MALDI Mass Spectrometry Imaging Reveals the Existence of an N-Acyl-homoserine Lactone Quorum Sensing System in Pseudomonas putida Biofilms
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strain and Reagents
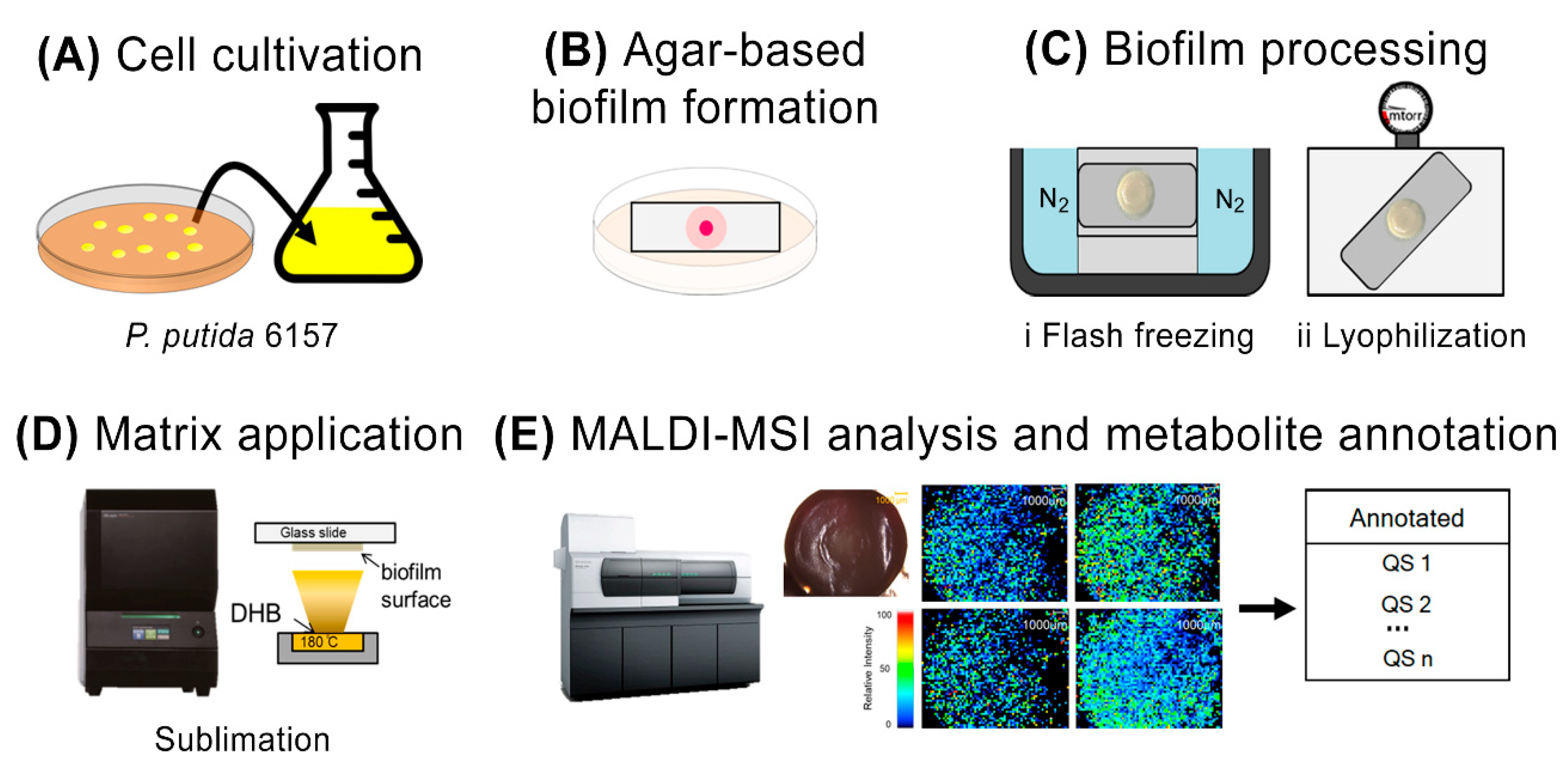
2.2. Process of Agar-Based Biofilm Cultivation
2.3. MALDI Matrix Application
2.4. MALDI-MSI Analysis
2.5. Metabolite Annotation
2.6. Genome Analysis
3. Results
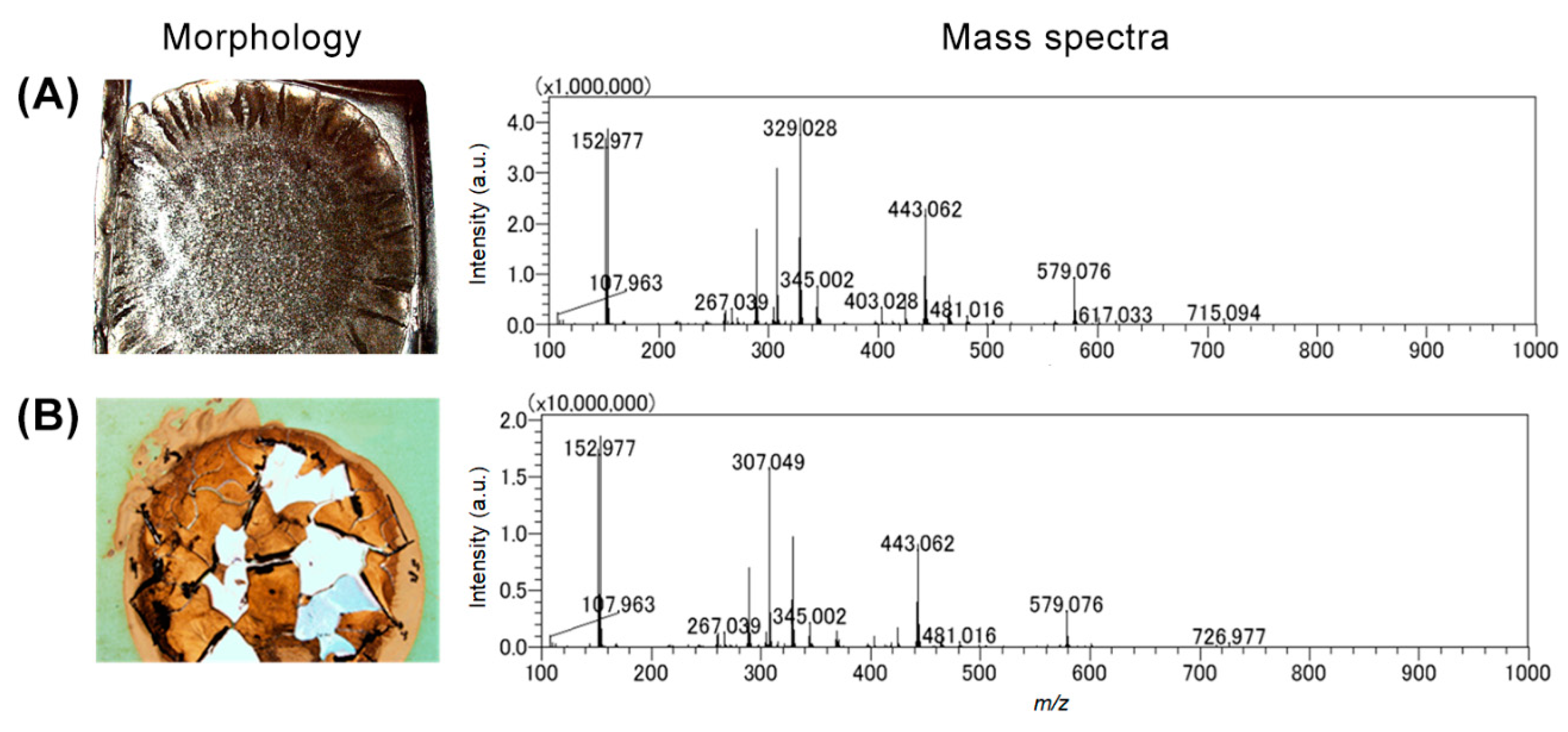
3.1. Modified Agar-Based Biofilms for MALDI-MSI
3.2. Correlation between the Production of AHL Metabolites and Biofilm Development in P. putida 6157
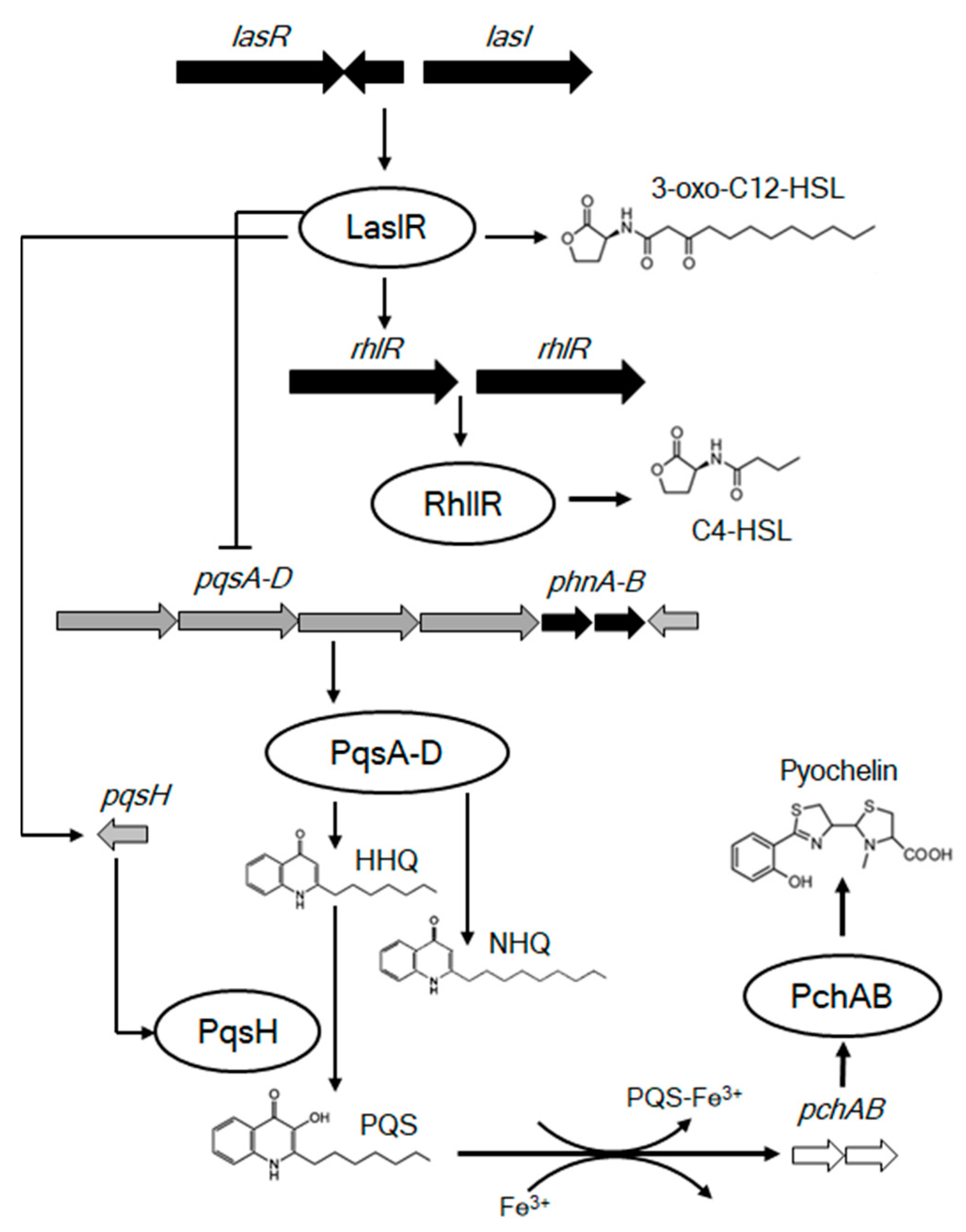
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 2004, 101, 16630–16635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria:the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, N.A.; Barnard, A.M.L.; Slater, H.; Simpson, N.J.L.; Salmond, G.P.C. Quorum-sensing in gram-negative bacteria. FEMS. Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef]
- Williams, P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 2007, 153, 3923–3938. [Google Scholar] [CrossRef] [Green Version]
