Isotopic Tracer for Absolute Quantification of Metabolites of the Pentose Phosphate Pathway in Bacteria
Abstract
1. Introduction
2. Pentose Phosphate Pathway
3. Isotopic Tracer
3.1. Studies of PPP Metabolism by Isotopic Tracer Method
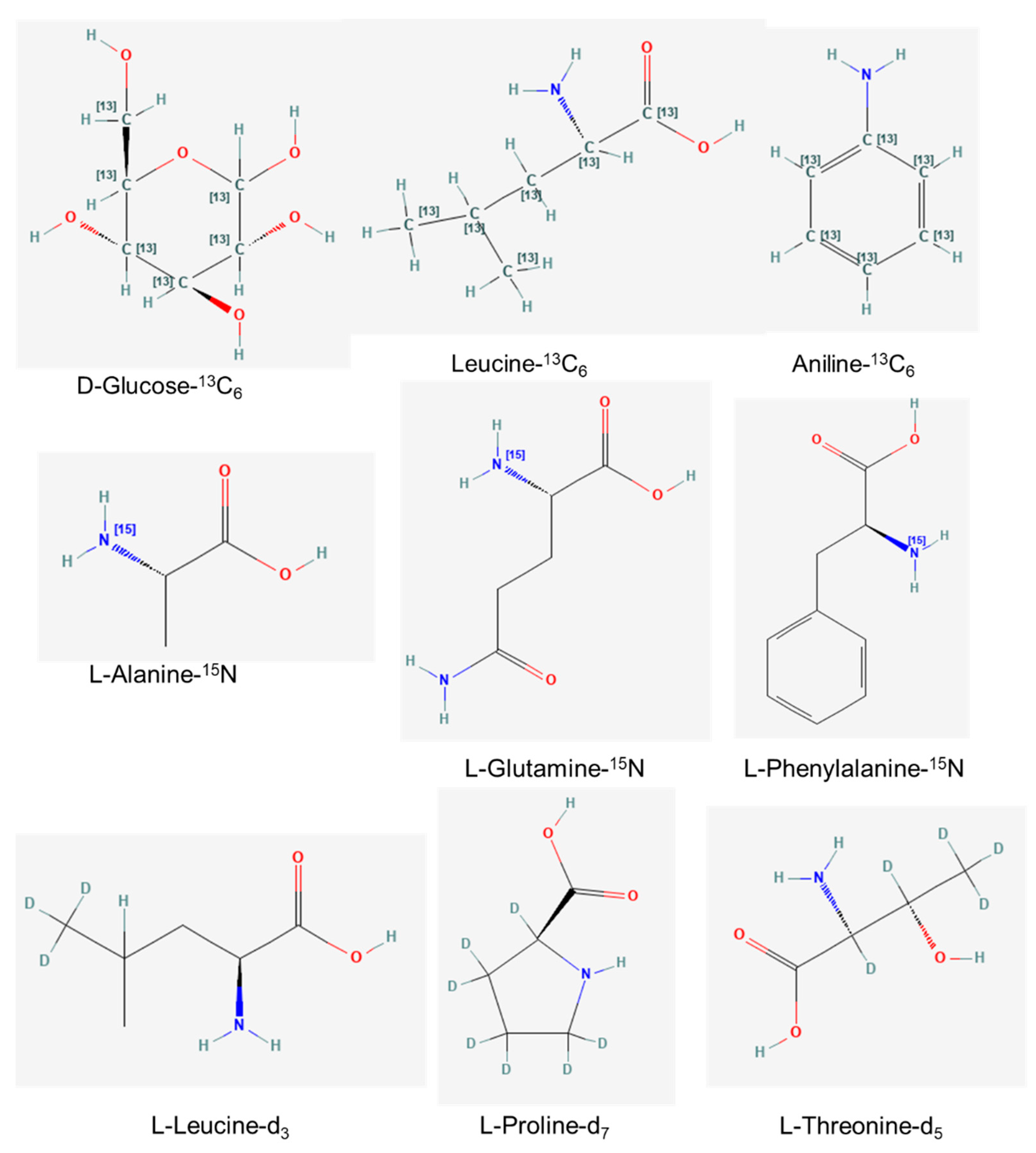
3.2. In Vivo Synthesis of Metabolite-Labeled Isotopes
3.3. In Vitro Synthesis of Metabolite-Labeled Isotopes
3.4. Aniline Tagging Method
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kovářová, J.; Barrett, M.P. The pentose phosphate pathway in parasitic Trypanosomatids. Trends Parasitol. 2016, 32, 622–634. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Bertels, L.K.; Murillo, L.F.; Heinisch, J.J. The pentose phosphate pathway in yeasts–more than a poor cousin of glycolysis. Biomolecules 2021, 11, 725. [Google Scholar] [CrossRef]
- Werner, C.; Doenst, T.; Schwarzer, M. Metabolic pathways and cycles. In The Scientist Guide to Cardiac Metabolism; Academic Press: Cambridge, MA, USA, 2016; pp. 39–55. [Google Scholar] [CrossRef]
- Lucarelli, G.; Galleggiante, V.; Rutigliano, M.; Sanguedolce, F.; Cagiano, S.; Bufo, P.; Lastilla, G.; Maiorano, E.; Ribatti, D.; Giglio, A.; et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget 2015, 6, 13371–13386. [Google Scholar] [CrossRef]
- Ge, T.; Yang, J.; Zhou, S.; Wang, Y.; Li, Y.; Tong, X. The role of the pentose phosphate pathway in diabetes and cancer. Front. Endocrinol. 2020, 11, 365. [Google Scholar] [CrossRef]
- Luo, X.; Liu, J.; Wang, H.; Lu, H. Metabolomics identified new biomarkers for the precise diagnosis of pancreatic cancer and associated tissue metastasis. Pharmacol. Res. 2020, 156, 104805. [Google Scholar] [CrossRef]
- Maifiah, M.H.M.; Creek, D.J.; Nation, R.L.; Forrest, A.; Tsuji, B.T.; Velkov, T.; Li, J. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci. Rep. 2017, 7, srep45527. [Google Scholar] [CrossRef]
- Han, M.L.; Liu, X.; Velkov, T.; Lin, Y.W.; Zhu, Y.; Li, M.; Yu, H.H.; Zhou, Z.; Creek, D.; Zhang, J.; et al. Metabolic analyses revealed time-dependent synergistic killing by colistin and aztreonam combination against multidrug-resistant Acinetobacter baumannii. Front. Microbiol. 2018, 9, 2776. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, J.; Maifiah, M.H.M.; Velkov, T.; Schreiber, F.; Li, J. Metabolic responses to polymyxin treatment in Acinetobacter baumannii ATCC 19606: Integrating transcriptomics and metabolomics with genome-scale metabolic modeling. mSystems 2019, 4, e00157-18. [Google Scholar] [CrossRef]
- Lin, Y.W.; Han, M.L.; Zhao, J.; Zhu, Y.; Rao, G.; Forrest, A.; Song, J.; Kaye, K.S.; Hertzog, P.; Purcell, A.; et al. Synergistic combination of polymyxin B and enrofloxacin induced metabolic perturbations in extensive drug-resistant Pseudomonas aeruginosa. Front. Pharmacol. 2019, 10, 1146. [Google Scholar] [CrossRef]
- Zhao, S.; Li, L. Chemical derivatization in LC-MS-based metabolomics study. TrAC Trends Anal. Chem. 2020, 131, 115988. [Google Scholar] [CrossRef]
- Wu, L.; Mashego, M.R.; van Dam, J.C.; Proell, A.M.; Vinke, J.L.; Ras, C.; van Winden, W.A.; van Gulik, W.M.; Heijnen, J.J. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 2005, 336, 164–171. [Google Scholar] [CrossRef]
