Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients
Abstract
:1. Introduction
2. Results
2.1. LRRK2 PD Patient Sample Characteristics
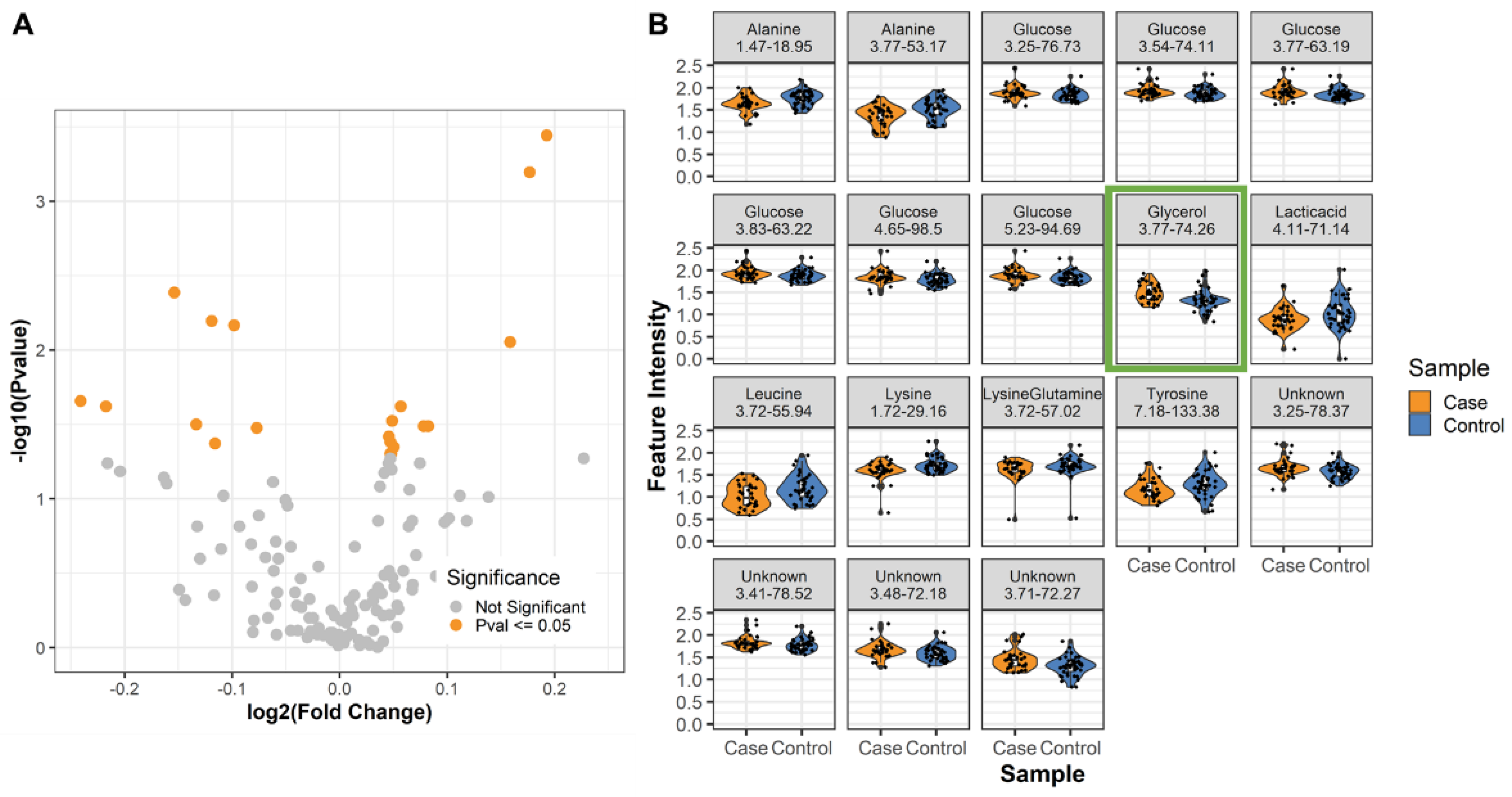
2.2. Differential Metabolites in Male LRRK2 PD
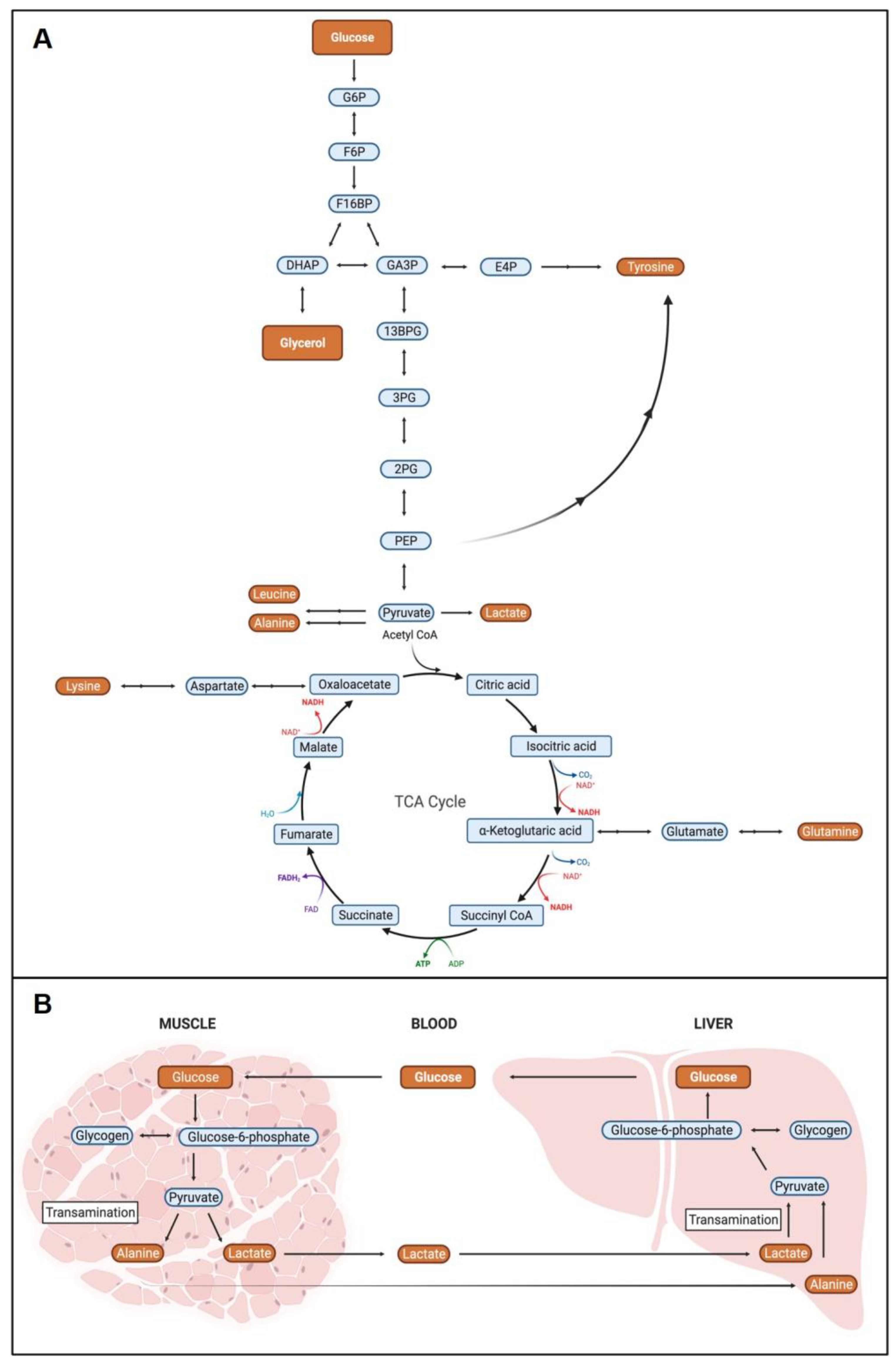
2.3. Metabolite Pathway Analysis
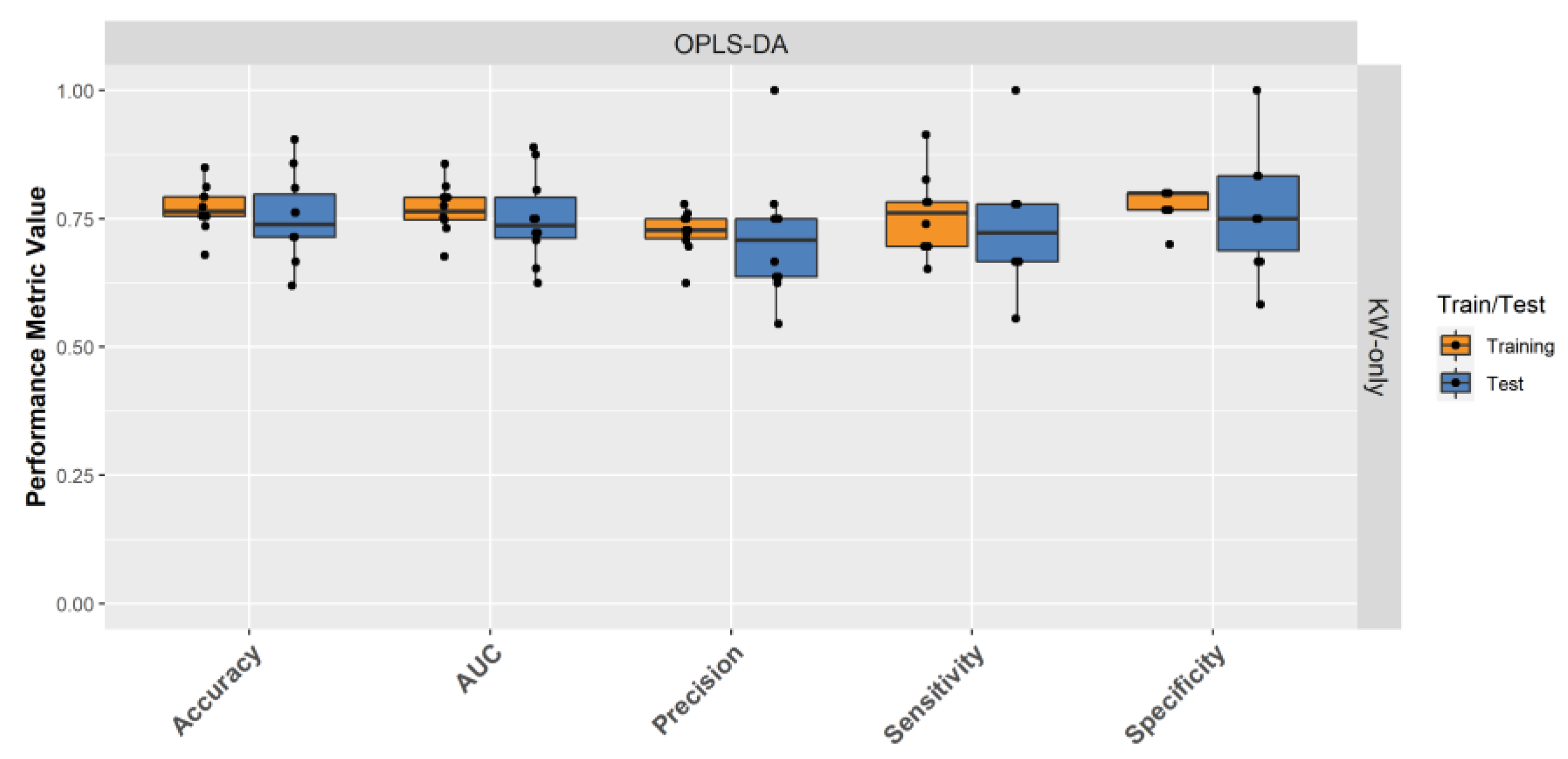
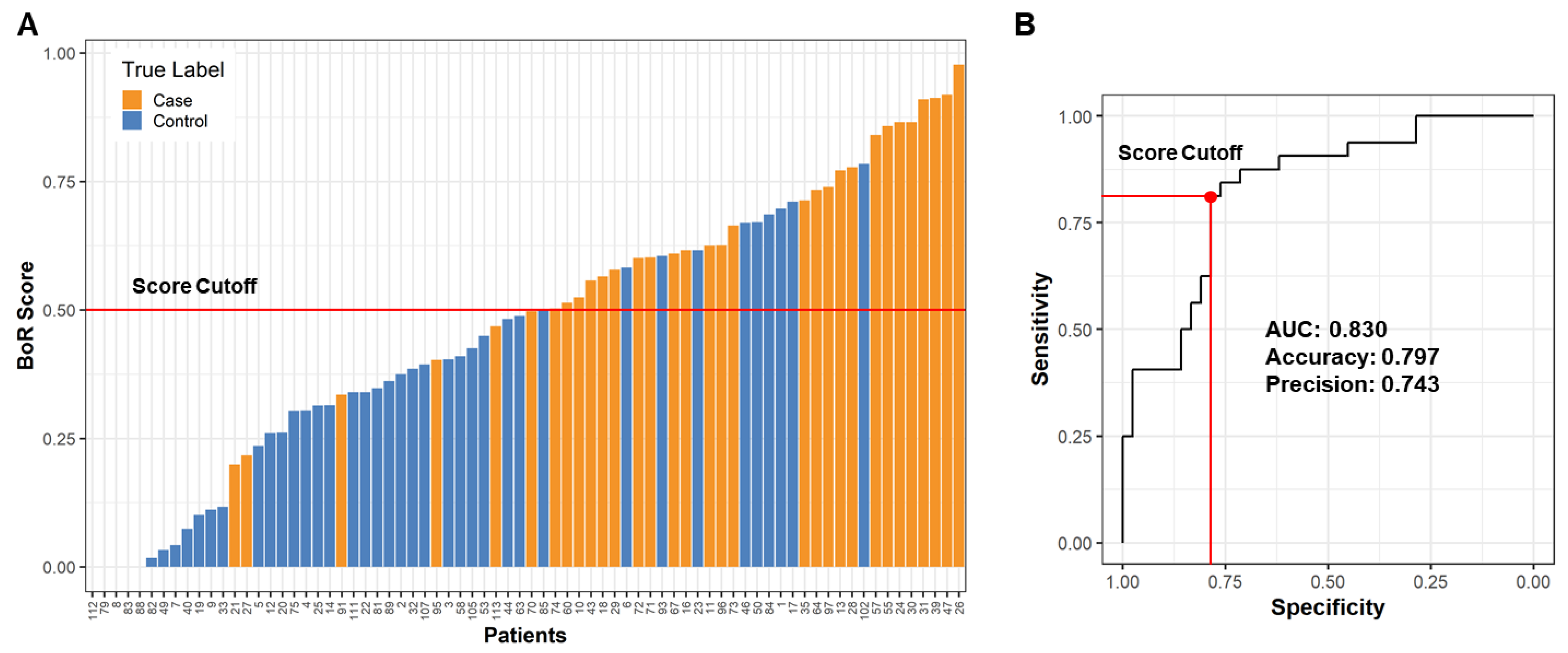
2.4. Building a Male LRRK2 PD Classifier
3. Discussion
4. Materials and Methods
4.1. Patient Enrollment and Plasma Collection
4.2. NMR Sample Preparation
4.3. NMR Data Collection and Processing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anandhan, A.; Jacome, M.S.; Lei, S.; Hernandez-Franco, P.; Pappa, A.; Panayiotidis, M.I.; Powers, R.; Franco, R. Metabolic Dysfunction in Parkinson’s Disease: Bioenergetics, Redox Homeostasis and Central Carbon Metabolism. Brain Res. Bull. 2017, 133, 12–30. [Google Scholar] [CrossRef]
- Hague, S.M.; Klaffke, S.; Bandmann, O. Neurodegenerative Disorders: Parkinson’s Disease and Huntington’s Disease. J. Neurol. Neurosurg. Psychiatry 2005, 76, 1058–1063. [Google Scholar] [CrossRef] [PubMed]
- Havelund, J.F.; Heegaard, N.H.H.; Færgeman, N.J.K.; Gramsbergen, J.B. Biomarker Research in Parkinson’s Disease Using Metabolite Profiling. Metabolites 2017, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zeng, F.; Jin, W.S.; Zhu, C.; Wang, Q.H.; Bu, X.L.; Luo, H.B.; Zou, H.Q.; Pu, J.; Zhou, Z.H.; et al. Comorbidity Burden of Patients with Parkinson’s Disease and Parkinsonism between 2003 and 2012: A Multicentre, Nationwide, Retrospective Study in China. Sci. Rep. 2017, 7, 1671. [Google Scholar] [CrossRef] [Green Version]
- Isaacson, J.R.; Brillman, S.; Chhabria, N.; Isaacson, S.H. Impact of DaTscan Imaging on Clinical Decision Making in Clinically Uncertain Parkinson’s Disease. J. Parkinsons Dis. 2021, 11, 885–889. [Google Scholar] [CrossRef]
- Lin, M.K.; Farrer, M.J. Genetics and Genomics of Parkinson’s Disease. Genome Med. 2014, 6, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.J.; Ben-Shlomo, Y.; Daniel, S.E.; Lees, A.J. What Features Improve the Accuracy of Clinical Diagnosis in Parkinson’s Disease: A Clinicopathologic Study. Neurology 1992, 42, 1142. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA—J. Am. Med. Assoc. 2020, 323, 548–560. [Google Scholar] [CrossRef]
