Differentiation of Glioblastoma and Brain Metastases by MRI-Based Oxygen Metabolomic Radiomics and Deep Learning
Abstract
:1. Introduction
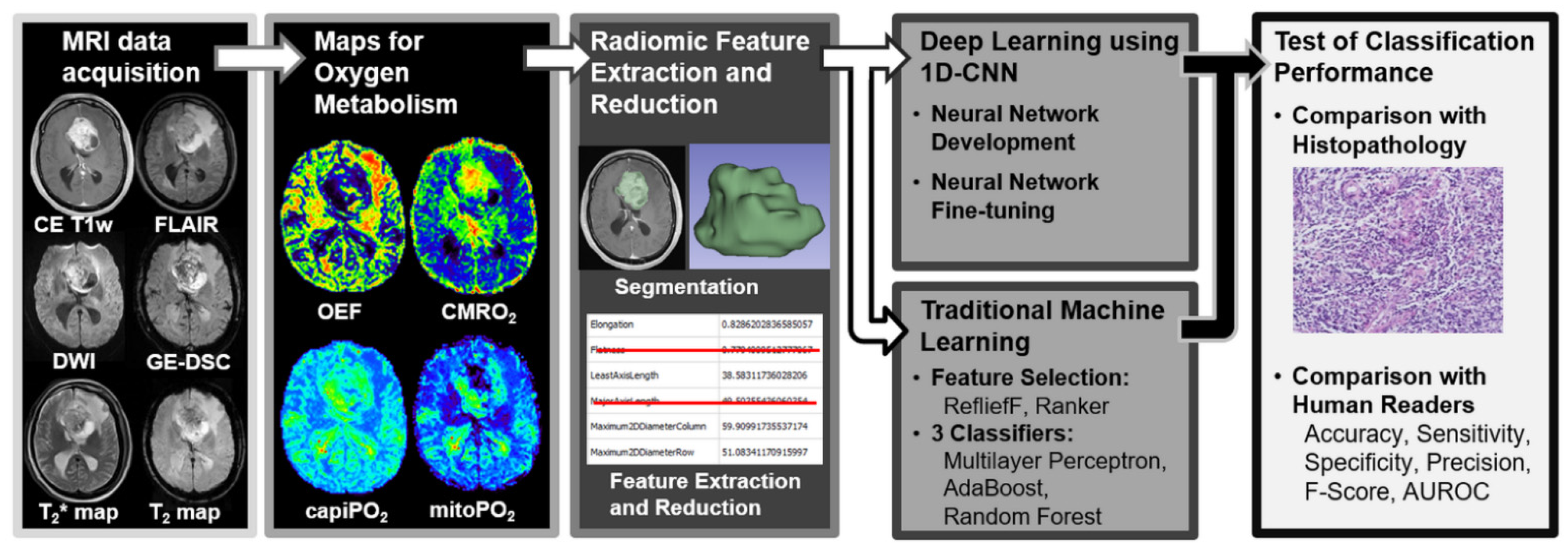
2. Materials and Methods
2.1. Ethics
2.2. Patient
2.3. MRI Data Acquisition
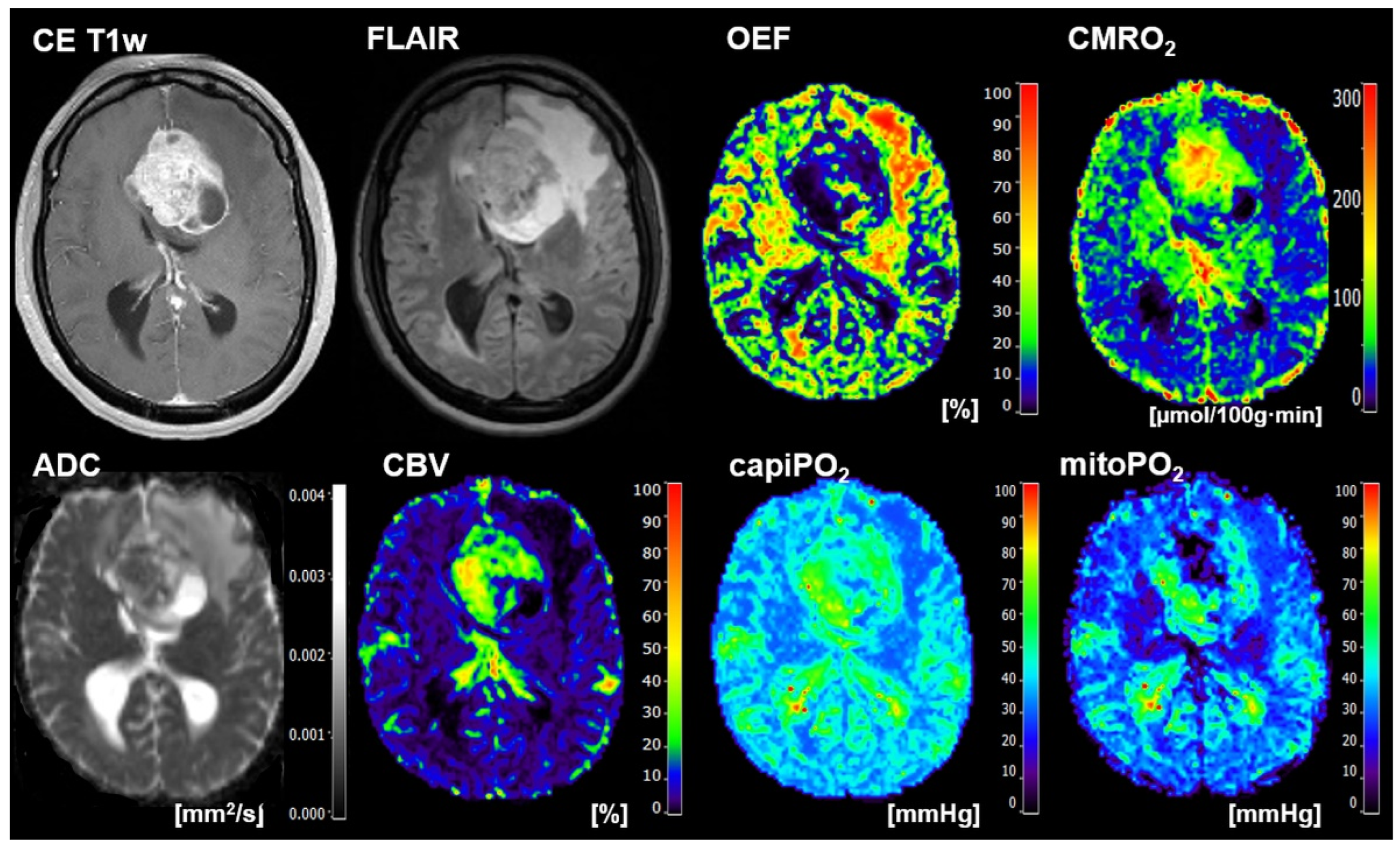
2.4. Calculation of MRI Biomarker Maps of Oxygen Metabolism
2.5. 3D Radiomic Feature Extraction of Oxygen Metabolism
2.6. Deep Learning
2.7. Traditional Machine Learning
2.8. Human Reading
2.9. Testing of the Models
3. Results
3.1. Patient Characteristics
3.2. The Selected Radiomic Features
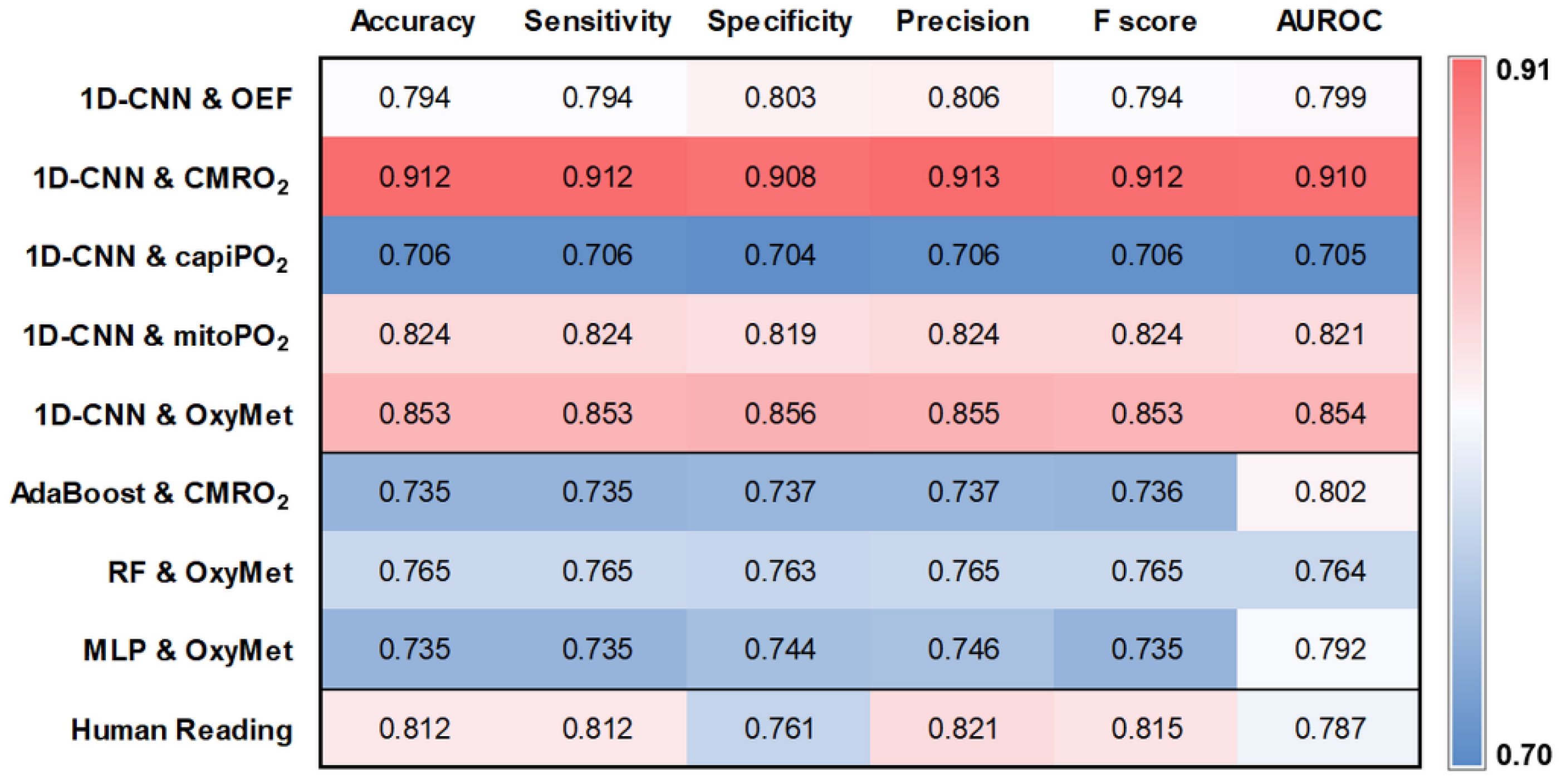
3.3. Performance of the Deep Learning Model
3.4. Comparison with Traditional Machine Learning Models and Human Readers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Delgado-López, P.D.; Corrales-García, E.M. Survival in glioblastoma: A review on the impact of treatment modalities. Clin. Transl. Oncol. 2016, 18, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Giordana, M.T.; Cordera, S.; Boghi, A. Cerebral metastases as first symptom of cancer: A clinico-pathologic study. J. Neurooncol. 2000, 50, 265–273. [Google Scholar] [CrossRef]
- Scoccianti, S.; Ricardi, U. Treatment of brain metastases: Review of phase III randomized controlled trials. Radiother. Oncol. 2012, 102, 168–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, M.; van den Bent, M.; Hopkins, K.; Tonn, J.C.; Stupp, R.; Falini, A.; Cohen-Jonathan-Moyal, E.; Frappaz, D.; Henriksson, R.; Balana, C.; et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014, 15, e395–e403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudy-Marqueste, C.; Carron, R.; Delsanti, C.; Loundou, A.; Monestier, S.; Archier, E.; Richard, M.A.; Regis, J.; Grob, J.J. On demand Gamma-Knife strategy can be safely combined with BRAF inhibitors for the treatment of melanoma brain metastases. Ann. Oncol. 2014, 25, 2086–2091. [Google Scholar] [CrossRef] [PubMed]
- Marko, N.F.; Weil, R.J.; Schroeder, J.L.; Lang, F.F.; Suki, D.; Sawaya, R.E. Extent of resection of glioblastoma revisited: Personalized survival modeling facilitates more accurate survival prediction and supports a maximum-safe-resection approach to surgery. J. Clin. Oncol. 2014, 32, 774–782. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Mizobuchi, Y.; Nakajima, K.; Otomi, Y.; Irahara, S.; Obama, Y.; Majigsuren, M.; Khashbat, D.; Kageji, T.; Nagahiro, S.; et al. Diagnosis of brain tumors using dynamic contrast-enhanced perfusion imaging with a short acquisition time. Springerplus 2015, 4, 88. [Google Scholar] [CrossRef] [Green Version]