- Mashego, M.R.; Wu, L.; van Dam, J.C.; Ras, C.; Vinke, J.L.; van Winden, W.A.; van Gulik, W.M.; Heijnen, J.J. MIRACLE: Mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 2004, 85, 620–628. [Google Scholar] [CrossRef]
- Huang, T.; Armbruster, M.R.; Coulton, J.B.; Edwards, J.L. Chemical tagging in mass spectrometry for systems biology. Anal. Chem. 2018, 91, 109–125. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Stupp, G.S.; Ajredini, R.; Lee-McMullen, B.; Beecher, C.; Edison, A.S. An overview of methods using 13C for improved compound identification in metabolomics and natural products. Front. Plant Sci. 2015, 6, 611. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, M.S.; Swarts, B.M.; Fox, D.M.; Lim, S.A.; Bertozzi, C.R. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev. 2015, 39, 184–202. [Google Scholar] [CrossRef]
- Srivastava, A.; Kowalski, G.M.; Callahan, D.L.; Meikle, P.J.; Creek, D.J. Strategies for extending metabolomics studies with stable isotope labelling and fluxomics. Metabolites 2016, 6, 32. [Google Scholar] [CrossRef]
- Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006, 2, 62. [Google Scholar] [CrossRef]
- Buescher, J.M.; Antoniewicz, M.R.; Boros, L.G.; Burgess, S.C.; Brunengraber, H.; Clish, C.B.; DeBerardinis, R.J.; Feron, O.; Frezza, C.; Ghesquiere, B.; et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015, 34, 189–201. [Google Scholar] [CrossRef]
- Jannasch, A.; Sedlak, M.; Adamec, J. Quantification of Pentose Phosphate Pathway (PPP) Metabolites by Liquid Chromatography-Mass Spectrometry. In Metabolic Profiling; Metz, T.O., Ed.; Humana Press: Totowa, NJ, USA, 2011; Volume 708, pp. 159–171. [Google Scholar] [CrossRef]
- Cadière, A.; Ortiz-Julien, A.; Camarasa, C.; Dequin, S. Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway. Metab. Eng. 2011, 13, 263–271. [Google Scholar] [CrossRef]
- Clasquin, M.F.; Melamud, E.; Singer, A.; Gooding, J.R.; Xu, X.; Dong, A.; Cui, H.; Campagna, S.R.; Savchenko, A.; Yakunin, A.F.; et al. Riboneogenesis in yeast. Cell 2011, 145, 969–980. [Google Scholar] [CrossRef]
- Zhang, Q.; Bartels, D. Octulose: A forgotten metabolite? J. Exp. Bot. 2017, 68, 5689–5694. [Google Scholar] [CrossRef]
- Jin, L.; Zhou, Y. Crucial role of the pentose phosphate pathway in malignant tumors (Review). Oncol. Lett. 2019, 17, 4213–4221. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Sayed, N.; Ditsworth, D.; Thompson, C.B. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008, 18, 54–61. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhu, Y.; Zhuge, T.; Li, B.; Gu, C. Metabolomics analysis discovers estrogen altering cell proliferation via the pentose phosphate pathway in infertility patient endometria. Front. Endocrinol. 2021, 12, 79114. [Google Scholar] [CrossRef]
- Bolaños, J.P.; Almeida, A.; Moncada, S. Glycolysis: A bioenergetic or a survival pathway? Trends Biochem. Sci. 2010, 35, 145–149. [Google Scholar] [CrossRef]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef]