- Selikhova, M.; Williams, D.R.; Kempster, P.A.; Holton, J.L.; Revesz, T.; Lees, A.J. A Clinico-Pathological Study of Subtypes in Parkinson’s Disease. Brain 2009, 132, 2947–2957. [Google Scholar] [CrossRef] [Green Version]
- Thenganatt, M.A.; Jankovic, J. Parkinson Disease Subtypes. JAMA Neurol. 2014, 71, 499–504. [Google Scholar] [CrossRef]
- Johansen, K.K.; Wang, L.; Aasly, J.O.; White, L.O.R.; Matson, W.R.; Henchcliffe, C.; Beal, M.F.; Bogdanov, M. Metabolomic Profiling in LRRK2-Related Parkinson’s Disease. PLoS ONE 2009, 4, e7551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, N.; Attal, F.; Amirat, Y.; Oukhellou, L.; Mohammed, S. Data-Driven Based Approach to Aid Parkinson’s Disease Diagnosis. Sensors 2019, 19, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolosa, E.; vander Borght, T.; Moreno, E. Accuracy of DaTSCAN (123I-Ioflupane) SPECT in Diagnosis of Patients with Clinically Uncertain Parkinsonism: 2-Year Follow-up of an Open-Label Study. Mov. Disord. 2007, 22, 2346–2351. [Google Scholar] [CrossRef] [PubMed]
- Salašová, A.; Yokota, C.; Potěšil, D.; Zdráhal, Z.; Bryja, V.; Arenas, E. A Proteomic Analysis of LRRK2 Binding Partners Reveals Interactions with Multiple Signaling Components of the WNT/PCP Pathway. Mol. Neurodegener. 2017, 12, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The Genetic Architecture of Parkinson’s Disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef]
- Price, A.; Manzoni, C.; Cookson, M.R.; Lewis, P.A. The LRRK2 Signalling System. Cell Tissue Res. 2018, 373, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.L.; Jain, S.; Lynch, J.M.; Pavese, N.; Abou-Sleiman, P.; Holton, J.L.; Healy, D.G.; Gilks, W.P.; Sweeney, M.G.; Ganguly, M.; et al. Mutations in the Gene LRRK2 Encoding Dardarin (PARK8) Cause Familial Parkinson’s Disease: Clinical, Pathological, Olfactory and Functional Imaging and Genetic Data. Brain 2005, 128, 2786–2796. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Brice, A. Parkinson’s Disease: From Monogenic Forms to Genetic Susceptibility Factors. Hum. Mol. Genet. 2009, 18, R48–R59. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.R.; van Netten, H.; Schulzer, M.; Mak, E.; Mckenzie, J.; Strongosky, A.; Sossi, V.; Ruth, T.J.; Lee, C.S.; Farrer, M.; et al. PET in LRRK2 Mutations: Comparison to Sporadic Parkinson’s Disease and Evidence for Presymptomatic Compensation. Brain 2005, 128, 2777–2785. [Google Scholar] [CrossRef] [Green Version]