- Mukundan, S.; Holder, C.; Olson, J.J. Neuroradiological assessment of newly diagnosed glioblastoma. J. Neurooncol. 2008, 89, 259–269. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlbauer, A.; Zimmermann, M.; Doerfler, A.; Oberndorfer, S.; Buchfelder, M.; Coras, R.; Kitzwögerer, M.; Roessler, K. Intratumoral heterogeneity of oxygen metabolism and neovascularization uncovers 2 survival-relevant subgroups of IDH1 wild-type glioblastoma. Neuro Oncol. 2018, 20, 1536–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlbauer, A.; Zimmermann, M.; Heinz, G.; Oberndorfer, S.; Doerfler, A.; Buchfelder, M.; Rössler, K. Magnetic resonance imaging biomarkers for clinical routine assessment of microvascular architecture in glioma. J. Cereb. Blood Flow Metab. 2017, 37, 632–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlbauer, A.; Zimmermann, M.; Kitzwögerer, M.; Oberndorfer, S.; Rössler, K.; Dörfler, A.; Buchfelder, M.; Heinz, G. MR Imaging–derived Oxygen Metabolism and Neovascularization Characterization for Grading and IDH Gene Mutation Detection of Gliomas. Radiology 2017, 283, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlbauer, A.; Mouridsen, K.; Doerfler, A.; Hansen, M.B.; Oberndorfer, S.; Zimmermann, M.; Buchfelder, M.; Heinz, G.; Roessler, K. Recurrence of glioblastoma is associated with elevated microvascular transit time heterogeneity and increased hypoxia. J. Cereb. Blood Flow Metab. 2018, 38, 422–432. [Google Scholar] [CrossRef] [Green Version]
- Stadlbauer, A.; Zimmermann, M.; Bennani-Baiti, B.; Helbich, T.H.; Baltzer, P.; Clauser, P.; Kapetas, P.; Bago-Horvath, Z.; Pinker, K. Development of a Non-invasive Assessment of Hypoxia and Neovascularization with Magnetic Resonance Imaging in Benign and Malignant Breast Tumors: Initial Results. Mol. Imaging Biol. 2019, 21, 758–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennani-Baiti, B.; Pinker, K.; Zimmermann, M.; Helbich, T.H.; Baltzer, P.A.; Clauser, P.; Kapetas, P.; Bago-Horvath, Z.; Stadlbauer, A. Non-Invasive Assessment of Hypoxia and Neovascularization with MRI for Identification of Aggressive Breast Cancer. Cancers 2020, 12, 2024. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef] [PubMed]
- Jana, G.C.; Sharma, R.; Agrawal, A. A 1D-CNN-Spectrogram Based Approach for Seizure Detection from EEG Signal. Procedia Comput. Sci. 2020, 167, 403–412. [Google Scholar] [CrossRef]