- Bassler, B.L.; Losick, R. Bacterially speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Nikel, P.I.; Martinez-Garcia, E.; de Lorenzo, V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014, 12, 368–379. [Google Scholar] [CrossRef]
- Vlamakis, H.; Aguilar, C.; Losick, R.; Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes. Dev. 2008, 22, 945–953. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.; Winzer, K.; Chan, W.C.; Cámara, M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007, 362, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Bolton, N.; Critchley, M.; Fabien, R.; Cromar, N.; Fallowfield, H. Microbially influenced corrosion of galvanized steel pipes in aerobic water systems. J. Appl. Microbiol. 2010, 109, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Dunham, S.J.B.; Ellis, J.F.; Li, B.; Sweedler, J.V. Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res. 2017, 50, 96–104. [Google Scholar] [CrossRef]
- Lanni, E.J.; Masyuko, R.N.; Driscoll, C.M.; Aerts, J.T.; Shrout, J.D.; Bohn, P.W.; Sweedler, J.V. MALDI-guided SIMS: Multiscale imaging of metabolites in bacterial biofilms. Anal. Chem. 2014, 86, 9139–9145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef]
- Norris, J.L.; Caprioli, R.M. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 2013, 113, 2309–2342. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.L.; Caprioli, R.M.; Skaar, E.P. Advanced mass spectrometry technologies for the study of microbial pathogenesis. Curr. Opin. Microbiol. 2014, 19, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.L.; Xu, Y.; Straight, P.D.; Dorrestein, P.C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, A.M.S.; Lami, R.; Escoubeyrou, K.; Intertaglia, L.; Mazurek, C.; Doberva, M.; Pérez-Ferrer, P.; Stien, D. Straightforward N-acyl homoserine lactone discovery and annotation by LC-MS/MS-based molecular networking. J. Proteome. Res. 2022, 21, 635–642. [Google Scholar] [CrossRef]
- Davies, S.K.; Fearn, S.; Allsopp, L.P.; Harrison, F.; Ware, E.; Diggle, S.P.; Filloux, A.; McPhail, D.S.; Bundy, J.G. Visualizing antimicrobials in bacterial biofilms: Three dimensional biochemical imaging using TOF-SIMS. mSphere. 2017, 2, e00211-17. [Google Scholar] [CrossRef] [PubMed]
- Wakeman, C.A.; Moore, J.L.; Noto, M.J.; Zhang, Y.; Singleton, M.D.; Prentice, B.M.; Gilston, B.A.; Doster, R.S.; Gaddy, J.A.; Chazin, W.J.; et al. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 2016, 7, 11951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phelan, V.V.; Fang, J.; Dorrestein, P.C. Mass spectrometry analysis of Pseudomonas aeruginosa treated with azithromycin. J. Am. Soc. Mass. Spectrom. 2015, 26, 873–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diggle, S.P.; Lumjiaktase, P.; Dipilato, F.; Winzer, K.; Kunakorn, M.; Barrett, D.A.; Chhabra, S.R.; Camara, M.; Williams, P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2006, 13, 701–710. [Google Scholar] [CrossRef]
- Barbarossa, M.V.; Kuttler, C.; Fekete, A.; Rothballer, M. A delay model for quorum sensing of Pseudomonas putida. Biosystems 2010, 102, 148–156. [Google Scholar] [CrossRef]
- Li, B.; Dunham, S.J.B.; Ellis, J.F.; Lange, J.D.; Smith, J.R.; Yang, N.; King, T.L.; Amaya, K.R.; Arnett, C.M.; Sweedler, J.V. A versatile strategy for characterization and imaging of drip flow microbial biofilms. Anal. Chem. 2018, 90, 6725–6734. [Google Scholar] [CrossRef]
- Baig, N.F.; Dunham, S.J.B.; Morales-Soto, N.; Shrout, J.D.; Sweedler, J.V.; Bohn, P.W. Multimodal chemical imaging of molecular messengers in emerging Pseudomonas aeruginosa bacterial communities. Analyst 2015, 140, 6544–6552. [Google Scholar] [CrossRef] [Green Version]
