S.; Terrazos, M.A.; Menin, L.; Schurch, C.M.; McCoy, K.D.; Kuehne, S.A.; Minton, N.P.; et al. Functional Intestinal Bile Acid 7alpha-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front. Cell. Infect. Microbiol. 2016, 6, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid. Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [Green Version]
- Funabashi, M.; Grove, T.L.; Wang, M.; Varma, Y.; McFadden, M.E.; Brown, L.C.; Guo, C.; Higginbottom, S.; Almo, S.C.; Fischbach, M.A. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 2020, 582, 566–570. [Google Scholar] [CrossRef]
- Solbach, P.; Chhatwal, P.; Woltemate, S.; Tacconelli, E.; Buhl, M.; Gerhard, M.; Thoeringer, C.K.; Vehreschild, M.; Jazmati, N.; Rupp, J.; et al. BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS ONE 2018, 13, e0196977. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile Acid 7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell. Chem. Biol. 2019, 26, 27.e4–34.e4. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Iwamoto, J.; Honda, A.; Tsuji, T.; Tamamushi, M.; Ueda, H.; Monma, T.; Konishi, N.; Yara, S.; Hirayama, T.; et al. Detection of Gut Dysbiosis due to Reduced Clostridium Subcluster XIVa Using the Fecal or Serum Bile Acid Profile. Inflamm. Bowel Dis. 2018, 24, 1035–1044. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [Green Version]
- Berkell, M.; Mysara, M.; Xavier, B.B.; van Werkhoven, C.H.; Monsieurs, P.; Lammens, C.; Ducher, A.; Vehreschild, M.; Goossens, H.; de Gunzburg, J.; et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 2021, 12, 2241. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vich Vila, A.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.; et al. Proton pump inhibitors affect the gut microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, E.G.; Milligan, J.; Frenette, C.; Lee, T.C. Continuous Proton Pump Inhibitor Therapy and the Associated Risk of Recurrent Clostridium difficile Infection. JAMA Intern. Med. 2015, 175, 784–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| CDI (+) (n = 12) | CDI (−) (n = 59) | p-Value | |
|---|---|---|---|
| Age (mean ± SEM) | 63.2 ± 26.5 | 69.0 ± 19.0 | NS |
| Gender (male:female) | 10:2 | 34:25 | NS |
| Underlying diseases | |||
| Hepato-biliary-pancreatic diseases a | 3 | 31 | NS |
| Gastrointestinal diseases b | 2 | 19 | NS |
| Inflammatory bowel disease c | 4 | 1 | p < 0.01 |
| Others d | 3 | 8 | NS |
| Medications | |||
| Antibiotics (on admission) | 0 | 0 | NS |
| Antibiotics (after admission) e | 7 | 59 | p < 0.001 |
| PPIs including vonoprazan (regular use) | 7 | 26 f | NS |
| CDI (+) (n = 12) | CDI (−) (n = 59) | Controls (n = 46) | |
|---|---|---|---|
| % | % | % | |
| Unconjugated BAs | 32.51 ± 7.05 † | 35.64 ± 4.10† | 55.97 ± 3.65 |
| CA | 11.93 ± 2.95 ** † | 4.36 ± 0.65 | 6.02 ± 0.85 |