- Smith, A.M.; Grandin, C.B.; Duprez, T.; Mataigne, F.; Cosnard, G. Whole brain quantitative CBF, CBV, and MTT measurements using MRI bolus tracking: Implementation and application to data acquired from hyperacute stroke patients. J. Magn. Reson. Imaging 2000, 12, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Bjørnerud, A.; Emblem, K.E. A Fully Automated Method for Quantitative Cerebral Hemodynamic Analysis Using DSC–MRI. J. Cereb. Blood Flow Metab. 2010, 30, 1066–1078. [Google Scholar] [CrossRef] [Green Version]
- Preibisch, C.; Volz, S.; Anti, S.; Deichmann, R. Exponential excitation pulses for improved water content mapping in the presence of background gradients. Magn. Reson. Med. 2008, 60, 908–916. [Google Scholar] [CrossRef]
- Prasloski, T.; Mädler, B.; Xiang, Q.-S.; MacKay, A.; Jones, C. Applications of stimulated echo correction to multicomponent T 2 analysis. Magn. Reson. Med. 2012, 67, 1803–1814. [Google Scholar] [CrossRef]
- Vafaee, M.S.; Vang, K.; Bergersen, L.H.; Gjedde, A. Oxygen Consumption and Blood Flow Coupling in Human Motor Cortex during Intense Finger Tapping: Implication for a Role of Lactate. J. Cereb. Blood Flow Metab. 2012, 32, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Kennan, R.P.; Zhong, J.; Gore, J.C. Intravascular susceptibility contrast mechanisms in tissues. Magn. Reson. Med. 1994, 31, 9–21. [Google Scholar] [CrossRef]
- Gjedde, A. Cerebral Blood Flow Change in Arterial Hypoxemia Is Consistent with Negligible Oxygen Tension in Brain Mitochondria. Neuroimage 2002, 17, 1876–1881. [Google Scholar] [CrossRef]
- Vafaee, M.S.; Gjedde, A. Model of Blood–Brain Transfer of Oxygen Explains Nonlinear Flow-Metabolism Coupling During Stimulation of Visual Cortex. J. Cereb. Blood Flow Metab. 2000, 20, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Mao, Y.; Li, H.; Yu, G.; Wan, H.; Li, B. Differentiating brain metastases from different pathological types of lung cancers using texture analysis of T1 postcontrast MR. Magn. Reson. Med. 2016, 76, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, D.R.; Awwad, A.; Meijer, L.; Manita, M.; Jaspan, T.; Dineen, R.A.; Grundy, R.G.; Auer, D.P. Metrics and textural features of MRI diffusion to improve classification of pediatric posterior fossa tumors. AJNR Am. J. Neuroradiol. 2014, 35, 1009–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Thibault, G.; Fertil, B.; Navarro, C.; Pereira, S.; Cau, P.; Levy, N.; Sequeira, J.; Mari, J.