Targeted Metabolome Profiling of Indonesian Shallots and Japanese Long-Day/Short-Day Bulb Onions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Metabolome Analysis
2.3. Data Analysis
3. Results
3.1. Characteristic Phenotyping of Shallots and Bulb Onions
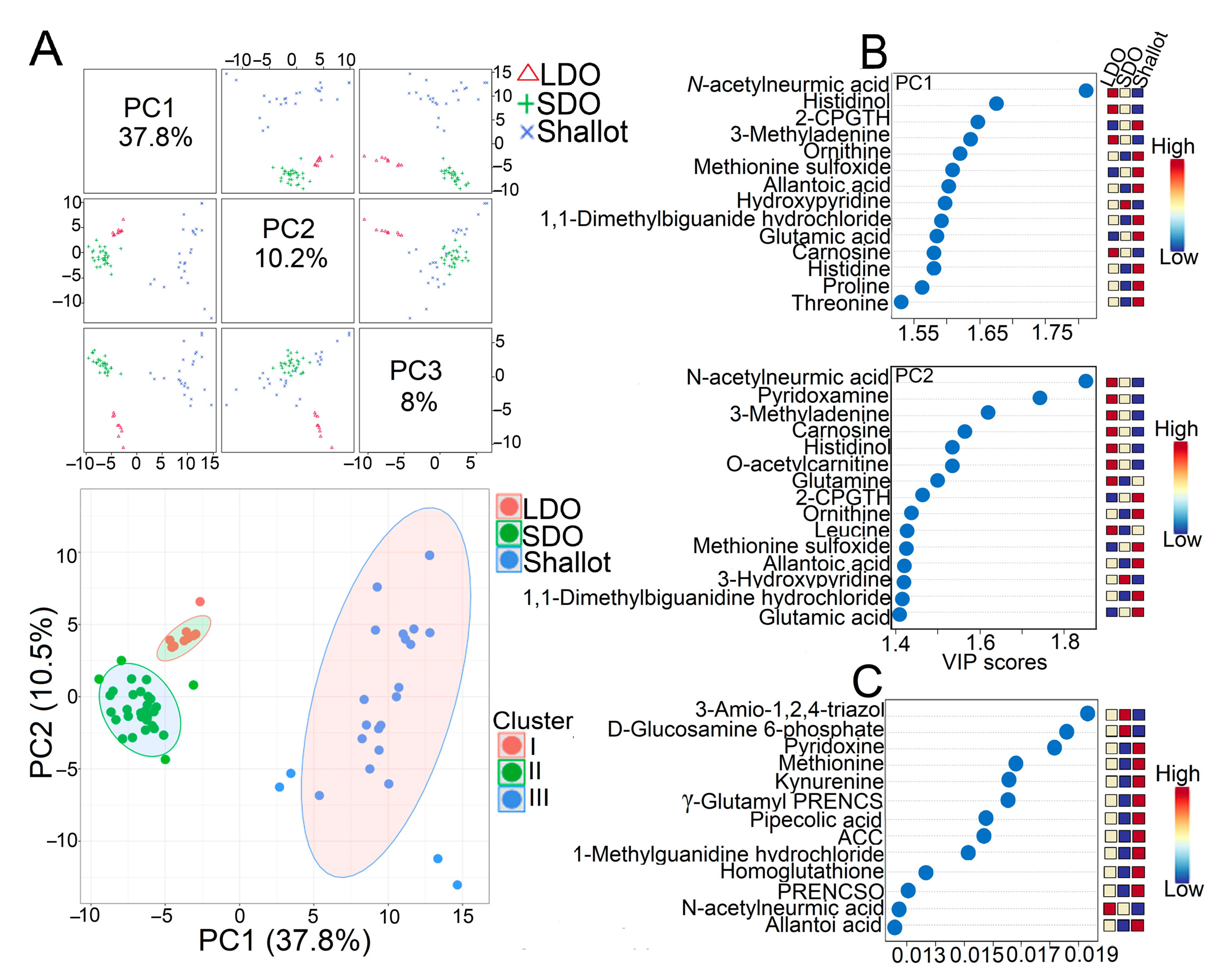
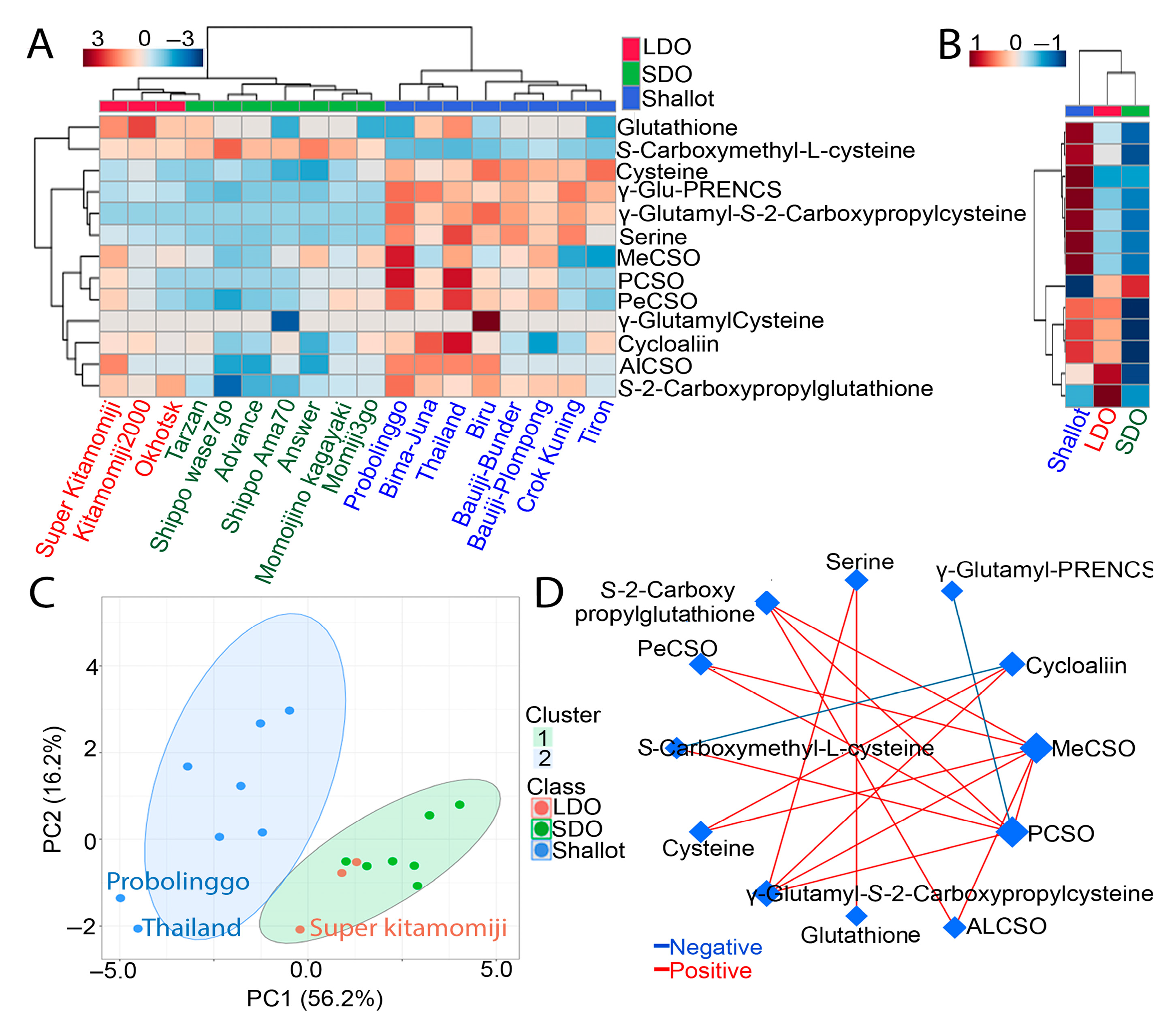
3.2. Comprehensive Metabolite Profiling of Shallots and Bulb Onions
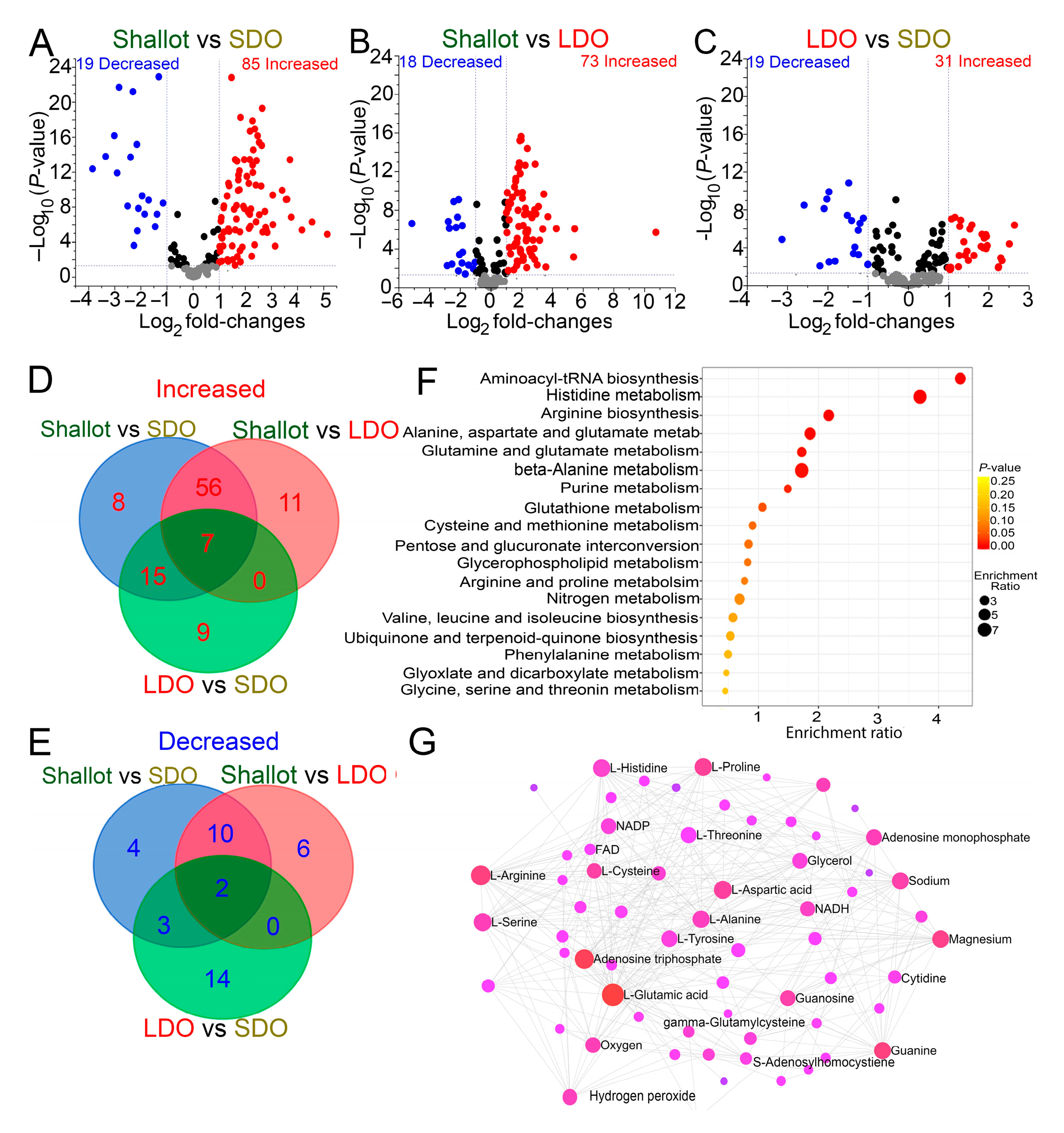
3.3. Genotype Correlation and Venn Diagram Analysis of Shallots and Bulb Onions
3.4. Metabolic Pathways Associated with Shallots and Bulb Onions
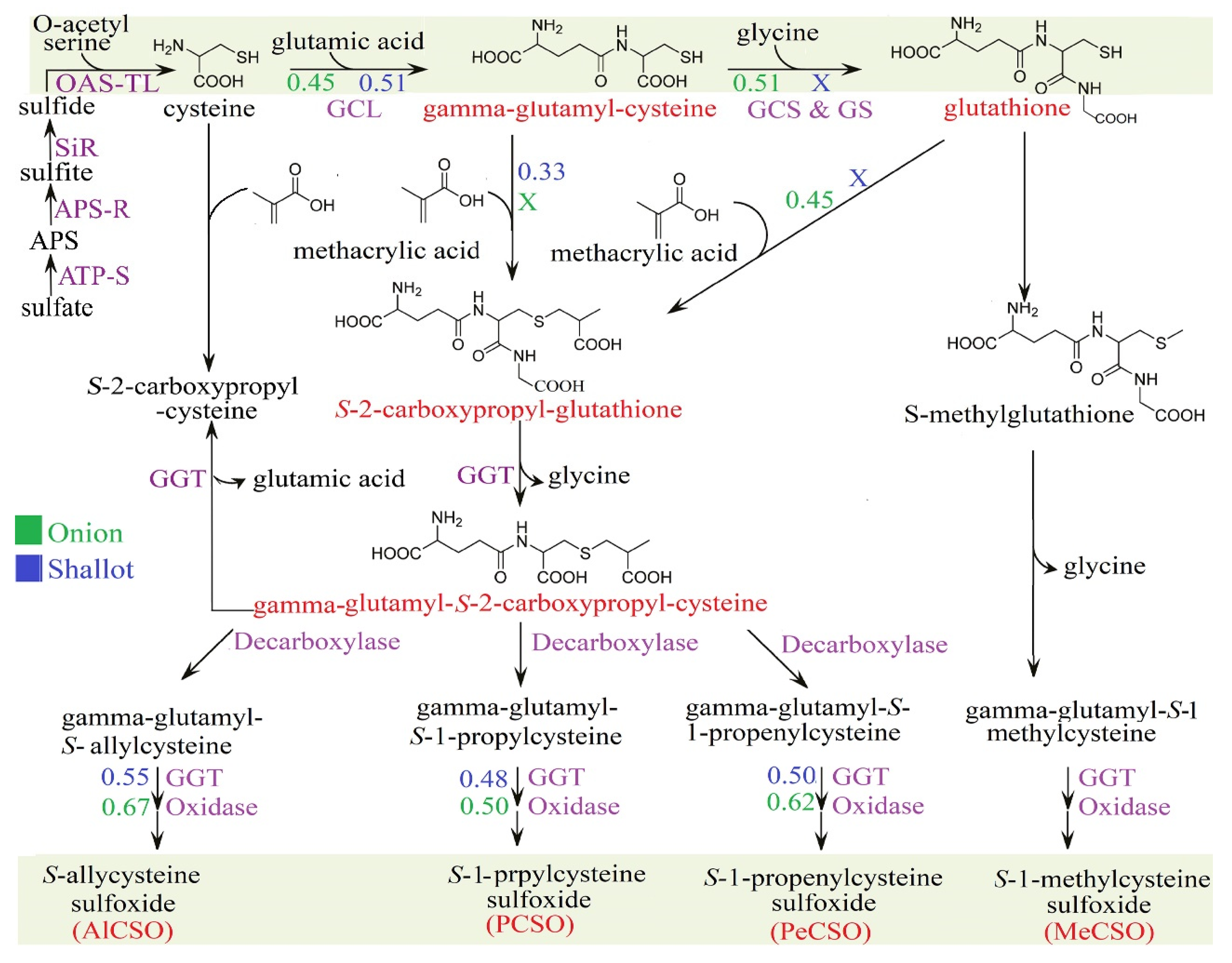
3.5. Comparative CSO Biosynthesis Pathway in Shallots and Bulb Onions
4. Discussion
4.1. Comparative Metabolome Analyses of Shallot and Bulb Onions
4.2. Comparative Analysis of the Sulfur Assimilation Pathway in Shallots and Bulb Onions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huo, Y.; Gao, L.; Liu, B.; Yang, Y.Y.; Kong, S.P.; Sun, Y.Q.; Yang, Y.H.; Wu, X. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 2019, 9, 12250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkers, R.; van Kaauwen, M.; Ament, K.; Burger-Meijer, K.; Egging, R.; Huits, H.; Kodde, L.; Kroon, L.; Shigyo, M.; Sato, S.; et al. Insights from the first genome assembly of onion (Allium cepa). G3 (Bethesda) 2021, 11, jkab243. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Hirata, S.; Mukae, T.; Yamada, T.; Sawada, Y.; El-Syaed, M.; Yamada, Y.; Sato, M.; Hirai, M.Y.; Shigyo, M. Comprehensive metabolite profiling in genetic resources of garlic (Allium sativum L.) collected from different geographical regions. Molecules 2021, 26, 1415. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Hirata, S.; Sawada, Y.; Hirai, M.Y.; Sato, S.; Hirakawa, H.; Mine, Y.; Tanaka, K.; Shigyo, M. Widely targeted metabolome and transcriptome landscapes of Allium fistulosum–A. cepa chromosome addition lines revealed a flavonoid hot spot-on chromosome 5A. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelrahman, M.; Sawada, Y.; Nakabayashi, R.; Sato, S.; Hirakawa, H.; El-Sayed, M.; Hirai, M.Y.; Saito, K.; Yamauchi, N.; Shigyo, M. Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Major, N.; Goreta Ban, S.; Urlić, B.; Ban, D.; Dumičić, G.; Perković, J. Morphological and biochemical diversity of shallot landraces preserved along the Croatian Coast. Front. Plant Sci. 2018, 9, 1749. [Google Scholar] [CrossRef] [PubMed]
- Perković, J.; Major., N.; Ban., D.; Cvitan., D.; Ban., S.G. Shallot species and subtypes discrimination based on morphology descriptors. Plants 2020, 10, 60. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Fernandes, Â.; Barros, L.; Ferreira, I.C.F.R.; Ntatsi, G. Morphological, nutritional and chemical description of “Vatikiotiko”, an onion local landrace from Greece. Food Chem. 2015, 182, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Abdelrahman, M.; Ariyanti, N.A.; Sawada, Y.; Tsuji, F.; Hirata, S.; Hang, T.T.M.; Okamoto, M.; Yamada, Y.; Tsugawa, H.; Hirai, M.Y.; et al. Metabolome-based discrimination analysis of shallot landraces and bulb onion cultivars associated with differences in the amino acid and flavonoid profiles. Molecules 2020, 25, 5300. [Google Scholar] [CrossRef]