- Kay, D.M.; Zabetian, C.P.; Factor, S.A.; Nutt, J.G.; Samii, A.; Griffith, A.; Bird, T.D.; Kramer, P.; Higgins, D.S.; Payami, H. Parkinson’s Disease and LRRK2: Frequency of a Common Mutation in U.S. Movement Disorder Clinics. Mov. Disord. 2006, 21, 519–523. [Google Scholar] [CrossRef]
- Erb, M.L.; Moore, D.J. LRRK2 and the Endolysosomal System in Parkinson’s Disease. J. Parkinsons Dis. 2020, 10, 1271–1291. [Google Scholar] [CrossRef]
- Seegobin, S.P.; Heaton, G.R.; Liang, D.; Choi, I.; Blanca Ramirez, M.; Tang, B.; Yue, Z. Progress in LRRK2-Associated Parkinson’s Disease Animal Models. Front. Neurosci. 2020, 14, 674. [Google Scholar] [CrossRef]
- Di Maio, R.; Hoffman, E.K.; Rocha, E.M.; Keeney, M.T.; Sanders, L.H.; de Miranda, B.R.; Zharikov, A.; van Laar, A.; Stepan, A.F.; Lanz, T.A.; et al. LRRK2 Activation in Idiopathic Parkinson’s Disease. Sci. Transl. Med. 2018, 10, eaar5429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakshi, R.; Macklin, E.A.; Logan, R.; Zorlu, M.M.; Xia, N.; Crotty, G.F.; Zhang, E.; Chen, X.; Ascherio, A.; Schwarzschild, M.A. Higher Urate in LRRK2 Mutation Carriers Resistant to Parkinson Disease. Ann. Neurol. 2019, 85, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, L.; O’Day, E.M. Perspective: A Potential Role for NUS in Metabolite-Based in Vitro Diagnostics. Magn. Reson. Chem. 2021, 59, 257–263. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The Apogee of the Omics Trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Le, W. Recent Advances and Perspectives of Metabolomics-Based Investigations in Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 3. [Google Scholar] [CrossRef] [Green Version]
- Okuzumi, A.; Hatano, T.; Ueno, S.I.; Ogawa, T.; Saiki, S.; Mori, A.; Koinuma, T.; Oji, Y.; Ishikawa, K.I.; Fujimaki, M.; et al. Metabolomics-Based Identification of Metabolic Alterations in PARK2. Ann. Clin. Transl. Neurol. 2019, 6, 525–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, K.B.; Moehle, M.S.; Alcalay, R.N.; West, A.B. Urinary LRRK2 Phosphorylation Predicts Parkinsonian Phenotypes in G2019S LRRK2 Carriers. Neurology 2016, 86, 994–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cipriani, S.; Chen, X.; Schwarzschild, M.A. Urate: A Novel Biomarker of Parkinsons Disease Risk, Diagnosis and Prognosis. Biomark. Med. 2010, 4, 701–712. [Google Scholar] [CrossRef] [Green Version]