-L. Shape and Texture Indexes Application to Cell Nuclei Classification. Int. J. Pattern Recognit. Artif. Intell. 2013, 27, 1357002. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man. Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Baessler, B.; Nestler, T.; Pinto dos Santos, D.; Paffenholz, P.; Zeuch, V.; Pfister, D.; Maintz, D.; Heidenreich, A. Radiomics allows for detection of benign and malignant histopathology in patients with metastatic testicular germ cell tumors prior to post-chemotherapy retroperitoneal lymph node dissection. Eur. Radiol. 2020, 30, 2334–2345. [Google Scholar] [CrossRef] [PubMed]
- Moradmand, H.; Aghamiri, S.M.R.; Ghaderi, R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020, 21, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Mannil, M.; Burgstaller, J.M.; Held, U.; Farshad, M.; Guggenberger, R. Correlation of texture analysis of paraspinal musculature on MRI with different clinical endpoints: Lumbar Stenosis Outcome Study (LSOS). Eur. Radiol. 2019, 29, 22–30. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- An, J.Y.; Seo, H.; Kim, Y.-G.; Lee, K.E.; Kim, S.; Kong, H.-J. Codeless Deep Learning of COVID-19 Chest X-Ray Image Dataset with KNIME Analytics Platform. Healthc. Inform. Res. 2021, 27, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Weigert, M.; Schmidt, U.; Boothe, T.; Müller, A.; Dibrov, A.; Jain, A.; Wilhelm, B.; Schmidt, D.; Broaddus, C.; Culley, S.; et al. Content-aware image restoration: Pushing the limits of fluorescence microscopy. Nat. Methods 2018, 15, 1090–1097. [Google Scholar] [CrossRef] [Green Version]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME-the Konstanz information miner: Version 2.0 and beyond. ACM SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Kiranyaz, S.; Avci, O.; Abdeljaber, O.; Ince, T.; Gabbouj, M.; Inman, D.J. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 2021, 151, 107398. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Ng, W.; Minasny, B.; Montazerolghaem, M.; Padarian, J.; Ferguson, R.; Bailey, S.; McBratney, A.B. Convolutional neural network for simultaneous prediction of several soil properties using visible/near-infrared, mid-infrared, and their combined spectra. Geoderma 2019, 352, 251–267. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Marhold, F.; Oberndorfer, S.; Heinz, G.; Buchfelder, M.; Kinfe, T.M.; Meyer-Bäse, A. Radiophysiomics: Brain Tumors Classification by Machine Learning and Physiological MRI Data. Cancers 2022, 14, 2363. