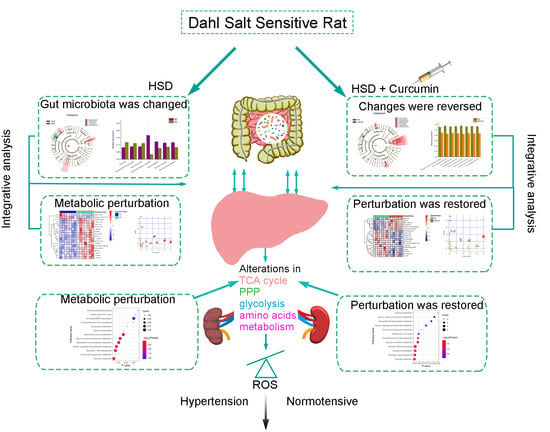
Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Treatment and Sample Collection
2.3. Blood Pressure (BP) Measurement
2.4. Metabolomics Analysis of the Liver and Kidney Tissues
2.5. 16S Ribosomal RNA Sequencing
2.6. Omics Dataset Pretreatment
2.7. Biochemical Analysis
2.8. Statistical Analysis
3. Results
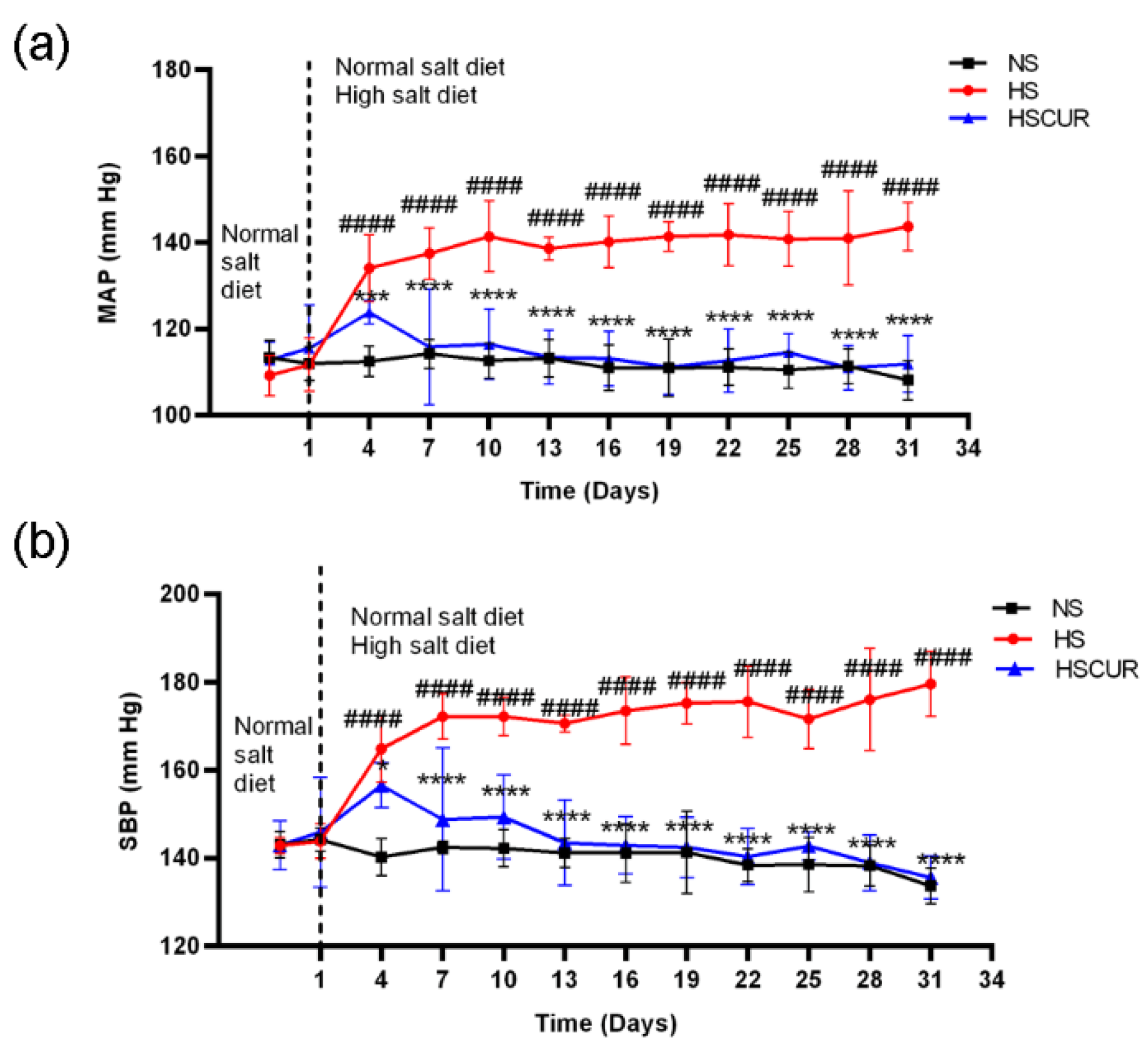
3.1. Characteristics of SS Rats after 5 Weeks of Treatment