- Igoillo-Esteve, M.; Maugeri, D.; Stern, A.L.; Beluardi, P.; Cazzulo, J.J. The pentose phosphate pathway in Trypanosoma cruzi: A potential target for the chemotherapy of Chagas disease. An. Acad. Bras. Cien. 2007, 79, 649–663. [Google Scholar] [CrossRef]
- Maugeri, D.A.; Cazzulo, J.J.; Burchmore, R.J.S.; Barrett, M.P.; Ogbunude, P.O.J. Pentose phosphate metabolism in Leishmania mexicana. Mol. Biochem. Parasitol. 2003, 130, 117–125. [Google Scholar] [CrossRef]
- Taylor, P.L.; Blakely, K.M.; De Leon, G.P.; Walker, J.R.; McArthur, F.; Evdokimova, E.; Zhang, K.; Valvano, M.; Wright, G.; Junop, M.S. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J. Biol. Chem. 2008, 283, 2835–2845. [Google Scholar] [CrossRef]
- Alteri, C.J.; Mobley, H.L.T. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 2012, 15, 3–9. [Google Scholar] [CrossRef]
- Hussein, M.; Han, M.L.; Zhu, Y.; Zhou, Q.; Lin, Y.W.; Hancock, R.E.W.; Hoyer, D.; Creek, D.J.; Li, J.; Velkov, T. Metabolomics study of the synergistic killing of polymyxin B in combination with amikacin against polymyxin-susceptible and -resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2020, 64, e01587-19. [Google Scholar] [CrossRef]
- Hussein, M.; Hu, X.; Paulin, O.K.A.; Crawford, S.; Zhou, Q.T.; Baker, M.; Schneider-Futschik, E.K.; Zhu, Y.; Li, J.; Velkov, T. Polymyxin B combinations with FDA-approved non-antibiotic phenothiazine drugs targeting multi-drug resistance of Gram-negative pathogens. Comput. Struct. Biotechnol. J. 2020, 18, 2247–2258. [Google Scholar] [CrossRef]
- Han, M.L.; Liu, X.; Velkov, T.; Lin, Y.W.; Zhu, Y.; Creek, D.J.; Barlow, C.K.; Yu, H.H.; Zhou, Z.; Zhang, J.; et al. Comparative metabolomics reveals key pathways associated with the synergistic killing of colistin and sulbactam combination against multidrug-resistant Acinetobacter baumannii. Front. Pharmacol. 2019, 10, 754. [Google Scholar] [CrossRef]
- Abdul Rahim, N.; Zhu, Y.; Cheah, S.E.; Johnson, M.D.; Yu, H.H.; Sidjabat, H.E.; Butler, M.S.; Cooper, M.A.; Fu, J.; Paterson, D.L.; et al. Synergy of the polymyxin-chloramphenicol combination against New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae is predominately driven by chloramphenicol. ACS Infect. Dis. 2021, 7, 1584–1595. [Google Scholar] [CrossRef]
- Hussein, M.; Han, M.L.; Zhu, Y.; Schneider-Futschik, E.K.; Hu, X.; Zhou, Q.T.; Lin, Y.-W.; Anderson, D.; Creek, D.; Hoyer, D.; et al. Mechanistic insights from global metabolomics studies into synergistic bactericidal effect of a polymyxin B combination with tamoxifen against cystic fibrosis MDR Pseudomonas aeruginosa. Comput. Struct. Biotechnol. J. 2018, 16, 587–599. [Google Scholar] [CrossRef]
- Creek, D.J.; Mazet, M.; Achcar, F.; Anderson, J.; Kim, D.H.; Kamour, R.; Morand, P.; Millerioux, Y.; Biran, M.; Kerkhoven, E.J.; et al. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015, 11, e1004689. [Google Scholar] [CrossRef]
- Wushensky, J.A.; Youngster, T.; Mendonca, C.M.; Aristilde, L. Flux connections between gluconate pathway, glycolysis, and pentose-phosphate pathway during carbohydrate metabolism in Bacillus megaterium QM B1551. Front. Microbiol. 2018, 9, 2789. [Google Scholar] [CrossRef]