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Faehndrich, J.; Weidauer, S.; Pilatus, U.; Oszvald, A.; Zanella, F.E.; Hattingen, E. Neuroradiological Viewpoint on the Diagnostics of Space-Occupying Brain Lesions. Clin. Neuroradiol. 2011, 21, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.H.; Erly, W.; Moser, F.G.; Maya, M.; Nael, K. Differentiation of solitary brain metastasis from glioblastoma multiforme: A predictive multiparametric approach using combined MR diffusion and perfusion. Neuroradiology 2015, 57, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Suh, C.H.; Kim, H.S.; Jung, S.C.; Choi, C.G.; Kim, S.J. Perfusion MRI as a diagnostic biomarker for differentiating glioma from brain metastasis: A systematic review and meta-analysis. Eur. Radiol. 2018, 28, 3819–3831. [Google Scholar] [CrossRef]
- Kamimura, K.; Nakajo, M.; Yoneyama, T.; Fukukura, Y.; Hirano, H.; Goto, Y.; Sasaki, M.; Akamine, Y.; Keupp, J.; Yoshiura, T. Histogram analysis of amide proton transfer-weighted imaging: Comparison of glioblastoma and solitary brain metastasis in enhancing tumors and peritumoral regions. Eur. Radiol. 2019, 29, 4133–4140. [Google Scholar] [CrossRef]
- Romano, A.; Moltoni, G.; Guarnera, A.; Pasquini, L.; Di Napoli, A.; Napolitano, A.; Espagnet, M.C.R.; Bozzao, A. Single brain metastasis versus glioblastoma multiforme: A VOI-based multiparametric analysis for differential diagnosis. Radiol. Med. 2022, 127, 490–497. [Google Scholar] [CrossRef]
- Gadda, D.; Mazzoni, L.N.; Pasquini, L.; Busoni, S.; Simonelli, P.; Giordano, G.P. Relationship between Apparent Diffusion Coefficients and MR Spectroscopy Findings in High-Grade Gliomas. J. Neuroimaging 2017, 27, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Stadlbauer, A.; Marhold, F.; Oberndorfer, S.; Heinz, G.; Zimmermann, M.; Buchfelder, M.; Heynold, E.; Kinfe, T.M. Metabolic Tumor Microenvironment Characterization of Contrast Enhancing Brain Tumors Using Physiologic MRI. Metabolites 2021, 11, 668. [Google Scholar] [CrossRef]