- Soorni, A.; Akrami, A.M.; Abolghasemi, R.; Vahedi, M. Transcriptome and phytochemical analyses provide insights into the organic sulfur pathway in Allium hirtifolium. Sci. Rep. 2021, 11, 768. [Google Scholar] [CrossRef]
- Ariyanti, N.A.; Torikai, K.; Kirana, R.P.; Hirata, S.; Sulistyaningsih, E.; Ito, S.-I.; Yamauchi, N.; Kobayashi, N.; Shigyo, M. Comparative study on phytochemical variations in Japanese F1 varieties of bulb onions and South-East Asian shallot landraces. Horticult. J. 2018, 87, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Fattorusso, E.M.; Iorizzi, M.; Lanzotti, V.; Taglialatela-Scafati, O. Chemical composition of shallot (Allium ascalonicum Hort.). J. Agric. Food Chem. 2002, 50, 5686–5690. [Google Scholar] [CrossRef]
- Benkeblia, N. Antimicrobial activity of essential oil extracts of various onions (Allium cepa) and garlic (Allium sativum). LWT-Food Sci. Technol. 2004, 37, 263–268. [Google Scholar] [CrossRef]
- Shigyo, M.; Kik, C. Onion. In Handbook of Plant Breeding, Vegetables; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; Volume II, pp. 121–159. [Google Scholar]
- Brewster, J.L. Onions and Other Vegetable Alliums, 2nd ed.; CABI: Wallingford, UK, 2008; pp. 1–372. [Google Scholar]
- Sawada, Y.; Tsukaya, H.; Li, Y.; Sato, M. A novel method for single-grain-based metabolic profiling of Arabidopsis seed. Metabolomics 2017, 13, 75. [Google Scholar] [CrossRef]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, 684–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Kumagai, H. Characteristics, biosynthesis, decomposition, metabolism and functions of the garlic odour precursor, S-allyl-L-cysteine sulfoxide. Exp. Ther. Med. 2020, 2, 1528–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, N.; Yabe, A.; Sugino, Y.; Murakami, S.; Sai-Ngam, N.; Sumi, S.; Tsuneyoshi, T.; Saito, K. Garlic γ-glutamyl transpeptidases that catalyze deglutamylation of biosynthetic intermediate of Alliin. Front Plant Sci. 2015, 5, 758. [Google Scholar] [CrossRef] [Green Version]