| CDCA | 5.33 ± 1.08 | 6.64 ± 1.27 | 10.71 ± 1.27 |
| DCA | 5.51 ± 2.38 | 7.77 ± 1.30 † | 12.88 ± 1.52 |
| LCA | 1.30 ± 0.47 | 0.63 ± 0.09 † | 1.33 ± 0.24 |
| UDCA | 1.26 ± 0.71 | 3.57 ± 0.89 | 2.99 ± 0.36 |
| UCA | 0.23 ± 0.09 | 0.11 ± 0.02 | 0.19 ± 0.03 |
| 3-epi-CA | 0.30 ± 0.09 | 0.26 ± 0.06 | 0.20 ± 0.04 |
| 12-epi-CA | 0.11 ± 0.05 | 0.03 ± 0.01 † | 0.11 ± 0.02 |
| 3-epi-DCA&CDCA | 1.58 ± 0.80 † | 1.76 ± 0.29 †† | 4.12 ± 0.39 |
| 3-epi-UDCA | 1.814 ± 0.85 † | 7.27 ± 1.33 | 12.10 ± 1.93 |
| 3-epi-LCA | 0.35 ± 0.11 | 0.15 ± 0.03 †† | 0.60 ± 0.09 |
| 3-dehydro-CA | 0.32 ± 0.13 * † | 0.10 ± 0.02 | 0.15 ± 0.03 |
| 7-oxo-DCA | 0.35 ± 0.13 | 0.34 ± 0.11 | 0.23 ± 0.08 |
| 12-oxo-CDCA | 0.23 ± 0.11 | 0.14 ± 0.03 | 0.14 ± 0.03 |
| 3-dehydro-CDCA | 0.34 ± 0.19 † | 0.22 ± 0.05 †† | 0.97 ± 0.14 |
| 3-dehydro-DCA | 0.63 ± 0.22 | 0.72 ± 0.17 | 0.65 ± 0.11 |
| 3-dehydro-UDCA | 0.22 ± 0.09 | 0.41 ± 0.10 | 0.37 ± 0.06 |
| 7-oxo-LCA | 0.22 ± 0.09 | 0.25 ± 0.05 † | 0.63 ± 0.10 |
| 12-oxo-LCA | 0.29 ± 0.12 | 0.78 ± 0.25 | 0.98 ± 0.18 |
| 3-dehydro-LCA | 0.18 ± 0.09 † | 0.15 ± 0.04 †† | 0.61 ± 0.08 |
| Glycine conjugated BAs | 59.83 ± 8.08 † | 51.53 ± 3.19 † | 40.45 ± 3.07 |
| GCA | 14.84 ± 4.59 | 13.91 ± 2.23 † | 7.02 ± 1.44 |
| GCDCA | 32.17 ± 5.97 * † | 21.69 ± 1.64 | 20.03 ± 1.59 |
| GDCA | 3.81 ± 1.46 | 6.90 ± 1.06 | 8.21 ± 1.08 |
| GLCA | 0.02 ± 0.01 | 0.21 ± 0.07 | 0.10 ± 0.03 |
| GUDCA | 8.99 ± 6.53 | 8.83 ± 1.51 | 5.10 ± 0.74 |
| Taurine conjugated BAs | 7.66 ± 2.79 | 12.83 ± 2.03 †† | 3.58 ± 0.88 |
| TCA | 0.99 ± 0.32 | 2.58 ± 0.61 | 1.09 ± 0.36 |
| TCDCA | 4.06 ± 1.28 | 8.43 ± 1.44 †† | 1.90 ± 0.47 |
| TDCA | 2.30 ± 1.85 | 1.56 ± 0.32 | 0.53 ± 0.15 |
| TLCA | 0.05 ± 0.02 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| TUDCA | 0.26 ± 0.14 | 0.23 ± 0.08 | 0.03 ± 0.01 |
| Total BAs | 100 | 100 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monma, T.; Iwamoto, J.; Honda, A.; Ueda, H.; Kakizaki, F.; Yara, S.; Miyazaki, T.; Ikegami, T. Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile. Metabolites 2022, 12, 331. https://doi.org/10.3390/metabo12040331
Monma T, Iwamoto J, Honda A, Ueda H, Kakizaki F, Yara S, Miyazaki T, Ikegami T. Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile. Metabolites. 2022; 12(4):331. https://doi.org/10.3390/metabo12040331
Chicago/Turabian StyleMonma, Tadakuni, Junichi Iwamoto, Akira Honda, Hajime Ueda, Fumio Kakizaki, Shoichiro Yara, Teruo Miyazaki, and Tadashi Ikegami. 2022. "Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile" Metabolites 12, no. 4: 331. https://doi.org/10.3390/metabo12040331
APA StyleMonma, T., Iwamoto, J., Honda, A., Ueda, H., Kakizaki, F., Yara, S., Miyazaki, T., & Ikegami, T. (2022). Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile. Metabolites, 12(4), 331. https://doi.org/10.3390/metabo12040331