- Yang, W.C.; Sedlak, M.; Regnier, F.E.; Mosier, N.; Ho, N.; Adamec, J. Simultaneous quantification of metabolites involved in central carbon and energy metabolism using reversed-phase liquid chromatography-mass spectrometry and in vitro 13C labeling. Anal. Chem. 2008, 80, 9508–9516. [Google Scholar] [CrossRef]
- Vilkhovoy, M.; Dai, D.; Vadhin, S.; Adhikari, A.; Varner, J.D. Absolute quantification of cell-free protein synthesis metabolism by reversed-phase liquid chromatography-mass spectrometry. J. Vis. Exp. 2019, 2019, e60329. [Google Scholar] [CrossRef]
- Katz, J.; Wood, H.G. The use of C14O2 yields from glucose-1- and -6-C14 for the evaluation of the pathways of glucose metabolism. J. Biol. Chem. 1963, 238, 517–523. [Google Scholar] [CrossRef]
- Sable, H.Z. Pentose metabolism in extracts of yeast and mammalian tissues. Biochim. Biophys. Acta 1952, 8, 687–697. [Google Scholar] [CrossRef]
- Novello, F.; McLean, P. The pentose phosphate pathway of glucose metabolism. Measurement of the non-oxidative reactions of the cycle. Biochem. J. 1968, 107, 775–791. [Google Scholar] [CrossRef]
- Becker, M.A. Patterns of phosphoribosylpyrophosphate and ribose 5 phosphate concentration and generation in fibroblasts from patients with gout and purine overproduction. J. Clin. Investig. 1976, 57, 308–318. [Google Scholar] [CrossRef]
- King, M.T.; Passonneau, J.V.; Veech, R.L. Radiometric measurement of phosphoribosylpyrophosphate and ribose 5-phosphate by enzymatic procedures. Anal. Biochem. 1990, 187, 179–186. [Google Scholar] [CrossRef]
- Shih, Y.C.; Hsiao, J.T.; Sheu, F. Molecules feasibility of utilizing stable-isotope dimethyl labeling in liquid chromatography-tandem mass spectrometry-based determination for food allergens-case of Kiwifruit. Molecules 2019, 24, 1920. [Google Scholar] [CrossRef]
- Weindl, D.; Wegner, A.; Hiller, K. Metabolome-wide analysis of stable isotope labeling-Is it worth the effort? Front. Physiol. 2015, 6, 344. [Google Scholar] [CrossRef]
- Chokkathukalam, A.; Kim, D.-H.; Barrett, M.P.; Breitling, R.; Creek, D.J. Stable isotope-labeling studies in metabolomics: New insights into structure and dynamics of metabolic networks. Bioanalysis 2014, 6, 511–524. [Google Scholar] [CrossRef]
- Duckwall, C.; Murphy, T.; Young, J. Mapping cancer cell metabolism with 13C flux analysis: Recent progress and future challenges. J. Carcinog. 2013, 12, 13. [Google Scholar] [CrossRef]
- Niedenführ, S.; Wiechert, W.; Nöh, K. How to measure metabolic fluxes: A taxonomic guide for 13C fluxomics. Curr. Opin. Biotechnol. 2015, 34, 82–90. [Google Scholar] [CrossRef]
- Kim, I.-Y.; Suh, S.-H.; Lee, I.-K.; Wolfe, R.R. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp. Mol. Med. 2016, 48, e203. [Google Scholar] [CrossRef] [PubMed]
- Triebl, A.; Wenk, M.R. Biomolecules analytical considerations of stable isotope labelling in lipidomics. Biomolecules 2018, 8, 151. [Google Scholar] [CrossRef] [PubMed]