3.2. Curcumin Alleviated Salt-Induced Hypertension in SS Rats
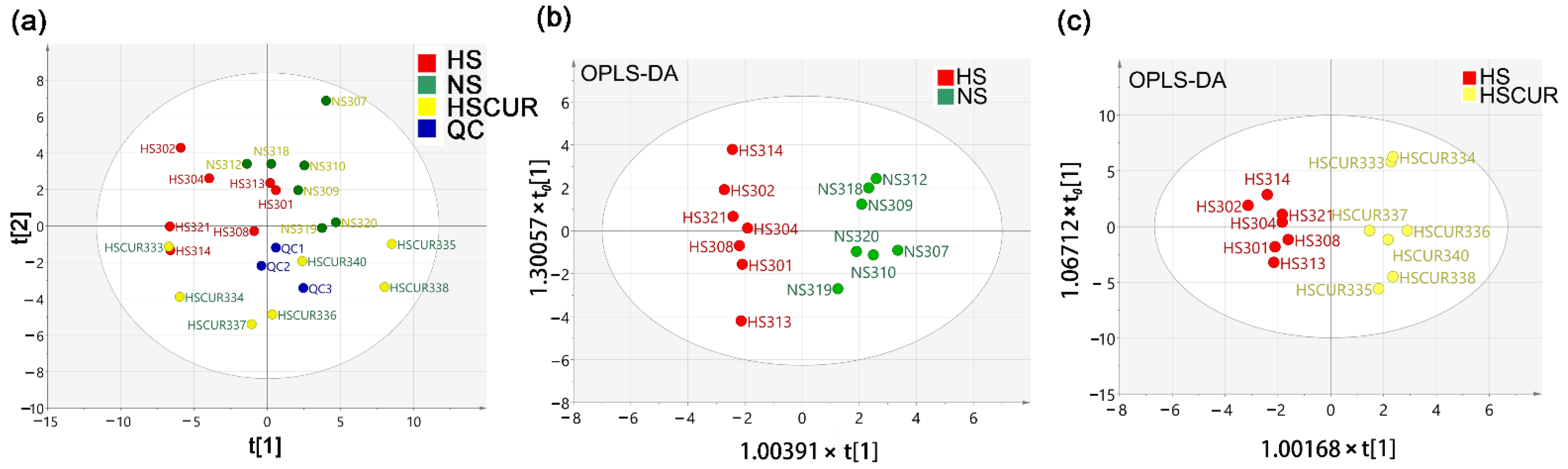
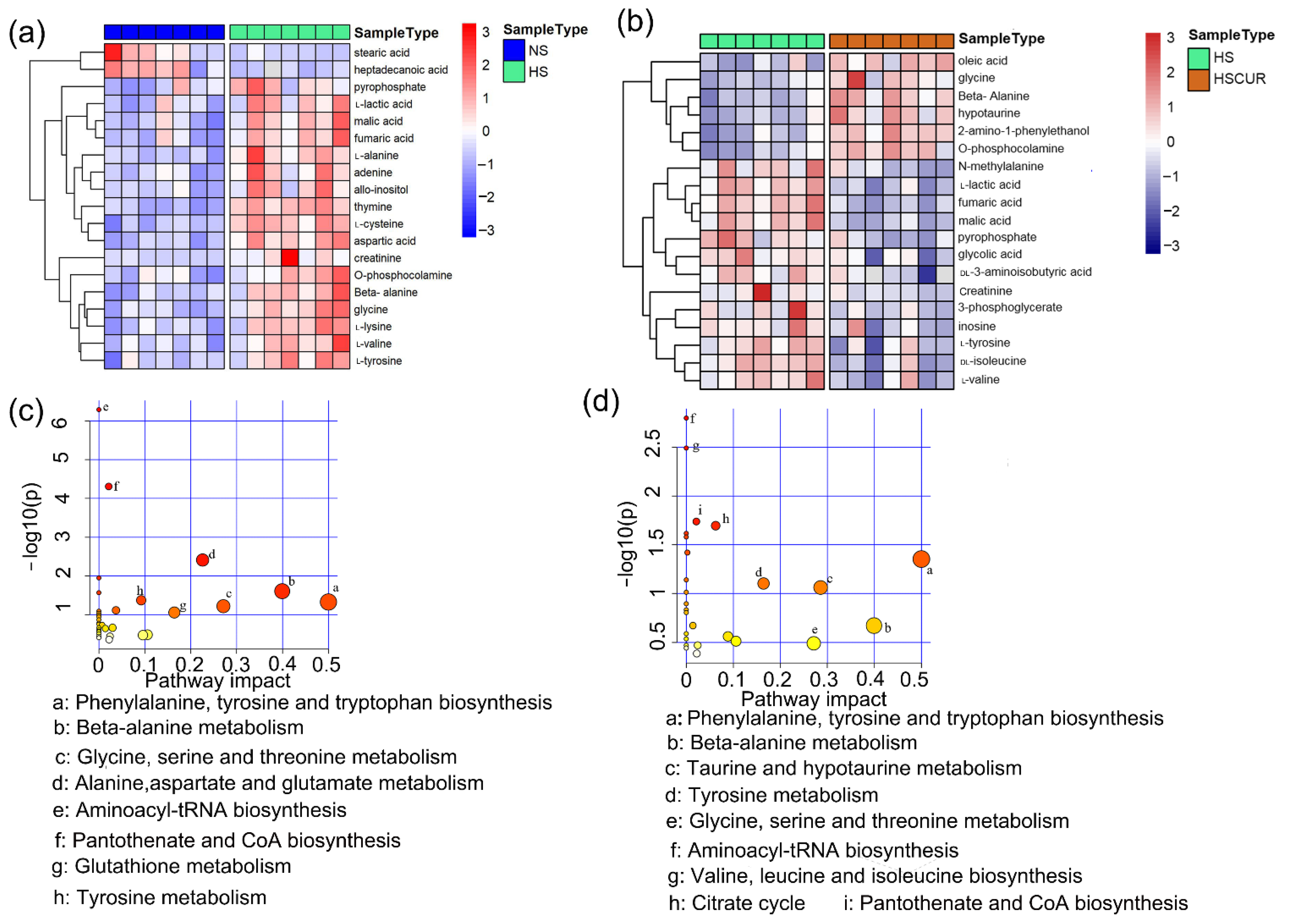