- The Parkinson Study Group SURE-PD3 Investigators. Effect of Urate-Elevating Inosine on Early Parkinson Disease Progression: The SURE-PD3 Randomized Clinical Trial. JAMA 2021, 326, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Warner, J.; Pinto, C.; Juric, D.; ODay, E. NMR-Metabolite-Resonance Signature to Predict HR+ Breast Cancer Patient Response to CDK4/6 Inhibitors. J. Clin. Oncol. 2019, 37, 3043. [Google Scholar] [CrossRef]
- Honrao, C.; Rao, S.R.; Teissier, N.; Call, S.G.; ODay, E.M.; Janku, F. Abstract LB031: Plasma Based Metabolic Profiling in Metastatic Gastrointestinal Stromal Tumors (GIST). In Proceedings of the AACR Annual Meeting 2021, Philadelphia, PA, USA, 17–21 May 2021. [Google Scholar]
- O’Day, E.; Leitzel, K.; Ali, S.M.; Zhang, B.; Dong, C.; Gu, H.; Shi, X.; Drabick, J.J.; Cream, L.; Vasekar, M.; et al. Abstract P4-10-25: Pretreatment serum metabolome predicts PFS in first-line trastuzumab-treated metastatic breast cancer. In SABCS, Proceedings of the 2019 San Antonio Breast Cancer Symposium, San Antonio, TX, USA, 10–14 December 2019; AACR Publications: Philadelphia, PA, USA, 2020. [Google Scholar]
- Seol, W.; Kim, H.; Son, I. Urinary Biomarkers for Neurodegenerative Diseases. Exp. Neurobiol. 2020, 29, 325–333. [Google Scholar] [CrossRef]
- Tzoulaki, I.; Ebbels, T.M.D.; Valdes, A.; Elliott, P.; Ioannidis, J.P.A. Design and Analysis of Metabolomics Studies in Epidemiologic Research: A Primer on -Omic Technologies. Am. J. Epidemiol. 2014, 180, 129–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Gonzalez, F.J. Challenges and Opportunities of Metabolomics. J. Cell. Physiol. 2012, 227, 2975–2981. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chong, N.; Lewis, N.E.; Jia, W.; Xie, G.; Garmire, L.X. Novel Personalized Pathway-Based Metabolomics Models Reveal Key Metabolic Pathways for Breast Cancer Diagnosis. Genome Med. 2016, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, L.; Gibbons, H. Sex Matters: A Focus on the Impact of Biological Sex on Metabolomic Profiles and Dietary Interventions. Proc. Nutr. Soc. 2020, 79, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Golbs, A.; Neuber, S.; Kamlage, B.; Christiansen, N.; Bethan, B.; Rennefahrt, U.; Schatz, P.; Lind, L. Effects of Long-Term Storage at −80 °C on the Human Plasma Metabolome. Metabolites 2019, 9, 99. [Google Scholar] [CrossRef] [Green Version]