- Hardee, M.E.; Zagzag, D. Mechanisms of glioma-associated neovascularization. Am. J. Pathol. 2012, 181, 1126–1141. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, A.; Singh, S.K.; Agnihotri, S.; Jalali, S.; Burrell, K.; Aldape, K.D.; Zadeh, G. GBM’s multifaceted landscape: Highlighting regional and microenvironmental heterogeneity. Neuro. Oncol. 2014, 16, 1167–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tateishi, M.; Nakaura, T.; Kitajima, M.; Uetani, H.; Nakagawa, M.; Inoue, T.; Kuroda, J.-I.; Mukasa, A.; Yamashita, Y. An initial experience of machine learning based on multi-sequence texture parameters in magnetic resonance imaging to differentiate glioblastoma from brain metastases. J. Neurol. Sci. 2020, 410, 116514. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, X.; Rui, W.; Pang, H.; Qiu, T.; Wang, J.; Xie, Q.; Jin, T.; Zhang, H.; Chen, H.; et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J. Magn. Reson. Imaging 2019, 49, 808–817. [Google Scholar] [CrossRef]
- Bae, S.; An, C.; Ahn, S.S.; Kim, H.; Han, K.; Kim, S.W.; Park, J.E.; Kim, H.S.; Lee, S.-K. Robust performance of deep learning for distinguishing glioblastoma from single brain metastasis using radiomic features: Model development and validation. Sci. Rep. 2020, 10, 12110. [Google Scholar] [CrossRef]
- Ortiz-Ramón, R.; Ruiz-España, S.; Mollá-Olmos, E.; Moratal, D. Glioblastomas and brain metastases differentiation following an MRI texture analysis-based radiomics approach. Phys. Medica 2020, 76, 44–54. [Google Scholar] [CrossRef]
- Tariciotti, L.; Caccavella, V.M.; Fiore, G.; Schisano, L.; Carrabba, G.; Borsa, S.; Giordano, M.; Palmisciano, P.; Remoli, G.; Remore, L.G.; et al. A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis and Primary Central Nervous System Lymphoma: A Pilot Study. Front. Oncol. 2022, 12, 816638. [Google Scholar] [CrossRef]
- Shin, I.; Kim, H.; Ahn, S.S.; Sohn, B.; Bae, S.; Park, J.E.; Kim, H.S.; Lee, S.-K. Development and Validation of a Deep Learning-Based Model to Distinguish Glioblastoma from Solitary Brain Metastasis Using Conventional MR Images. AJNR Am. J. Neuroradiol. 2021, 42, 838–844. [Google Scholar] [CrossRef]
- Baazaoui, H.; Hubertus, S.; Maros, M.E.; Mohamed, S.A.; Förster, A.; Schad, L.R.; Wenz, H. Artificial Neural Network-Derived Cerebral Metabolic Rate of Oxygen for Differentiating Glioblastoma and Brain Metastasis in MRI: A Feasibility Study. Appl. Sci. 2021, 11, 9928. [Google Scholar] [CrossRef]
- Sartoretti, E.; Sartoretti, T.; Wyss, M.; Reischauer, C.; van Smoorenburg, L.; Binkert, C.A.; Sartoretti-Schefer, S.; Mannil, M. Amide proton transfer weighted (APTw) imaging based radiomics allows for the differentiation of gliomas from metastases. Sci. Rep. 2021, 11, 5506. [Google Scholar] [CrossRef]