- Grocholska, P.; Leonidov Tsakovski, S. Trends in the Hydrogen−Deuterium exchange at the carbon centers. Preparation of Internal Standards for quantitative analysis by LC-MS. Molecules 2021, 26, 2989. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, S.; Raijmakers, R.; Heck, A.J.R.; Mohammed, S. Evaluation of the deuterium isotope effect in zwitterionic hydrophilic interaction liquid chromatography separations for implementation in a quantitative proteomic approach. Anal. Chem. 2011, 83, 8352–8356. [Google Scholar] [CrossRef]
- Zhao, S.; Li, L. Chemical isotope labeling LC-MS for metabolomics. In Cancer Metabolomics; Hu, S., Ed.; Springer: Cham, Switzerland, 2007; pp. 1–18. [Google Scholar]
- Ahn, W.S.; Crown, S.B.; Antoniewicz, M.R. Evidence for transketolase-like TKTL1 flux in CHO cells based on parallel labeling experiments and 13C-metabolic flux analysis. Metab. Eng. 2016, 37, 72–78. [Google Scholar] [CrossRef]
- Brekke, E.M.F.; Walls, A.B.; Schousboe, A.; Waagepetersen, H.S.; Sonnewald, U. Quantitative importance of the pentose phosphate pathway determined by incorporation of 13C from 2-13C and 3-13C glucose into TCA cycle intermediates and neurotransmitter amino acids in functionally intact neurons. J. Cereb. Blood Flow Metab. 2012, 32, 1788–1799. [Google Scholar] [CrossRef]
- Crown, S.B.; Indurthi, D.C.; Ahn, W.S.; Choi, J.; Papoutsakis, E.T.; Antoniewicz, M.R. Resolving the TCA cycle and pentose-phosphate pathway of Clostridium acetobutylicum ATCC 824 using isotopomer analysis, in vitro re-citrate synthase activities and expression analysis. Biotechnol. J. 2011, 6, 300–305. [Google Scholar] [CrossRef]
- Crown, S.B.; Ahn, W.S.; Antoniewicz, M.R. Rational design of 13C-labeling experiments for metabolic flux analysis in mammalian cells. BMC Syst. Biol. 2012, 6, 43. [Google Scholar] [CrossRef]
- Fan, J.; Ye, J.; Kamphorst, J.J.; Shlomi, T.; Thompson, C.B.; Rabinowitz, J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014, 510, 298–302. [Google Scholar] [CrossRef]
- Lee, W.N.P.; Boros, L.G.; Puigjaner, J.; Bassilian, S.; Lim, S.; Cascante, M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. Am. J. Physiol. Endocrinol. Metab. 1998, 274, E843–E851. [Google Scholar] [CrossRef]
- Nakahigashi, K.; Toya, Y.; Ishii, N.; Soga, T.; Hasegawa, M.; Watanabe, H.; Takai, Y.; Honma, M.; Mori, H.; Tomita, M. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 2009, 5, 306. [Google Scholar] [CrossRef] [PubMed]
- Antoniewicz, M.R. A guide to 13C metabolic flux analysis for the cancer biologist. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef]
- Lewis, C.A.; Parker, S.J.; Fiske, B.P.; McCloskey, D.; Gui, D.Y.; Green, C.R.; Vokes, N.I.; Feist, A.M.; Heiden, M.G.V.; Metallo, C.M. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 2014, 55, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.S.; Antoniewicz, M.R. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab. Eng. 2011, 13, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Metabolic labeling and chemoselective ligation|Thermo Fisher Scientific—MY. Available online: https://www.thermofisher.com/my/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/metabolic-labeling-chemoselective-ligation.html (accessed on 22 April 2022).