- Bingol, K.; Li, D.W.; Zhang, B.; Brüschweiler, R. Comprehensive Metabolite Identification Strategy Using Multiple Two-Dimensional NMR Spectra of a Complex Mixture Implemented in the COLMARm Web Server. Anal. Chem. 2016, 88, 12411–12418. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.S.; Santosh, W.; Kumar, S.; Christlet, H.T.T. Metabolic Profiling of Parkinson’s Disease: Evidence of Biomarker from Gene Expression Analysis and Rapid Neural Network Detection. J. Biomed. Sci. 2009, 16, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argilés, J.M.; Campos, N.; Lopez-Pedrosa, J.M.; Rueda, R.; Rodriguez-Mañas, L. Skeletal Muscle Regulates Metabolism via Interorgan Crosstalk: Roles in Health and Disease. J. Am. Med. Dir. Assoc. 2016, 17, 789–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta—A System for Feature Selection. Fundam. Inform. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Rajput, C.; Sarkar, A.; Sachan, N.; Rawat, N.; Singh, M.P. Is Gut Dysbiosis an Epicenter of Parkinson’s Disease? Neurochem. Res. 2021, 46, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Landi, G.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Urbani, A.; Bossola, M.; et al. Circulating Amino Acid Signature in Older People with Parkinson’s Disease: A Metabolic Complement to the EXosomes in PArkiNson Disease (EXPAND) Study. Exp. Gerontol. 2019, 128, 110766. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, S.P.; el Aidy, S. Bacterial Metabolites Mirror Altered Gut Microbiota Composition in Patients with Parkinson’s Disease. J. Parkinsons Dis. 2019, 9, S359–S370. [Google Scholar] [CrossRef] [Green Version]
- Peppard, R.F.; Martin, W.R.W.; Schulzer, M.; Guttman, M.; Tsui, J.K.C.; Carr, G.D.; Phillips, A.G.; Mcgeer, P.L.; Grochowski, E.; Calne, D.B. Cerebral Glucose Metabolism in Parkinson’s Disease With and Without Dementia. Arch. Neurol. 1992, 49, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Li, T.; Liu, Z.; Wang, X.; Xu, X.; Li, S.; Xu, G.; Le, W. Comprehensive Metabolic Profiling of Parkinson’s Disease by Liquid Chromatography-Mass Spectrometry. Mol. Neurodegener. 2021, 16, 4. [Google Scholar] [CrossRef]