- Cao, X.; Tan, D.; Liu, Z.; Liao, M.; Kan, Y.; Yao, R.; Zhang, L.; Nie, L.; Liao, R.; Chen, S.; et al. Differentiating solitary brain metastases from glioblastoma by radiomics features derived from MRI and 18F-FDG-PET and the combined application of multiple models. Sci. Rep. 2022, 12, 5722. [Google Scholar] [CrossRef] [PubMed]
- Dikaios, N. Deep learning magnetic resonance spectroscopy fingerprints of brain tumours using quantum mechanically synthesised data. NMR Biomed. 2021, 34, e4479. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]



| Type | Input Size 1 | No. of Filters | Kernel Size | Stride | Padding | Activation |
|---|---|---|---|---|---|---|
| Input | 74, 1 (296, 1) | |||||
| 1D Convolution Layer | 74, 1 (296, 1) | 8 | 2 | 1 | same | ReLU 2 |
| Max Pooling Layer | 74, 8 (296, 8) | 2 | 1 | same | ||
| 1D Convolution Layer | 74, 8 (296, 8) | 8 | 3 | 1 | same | ReLU |
| Drop out (rate 0.2) | 74, 8 (296, 8) | |||||
| Max Pooling Layer | 74, 8 (296, 8) | 2 | 1 | same | ||
| 1D Convolution Layer | 74, 8 (296, 8) | 8 | 3 | 1 | same | ReLU |
| Drop out (rate 0.2) | 74, 8 (296, 8) | |||||
| Max Pooling Layer | 74, 8 (296, 8) | 2 | 2 | same | ||
| Flatten Layer | 37, 8 (148, 8) | |||||
| Dense layer | 296 (1184) | ReLU | ||||
| Drop out (rate 0.2) | 296 (1184) | |||||
| Dense layer | 296 (1184) | Sigmoid |
| Features with High Reproducibility (ICC ≥ 0.8) | Excluded Features | |
|---|---|---|
| Shape Features | Elongation, Mesh Volume, Voxel Volume | Flatness, Least Axis Length, Major Axis Length, Maximum 2D Diameter Column, Maximum 2D Diameter Row, Maximum 2D Diameter Slice, Maximum 3D Diameter, Minor Axis Length, Sphericity, Surface Area, Surface Volume Ratio |
| First-Order Features | 10th Percentile, 90th Percentile, Energy, Entropy, Interquartile Range, Kurtosis, Maximum, Mean Absolute Deviation, Mean, Median, Minimum, Range, Robust Mean Absolute Deviation, Root Mean Squared, Skewness, Total Energy, Uniformity, Variance | |
| Texture Features | GLCM 1: Autocorrelation, Cluster Prominence, Cluster Shade, Cluster Tendency, Contrast, Correlation, Difference Average, Difference Entropy, Difference Variance, Imc1, Imc2, Inverse Variance, Joint Average, Joint Energy, Joint Entropy, Maximum Probability, Sum Average, Sum Entropy, Sum Squares GLDM 2: Dependence Entropy, Dependence Non-Uniformity, Dependence Variance, Gray Level Variance, High Gray Level Emphasis, Large Dependence Emphasis, Large Dependence Low Gray Level Emphasis, Small Dependence Emphasis, Small Dependence High Gray