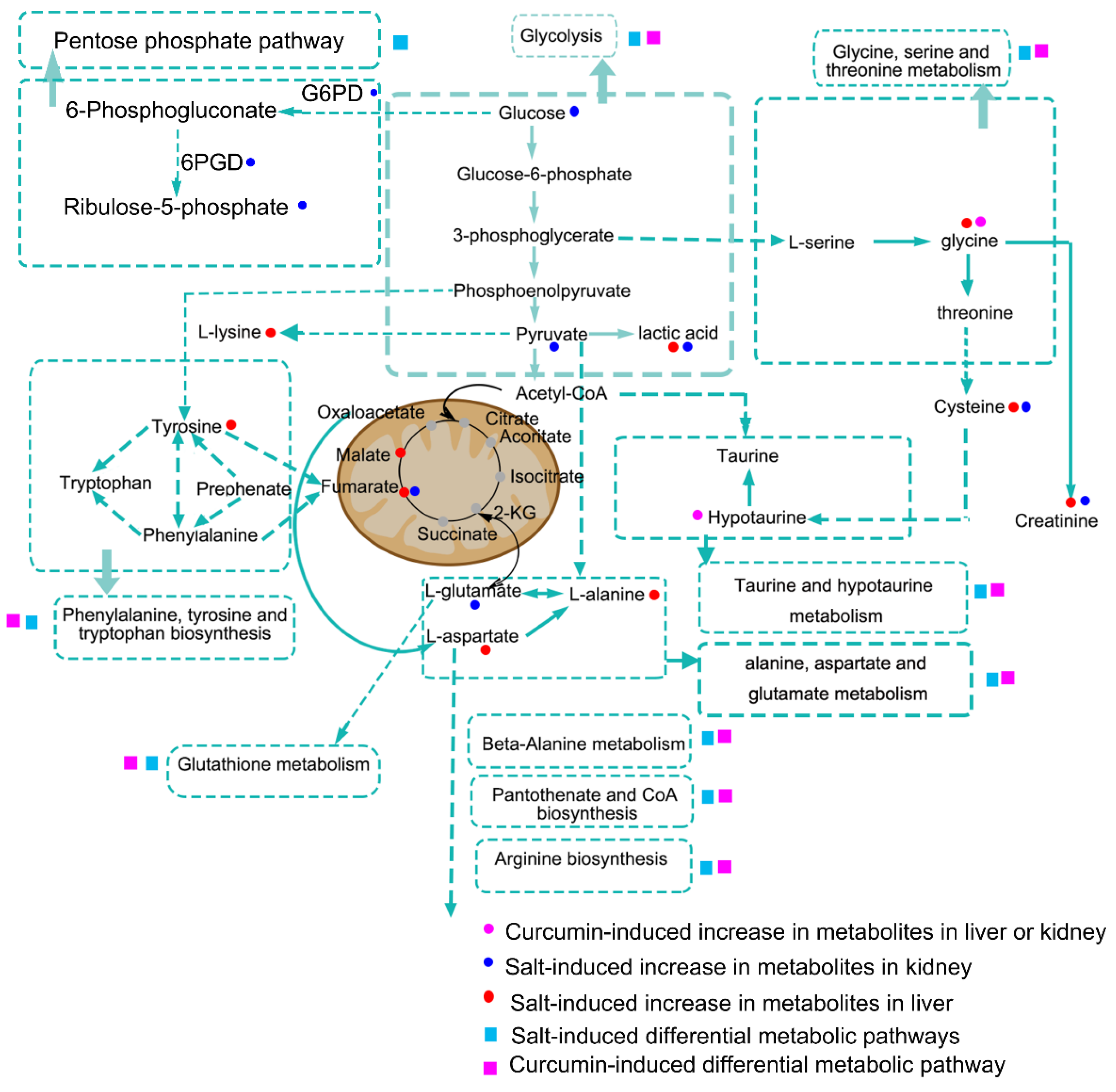
3.3. Curcumin Administration Restored the Salt-Induced Variations in Hepatic Metabolism
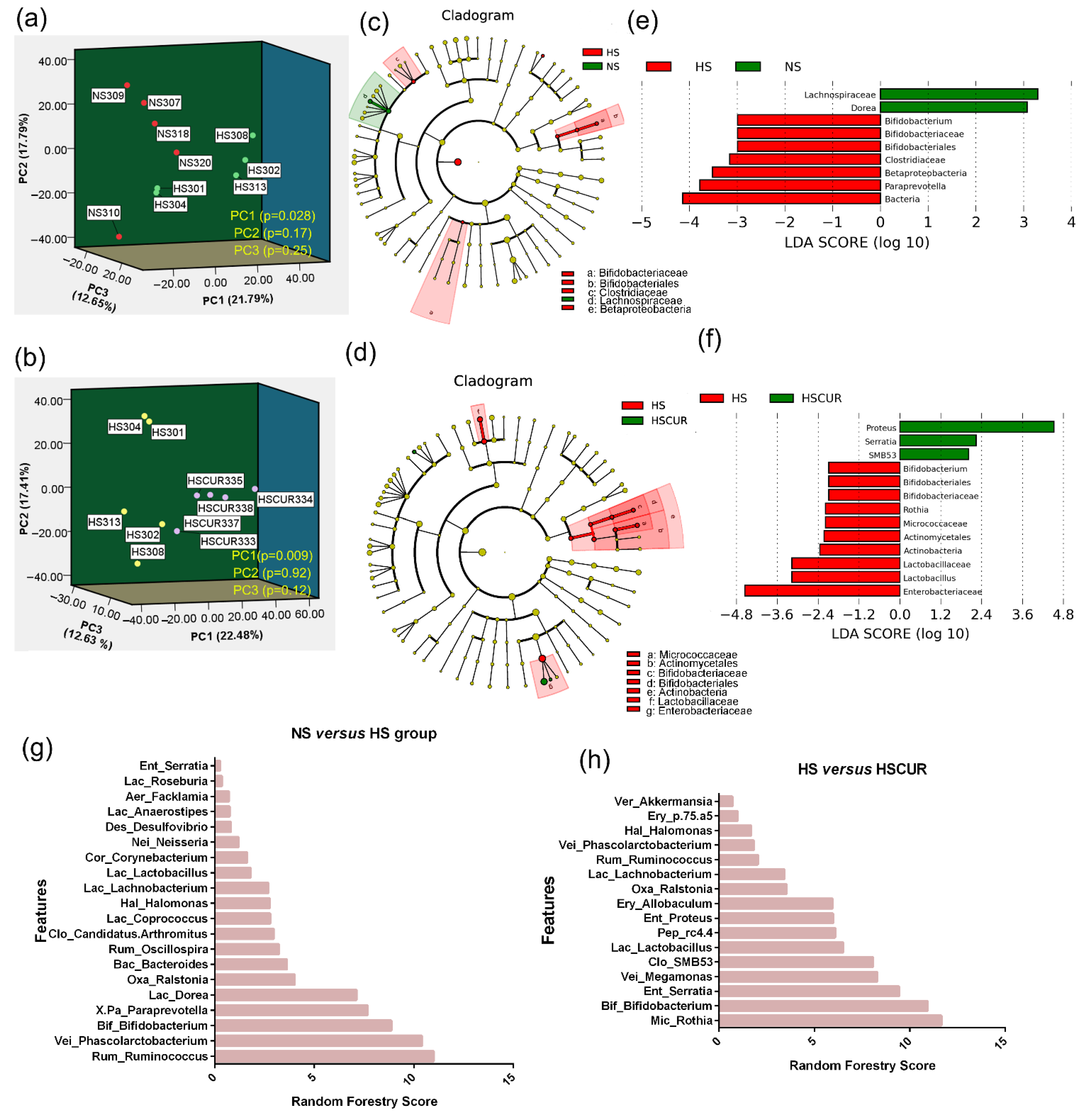