- Fritsch, R.M.; Friesen, N. Evolution, domestication, and taxonomy. In Allium Crop Science: Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CABI: Wallingford, UK, 2002; pp. 5–30. [Google Scholar]
- Cumo, C.E. Onion. In Foods that Changed History: How Foods Shaped Civilization from the Ancient World to the Present; ABC-CLIO: Santa Barbara, CA, USA, 2015; pp. 248–250. [Google Scholar]
- MAFF. Statistics of Bulb Onion Production; Ministry of Agriculture, Forestry and Fisheries: Tokyo, Japan, 2019; Available online: www.maff.go.jp/e/policies/env/sustainagri/promogap.htm (accessed on 15 September 2022).
- Tsuchiya, Y.; Nakamura, Y.; Mikami, T. Japanese bulb onion: Production, consumption, and cultivars. J. Hortic. Res. 2021, 29, 75–82. [Google Scholar] [CrossRef]
- Komochi, S. Bulb onion. In Cultivation History of Vegetable Crops in Hokkaido. Study Group on Vegetable Crops in Hokkaido; Kominami-Insatsu: Sapporo, Japan, 2015; pp. 232–249. [Google Scholar]
- Marotti, M.; Piccaglia, R. Characterization of flavonoids in different cultivars of onion (Allium cepa L.). J. Food Sci. 2002, 63, 1229–1232. [Google Scholar] [CrossRef]
- Rezvan, S.; Nezami, A.; Kafi, M.; Sanjani, S. Evaluation of freezing tolerance of three Persian shallot (Allium altissimum Regel) ecotypes under controlled condition. In Proceedings of the 9th International Plant Cold Hardiness Seminar, Luxembourg, 17–22 July 2011; pp. 7–17. [Google Scholar]