- Wuolikainen, A.; Jonsson, P.; Ahnlund, M.; Antti, H.; Marklund, S.L.; Moritz, T.; Forsgren, L.; Andersen, P.M.; Trupp, M. Multi-Platform Mass Spectrometry Analysis of the CSF and Plasma Metabolomes of Rigorously Matched Amyotrophic Lateral Sclerosis, Parkinson’s Disease and Control Subjects. Mol. BioSyst. 2016, 12, 1287–1298. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, C.; Zhao, N.; Li, W.; Yang, Z.; Liu, X.; Le, W.; Zhang, X. Potential Biomarkers of Parkinson’s Disease Revealed by Plasma Metabolic Profiling. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1081–1082, 101–108. [Google Scholar] [CrossRef]
- Öhman, A.; Forsgren, L. NMR Metabonomics of Cerebrospinal Fluid Distinguishes between Parkinson’s Disease and Controls. Neurosci. Lett. 2015, 594, 36–39. [Google Scholar] [CrossRef]
- Mally, J.; Szalai, G.; Stone, T.W. Changes in the Concentration of Amino Acids in Serum and Cerebrospinal Fluid of Patients with Parkinson’s Disease. J. Neurol. Sci. 1997, 151, 159–162. [Google Scholar] [CrossRef]
- Molina, J.A.; Jiménez-Jiménez, F.J.; Gómez, P.; Vargas, C.; Navarro, J.A.; Ortí-Pareja, M.; Gasalla, T.; Benito-León, J.; Bermejo, F.; Arenas, J. Decreased Cerebrospinal Fluid Levels of Neutral and Basic Amino Acids in Patients with Parkinson’s Disease. J. Neurol. Sci. 1997, 150, 123–127. [Google Scholar] [CrossRef]
- Wu, J.; Wuolikainen, A.; Trupp, M.; Jonsson, P.; Marklund, S.L.; Andersen, P.M.; Forsgren, L.; Öhman, A. NMR Analysis of the CSF and Plasma Metabolome of Rigorously Matched Amyotrophic Lateral Sclerosis, Parkinson’s Disease and Control Subjects. Metabolomics 2016, 12, 101. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut Microbiota and Metabolome Alterations Associated with Parkinson’s Disease. mSystems 2020, 5, e00561-20. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Liu, L.F.; Tang, Z.; Zhang, M.; Chua, K.K.; Song, J.X.; Mok, V.C.T.; Li, M.; Cai, Z. Comprehensive Urinary Metabolomic Profiling and Identification of Potential Noninvasive Marker for Idiopathic Parkinson’s Disease. Sci. Rep. 2015, 5, 13888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, S.; Goyal, V.; Kumaran, S.S.; Dwivedi, S.N.; Srivastava, A.; Jagannathan, N.R. Quantitative Metabolomics of Saliva Using Proton NMR Spectroscopy in Patients with Parkinson’s Disease and Healthy Controls. Neurol. Sci. 2020, 41, 1201–1210. [Google Scholar] [CrossRef]