- Ca’rcamo-Oyarce, G.; Lumjiaktase, P.; Kummerli, R.; Eberl, L. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat. Commun. 2015, 6, 5945–5953. [Google Scholar] [CrossRef] [Green Version]
- Bernabè, G.; Marzaro, G.; Di Pietra, G.; Otero, A.; Bellato, M.; Pauletto, A.; Scarpa, M.; Sut, S.; Chilin, A.; Dall’Acqua, S.; et al. A novel phenolic derivative inhibits AHL-dependent quorum sensing signaling in Pseudomonas aeruginosa. Front. Pharmacol. 2022, 13, 996871. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2014, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Passador, L.; Cook, J.M.; Gambello, M.J.; Rust, L.; Iglewski, B.H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 1993, 260, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
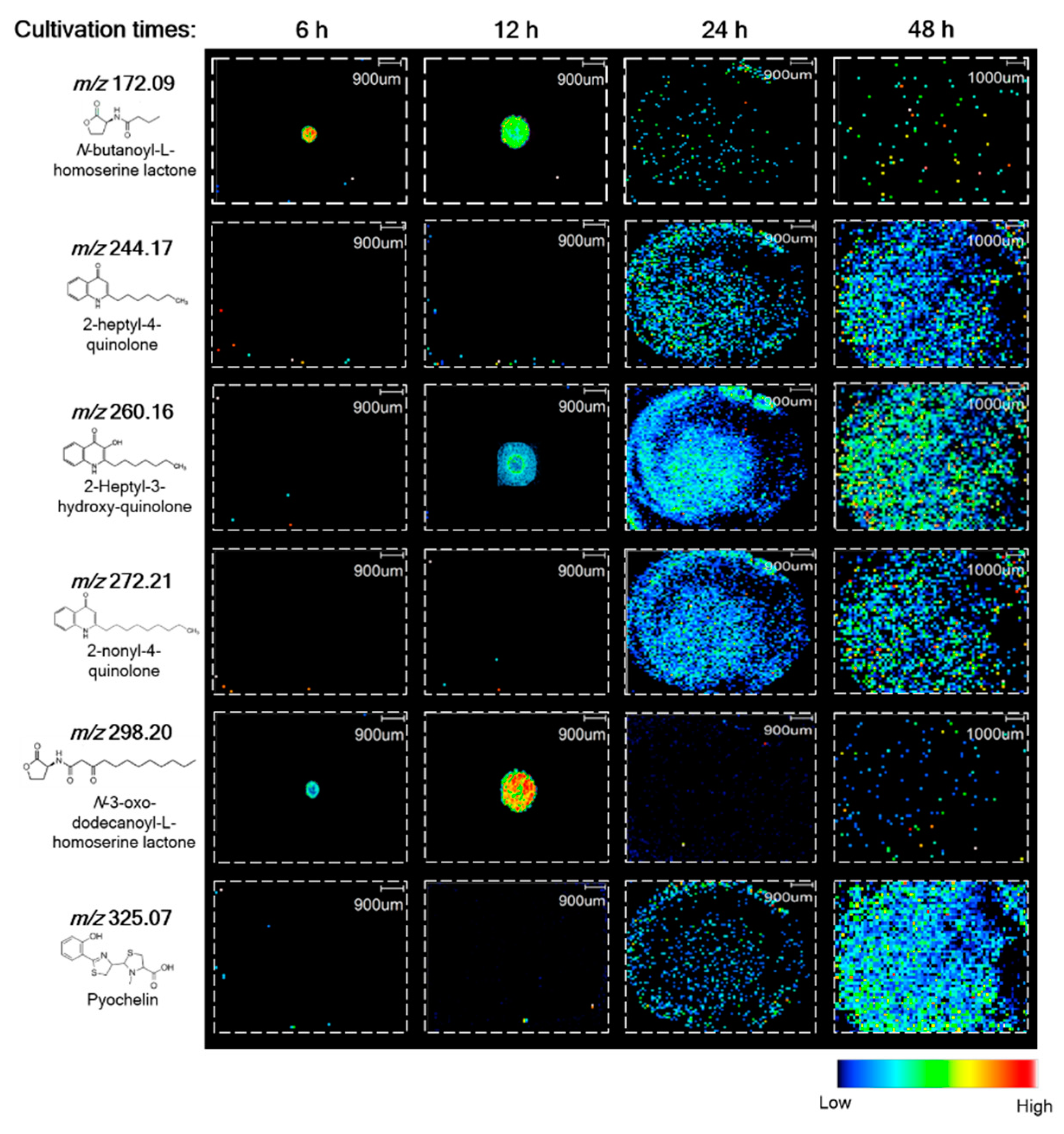
| Compound ID | Molecular Formula | [M + H]+ Theoretical | [M + H]+ Exp. | References |
|---|---|---|---|---|
| C4-HSL | C8H13NO3 | 172.09 | 172.09 | [20] |
| 3-oxo-C12-HSL | C16H27NO4 | 298.20 | 298.20 | [20] |
| HHQ | C16H21NO | 244.17 | 244.17 | [21] |
| PQS | C16H21NO2 | 260.17 | 260.16 | [21] |
| NHQ | C18H25NO | 272.21 | 272.21 | [21] |