Level Emphasis GLRLM 3: Gray Level Non-Uniformity Normalized, Gray Level Variance, High Gray Level Run Emphasis, Long Run Emphasis, Long Run High Gray Level Emphasis, Long Run Low Gray Level Emphasis, Run Entropy, Run Length Non-Uniformity, Run Variance, Short Run High Gray Level Emphasis GLSZM 4: Gray Level Non-Uniformity, Gray Level Non -Uniformity Normalized, Gray Level Variance, High Gray Level Zone Emphasis, Large Area Low Gray Level Emphasis, Size Zone Non-Uniformity, Size Zone Non-Uniformity Normalized, Small Area Emphasis, Small Area High Gray Level Emphasis, Zone Entropy, Zone Percentage NGTDM 5: Busyness, Complexity, Contrast, Strength | GLCM: Id, ldm, Idmn, Idn, MCC GLDM: Dependence Non-Uniformity Normalized, GLDM Gray Level Non- Uniformity, Large Dependence High Gray Level Emphasis, Low Gray Level Emphasis, Small Dependence Low Gray Level Emphasis GLRLM: Gray Level Non-Uniformity, Low Gray Level Run Emphasis, Run Length Non-Uniformity Normalized, Run Percentage, Short Run Emphasis, Short Run Low Gray Level Emphasis GLSZM: Large Area Emphasis, Large Area High Gray Level Emphasis, Low Gray Level Zone Emphasis, Small Area Low Gray Level Emphasis, Zone Variance NGTDM: Coarseness |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stadlbauer, A.; Heinz, G.; Marhold, F.; Meyer-Bäse, A.; Ganslandt, O.; Buchfelder, M.; Oberndorfer, S. Differentiation of Glioblastoma and Brain Metastases by MRI-Based Oxygen Metabolomic Radiomics and Deep Learning. Metabolites 2022, 12, 1264. https://doi.org/10.3390/metabo12121264
Stadlbauer A, Heinz G, Marhold F, Meyer-Bäse A, Ganslandt O, Buchfelder M, Oberndorfer S. Differentiation of Glioblastoma and Brain Metastases by MRI-Based Oxygen Metabolomic Radiomics and Deep Learning. Metabolites. 2022; 12(12):1264. https://doi.org/10.3390/metabo12121264
Chicago/Turabian StyleStadlbauer, Andreas, Gertraud Heinz, Franz Marhold, Anke Meyer-Bäse, Oliver Ganslandt, Michael Buchfelder, and Stefan Oberndorfer. 2022. "Differentiation of Glioblastoma and Brain Metastases by MRI-Based Oxygen Metabolomic Radiomics and Deep Learning" Metabolites 12, no. 12: 1264. https://doi.org/10.3390/metabo12121264
APA StyleStadlbauer, A., Heinz, G., Marhold, F., Meyer-Bäse, A., Ganslandt, O., Buchfelder, M., & Oberndorfer, S. (2022). Differentiation of Glioblastoma and Brain Metastases by MRI-Based Oxygen Metabolomic Radiomics and Deep Learning. Metabolites, 12(12), 1264. https://doi.org/10.3390/metabo12121264