- Kumari, S.; Kumaran, S.S.; Goyal, V.; Sharma, R.K.; Sinha, N.; Dwivedi, S.N.; Srivastava, A.K.; Jagannathan, N.R. Identification of Potential Urine Biomarkers in Idiopathic Parkinson’s Disease Using NMR. Clin. Chim. Acta 2020, 510, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Segura-Aguilar, J.; Paris, I.; Muñoz, P.; Ferrari, E.; Zecca, L.; Zucca, F.A. Protective and Toxic Roles of Dopamine in Parkinson’s Disease. J. Neurochem. 2014, 129, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Zhao, X.; Yang, S.; Cui, H.; Wang, G. Metabolomic Signature between Metabolically Healthy Overweight/Obese and Metabolically Unhealthy Overweight/Obese: A Systematic Review. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.; Lei, S.; Anandhan, A.; Marshall, D.D.; Worley, B.; Cerny, R.L.; Dodds, E.D.; Huang, Y.; Panayiotidis, M.I.; Pappa, A.; et al. Metabolic Investigations of the Molecular Mechanisms Associated with Parkinson’s Disease. Metabolites 2017, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Heo, H.Y.; Park, J.M.; Kim, C.H.; Han, B.S.; Kim, K.B.S.; Seol, W. LRRK2 Enhances Oxidative Stress-Induced Neurotoxicity via Its Kinase Activity. Exp. Cell Res. 2010, 316, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Hur, E.M.; Lee, B.D. Lrrk2 at the Crossroad of Aging and Parkinson’s Disease. Genes 2021, 12, 505. [Google Scholar] [CrossRef] [PubMed]
- Redenšek, S.; Dolžan, V.; Kunej, T. From Genomics to Omics Landscapes of Parkinson’s Disease: Revealing the Molecular Mechanisms. OMICS J. Integr. Biol. 2018, 22, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, R.B.; Perotte, A.J.; Zhou, B.; Liong, C.; Shorr, E.J.; Marder, K.B.S.; Kang, U.J.; Waters, C.H.; Levy, O.A.; Xu, Y.; et al. Elevated GM3 Plasma Concentration in Idiopathic Parkinson’s Disease: A Lipidomic Analysis. PLoS ONE 2017, 12, e0172348. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Levy, O.A.; Waters, C.C.; Fahn, S.; Ford, B.; Kuo, S.H.; Mazzoni, P.; Pauciulo, M.W.; Nichols, W.C.; Gan-Or, Z.; et al. Glucocerebrosidase Activity in Parkinson’s Disease with and without GBA Mutations. Brain 2015, 138, 2648–2658. [Google Scholar] [CrossRef] [Green Version]
- Hyberts, S.G.; Milbradt, A.G.; Wagner, A.B.; Arthanari, H.; Wagner, G. Application of Iterative Soft Thresholding for Fast Reconstruction of NMR Data Non-Uniformly Sampled with Multidimensional Poisson Gap Scheduling. J. Biomol. NMR 2012, 52, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; The R Core Team; et al. Caret: Classification and Regression Training. Available online: https://cran.r-project.org/package=caret (accessed on 13 December 2021).