- van den Brink, L.; Basuki, R.S. Production of true seed shallots in Indonesia. Acta Hortic. 2012, 958, 115–120. [Google Scholar] [CrossRef]
- Triharyanto, E.; Sudadi, S.; Rawandari, S. Adaptation of six shallots varieties to phosphate solubilizing bacteria on the flower formation, seeds formation, and yields on the lowland. IOP Conf. Ser. Earth Environ. Sci. 2018, 142, 012–067. [Google Scholar] [CrossRef]
- Kebede, W. Shallot (Allium cepa var. ascolonicum) Responses to Plant Nutrients and Soil Moisture in a Sub-Humid Tropical Climate. Ph.D. Dissertation, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2003. [Google Scholar]
- Askari-Khorasgani, O.; Pessarakli, M. Agricultural management and environmental requirements for production of true shallot seeds—A review. Adv. Plants Agric. Res. 2019, 9, 318–322. [Google Scholar]
- Kusano, M.; Jonsson, P.; Fukushima, A.; Gullberg, J.; Sjöström, M.; Trygg, J.; Moritz, T. Metabolite signature during short-day induced growth cessation in Populus. Front. Plant Sci. 2011, 2, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, M.; Cortleven, A.; Novak, O.; Schmülling, T. Root-derived trans-zeatin cytokinin protects Arabidopsis plants against photoperiod stress. Plant Cell Environ. 2020, 43, 2637–2649. [Google Scholar] [CrossRef] [PubMed]
- Roeber, V.M.; Schmülling, T.; Cortleven, A. The Photoperiod: Handling and causing stress in plants. Front. Plant Sci. 2022, 12, 781988. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, X.; Wang, W.; Wang, Y.; Ming, F. The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2013, 8, e73541. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Griebel, T.; Zeier, J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008, 147, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Randle, W.M. Increasing nitrogen concentration in hydroponic solutions affects onion flavor and bulb quality. J. Am. Soc. Hortic. Sci. 2000, 125, 254–259. [Google Scholar] [CrossRef]
- McCallum, J.; Pither-Joyce, M.; Shaw, M.; Kenel, F.; Davis, S.; Butler, R.; Scheffer, J.; Jakse, J.; Havey, M.J. Genetic mapping of sulfur assimilation genes reveals a QTL for onion bulb pungency. Theor. Appl. Genet. 2007, 114, 815–822. [Google Scholar] [CrossRef]
- Hell, R.; Wirtz, M. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. Arabidopsis Book 2011, 9, e0154. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.W.; Hang, L.F.; Ali, S.; Xu, X.Y.; Liu, Y.J.; Yan, Q.Q.; Luo, Q.Y.; Li, Y.; Lin, L.J.; Li, H.X.; et al. Biosynthesis and metabolism of garlic odor compounds in cultivated Chinese chives (Allium tuberosum) and wild Chinese chives (Allium hookeri). Int. J. Mol. Sci. 2022, 23, 7013. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.E.; Shaw, M.L. γ-Glutamyl peptides in the biosynthesis of S-alk(en)yl-L-cysteine sulfoxides (flavor precursors) in Allium. Phytochemistry 1989, 28, 455–460. [Google Scholar] [CrossRef]
- Randle, W.M.; Lancaster, J.E.; Shaw, M.L.; Sutton, K.H.; Hay, R.L.; Bussard, M.L. Quantifying onion flavor compounds responding to sulfur fertility—sulfur increases levels of alk(en)yl cysteine sulphoxides and biosynthetic intermediates. J. Am. Soc. Hortic. Sci. 1995, 120, 1075–1081. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Sugii, M.; Kakimoto, T. Metabolic incorporation of L-valine-C14 into S-(2-carboxypropyl) glutathione and S-(2-carboxypropyl) cysteine in garlic. Chem. Pharm. Bull. (Tokyo) 1962, 10, 328–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granroth, B. Biosynthesis and decomposition of cysteine derivatives in onion and other Allium species. Ann. Acad. Sci. Fenn. A 1970, 154, 1–71. [Google Scholar]
- Bhatwalkar, S.B.; Mondal, R.; Krishna, S.B.N.; Adam, J.K.; Govender, P.; Anupam, R. Antibacterial properties of organosulfur compounds of garlic (Allium sativum). Front. Microbiol. 2021, 12, 613077. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-X.; Lin, F.-J.; Li, H.; Li, H.-B.; Wu, D.-T.; Geng, F.; Ma, W.; Wang, Y.; Miao, B.-H.; Gan, R.-Y. Recent advances in bioactive compounds, health functions, and safety concerns of onion (Allium cepa L.). Front. Nutr. 2021, 8, 669805. [Google Scholar] [CrossRef]
- Block, E. The organosulfur chemistry of the genus Allium—implications for the organic chemistry of sulfur. Angew. Chem. Int. Ed. Engl. 1992, 31, 1135–1178. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.; Shin, D.; Yoo, M. Change in organosulfur compounds in onion (Allium cepa L.) during heat treatment. Food Sci. Biotechnol. 2016, 25, 115–119. [Google Scholar] [CrossRef]
- Lee, E.J.; Yoo, K.S.; Jifon, J.; Patil, B.S. Characterization of short-day onion cultivars of 3 pungency levels with flavor precursor, free amino acid, sulfur, and sugar contents. J. Food Sci. 2009, 74, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.S.; Pike, L.M. Determination of flavor precursor compound S-alk(en)yl-L-cysteine sulfoxides by an HPLC method and their distribution in Allium species. Sci. Hortic. 1998, 75, 1–10. [Google Scholar]