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Heinzle, E. Isotope labeling experiments in metabolomics and fluxomics. WIREs Syst. Biol. Med. 2012, 4, 261–272. [Google Scholar] [CrossRef]
- Paul Lee, W.N.; Wahjudi, P.N.; Xu, J.; Go, V.L. Tracer-based Metabolomics: Concepts and Practices. Clin. Biochem. 2010, 43, 1269–1277. [Google Scholar] [CrossRef][Green Version]
- Suchanek, M.; Radzikowska, A.; Thiele, C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2005, 2, 261–268. [Google Scholar] [CrossRef]
- Crivat, G.; Taraska, J.W. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 2012, 30, 8–16. [Google Scholar] [CrossRef]
- Rappsilber, J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 2011, 173, 530–540. [Google Scholar] [CrossRef]

| Isotopic Tracer | Sample | Method | PPP Metabolites | Reference |
|---|---|---|---|---|
| U-13C labeled medium | Saccharomyces cerevisiae | Rapid sampling and mix of chemostat labeled with 12C-labeled steady state glucose and 13C-labeled. | G6P, F6P | [14] |
| U-13C6-glucose | S. cerevisiae | Feed the culture with U-13C6-glucose and the sample is added to the unlabeled calibration standards as IS. | G6P, F6P | [13] |
| U-12C and U-13C-glucose | Trypanosoma brucei | Replacing the growth media with media containing 12C-labeled and 13C-labeled glucose. | R5P, F6P | [39] |
| U-13C6-glucose | Bacillus megaterium | Addition of sample into U-13C6-glucose agar plates, followed by the continuation of the culture at regular intervals for isotopic switches | G6P, F6P, 6PG, R5P, S7P, E4P | [40] |
| 12C6- aniline and 13C6-aniline | S. cerevisiae | Tagging of internal standards with 13C6-aniline and derivatization of compounds in the sample with 12C6-aniline | G6P, F6P, DR5P, G3P, DE4P, DR5P | [41] |
| S. cerevisiae | G6P, F6P, DR5P, G3P, 6PG, DE4P, DR5P, DS7P, X5P | [21] | ||
| Escherichia coli | G6P, F6P, DR5P, G3P, 6PG, DE4P, DR5P, DS7P | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohd Kamal, K.; Mahamad Maifiah, M.H.; Zhu, Y.; Abdul Rahim, N.; Hashim, Y.Z.H.-Y.; Abdullah Sani, M.S. Isotopic Tracer for Absolute Quantification of Metabolites of the Pentose Phosphate Pathway in Bacteria. Metabolites 2022, 12, 1085. https://doi.org/10.3390/metabo12111085
Mohd Kamal K, Mahamad Maifiah MH, Zhu Y, Abdul Rahim N, Hashim YZH-Y, Abdullah Sani MS. Isotopic Tracer for Absolute Quantification of Metabolites of the Pentose Phosphate Pathway in Bacteria. Metabolites. 2022; 12(11):1085. https://doi.org/10.3390/metabo12111085
Chicago/Turabian StyleMohd Kamal, Khairunnisa, Mohd Hafidz Mahamad Maifiah, Yan Zhu, Nusaibah Abdul Rahim, Yumi Zuhanis Has-Yun Hashim, and Muhamad Shirwan Abdullah Sani. 2022. "Isotopic Tracer for Absolute Quantification of Metabolites of the Pentose Phosphate Pathway in Bacteria" Metabolites 12, no. 11: 1085. https://doi.org/10.3390/metabo12111085
APA StyleMohd Kamal, K., Mahamad Maifiah, M. H., Zhu, Y., Abdul Rahim, N., Hashim, Y. Z. H.-Y., & Abdullah Sani, M. S. (2022). Isotopic Tracer for Absolute Quantification of Metabolites of the Pentose Phosphate Pathway in Bacteria. Metabolites, 12(11), 1085. https://doi.org/10.3390/metabo12111085