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2/3, 18–22. [Google Scholar]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. Available online: https://ggplot2.tidyversE.M.O.rg (accessed on 13 December 2021).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Darweesh, S.K.L.; Verlinden, V.J.A.; Stricker, B.H.; Hofman, A.; Koudstaal, P.J.; Ikram, M.A. Trajectories of Prediagnostic Functioning in Parkinson’s Disease. Brain 2017, 140, 429–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Case | Control | ||
|---|---|---|---|
| Number of samples | 32 | 42 | |
| Age at sample collection in years (SD) | 70.0 (8.1) | 69.5 (7.2) | |
| Age at onset in years (SD) | 58.9 (10.1) | - | |
| Sample collection time after PD Dx in years (SD) | 10.9 (8.3) | - | |
| MoCA (SD) | 25.4 (4.8) | 26.5 (2.5) | |
| BMI (SD) | 26.7 (2.6) | 28.3 (4.8) | |
| Ethnicity | White | 32 | 39 |
| Hispanic | 0 | 1 | |
| Black | 0 | 2 | |
| Annotation | 1H ppm | 13C ppm | Fold Change (Case/Control) | p-Value | FDR p-Value |
|---|---|---|---|---|---|
| Glycerol | 3.77 | 74.26 | 1.130 | 0.001 | 0.042 |
| Alanine | 3.77 | 53.17 | 0.899 | 0.004 | 0.178 |
| Lysine | 1.72 | 29.16 | 0.921 | 0.006 | 0.178 |
| Alanine | 1.47 | 18.95 | 0.934 | 0.007 | 0.178 |
| Unknown | 3.71 | 72.27 | 1.116 | 0.009 | 0.193 |
| Lactic acid | 4.11 | 71.14 | 0.846 | 0.022 | 0.302 |
| Leucine | 3.72 | 55.94 | 0.860 | 0.024 | 0.302 |
| Unknown | 3.41 | 78.52 | 1.040 | 0.024 | 0.302 |
| Glucose | 3.77 | 63.19 | 1.034 | 0.030 | 0.302 |
| Tyrosine | 7.18 | 133.38 | 0.912 | 0.032 | 0.302 |
| Unknown | 3.48 | 72.18 | 1.056 | 0.032 | 0.302 |
| Unknown | 3.25 | 78.37 | 1.059 | 0.032 | 0.302 |
| Lysine/Glutamine | 3.72 | 57.02 | 0.948 | 0.033 | 0.302 |
| Glucose | 3.54 | 74.11 | 1.032 | 0.038 | 0.302 |
| Glucose | 4.65 | 98.5 | 1.033 | 0.041 | 0.302 |
| Glucose | 3.25 | 76.73 | 1.035 | 0.045 | 0.302 |
| Glucose | 3.83 | 63.22 | 1.033 | 0.050 | 0.302 |
| Glucose | 5.23 | 94.69 | 1.034 | 0.050 | 0.302 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, C.; Honrao, C.; Rodrigues, L.O.; Wolf, J.; Sheehan, K.B.; Surface, M.; Alcalay, R.N.; O’Day, E.M. Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients. Metabolites 2022, 12, 149. https://doi.org/10.3390/metabo12020149
Dong C, Honrao C, Rodrigues LO, Wolf J, Sheehan KB, Surface M, Alcalay RN, O’Day EM. Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients. Metabolites. 2022; 12(2):149. https://doi.org/10.3390/metabo12020149
Chicago/Turabian StyleDong, Chen, Chandrashekhar Honrao, Leonardo O. Rodrigues, Josephine Wolf, Keri B. Sheehan, Matthew Surface, Roy N. Alcalay, and Elizabeth M. O’Day. 2022. "Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients" Metabolites 12, no. 2: 149. https://doi.org/10.3390/metabo12020149
APA StyleDong, C., Honrao, C., Rodrigues, L. O., Wolf, J., Sheehan, K. B., Surface, M., Alcalay, R. N., & O’Day, E. M. (2022). Plasma Metabolite Signature Classifies Male LRRK2 Parkinson’s Disease Patients. Metabolites, 12(2), 149. https://doi.org/10.3390/metabo12020149