- Saghir, A.R.; Mann, L.K.; Yamaguchi, M. Composition of volatiles in Allium as related to habitat, stage of growth, and plant part. Plant Physiol. 1965, 40, 681. [Google Scholar] [CrossRef] [PubMed]
- De Col, V.; Fuchs, P.; Nietzel, T.; Elsässer, M.; Voon, C.P.; Candeo, A.; Seeliger, I.; Fricker, M.D.; Grefen, C.; Møller, I.M.; et al. ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 2017, 6, e26770. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuse, K.; Abdelrahman, M.; Ariyanti, N.A.; Tsuji, F.; Hirata, S.; Nakajima, T.; Sato, M.; Hirai, M.Y.; Manochai, B.; Shigyo, M. Targeted Metabolome Profiling of Indonesian Shallots and Japanese Long-Day/Short-Day Bulb Onions. Metabolites 2022, 12, 1260. https://doi.org/10.3390/metabo12121260
Matsuse K, Abdelrahman M, Ariyanti NA, Tsuji F, Hirata S, Nakajima T, Sato M, Hirai MY, Manochai B, Shigyo M. Targeted Metabolome Profiling of Indonesian Shallots and Japanese Long-Day/Short-Day Bulb Onions. Metabolites. 2022; 12(12):1260. https://doi.org/10.3390/metabo12121260
Chicago/Turabian StyleMatsuse, Kanako, Mostafa Abdelrahman, Nur Aeni Ariyanti, Fumitada Tsuji, Sho Hirata, Tetsuya Nakajima, Muneo Sato, Masami Yokota Hirai, Benya Manochai, and Masayoshi Shigyo. 2022. "Targeted Metabolome Profiling of Indonesian Shallots and Japanese Long-Day/Short-Day Bulb Onions" Metabolites 12, no. 12: 1260. https://doi.org/10.3390/metabo12121260
APA StyleMatsuse, K., Abdelrahman, M., Ariyanti, N. A., Tsuji, F., Hirata, S., Nakajima, T., Sato, M., Hirai, M. Y., Manochai, B., & Shigyo, M. (2022). Targeted Metabolome Profiling of Indonesian Shallots and Japanese Long-Day/Short-Day Bulb Onions. Metabolites, 12(12), 1260. https://doi.org/10.3390/metabo12121260