| Pyochelin | C14H16N2O3S2 | 325.06 | 325.07 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitchapa, R.; Dissook, S.; Putri, S.P.; Fukusaki, E.; Shimma, S. MALDI Mass Spectrometry Imaging Reveals the Existence of an N-Acyl-homoserine Lactone Quorum Sensing System in Pseudomonas putida Biofilms. Metabolites 2022, 12, 1148. https://doi.org/10.3390/metabo12111148
Pitchapa R, Dissook S, Putri SP, Fukusaki E, Shimma S. MALDI Mass Spectrometry Imaging Reveals the Existence of an N-Acyl-homoserine Lactone Quorum Sensing System in Pseudomonas putida Biofilms. Metabolites. 2022; 12(11):1148. https://doi.org/10.3390/metabo12111148
Chicago/Turabian StylePitchapa, Rattanaburi, Sivamoke Dissook, Sastia Prama Putri, Eiichiro Fukusaki, and Shuichi Shimma. 2022. "MALDI Mass Spectrometry Imaging Reveals the Existence of an N-Acyl-homoserine Lactone Quorum Sensing System in Pseudomonas putida Biofilms" Metabolites 12, no. 11: 1148. https://doi.org/10.3390/metabo12111148
APA StylePitchapa, R., Dissook, S., Putri, S. P., Fukusaki, E., & Shimma, S. (2022). MALDI Mass Spectrometry Imaging Reveals the Existence of an N-Acyl-homoserine Lactone Quorum Sensing System in Pseudomonas putida Biofilms. Metabolites, 12(11), 1148. https://doi.org/10.3390/metabo12111148