3.4. HSD and Curcumin Administration Changed the Gut Microbial Community and Functional Potential
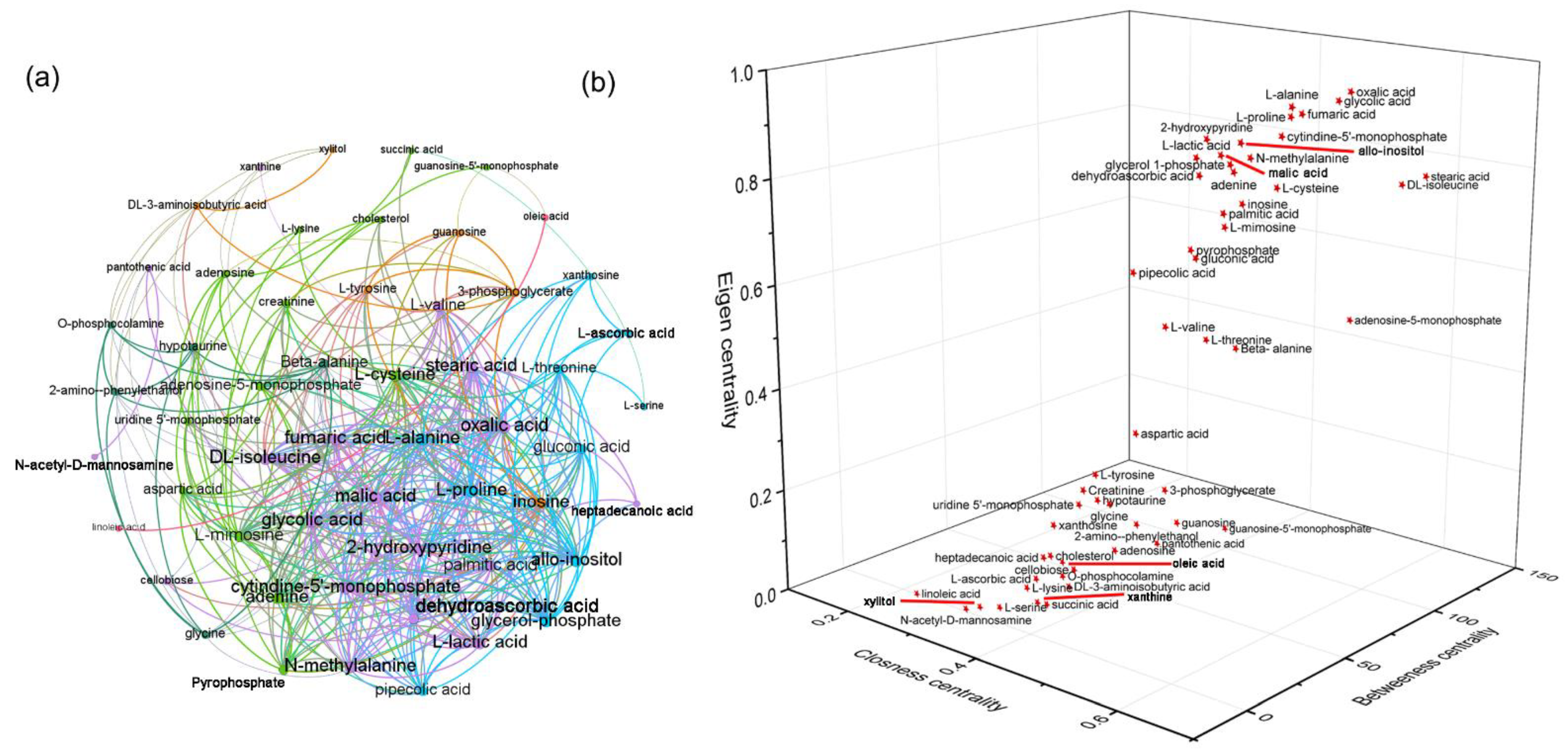
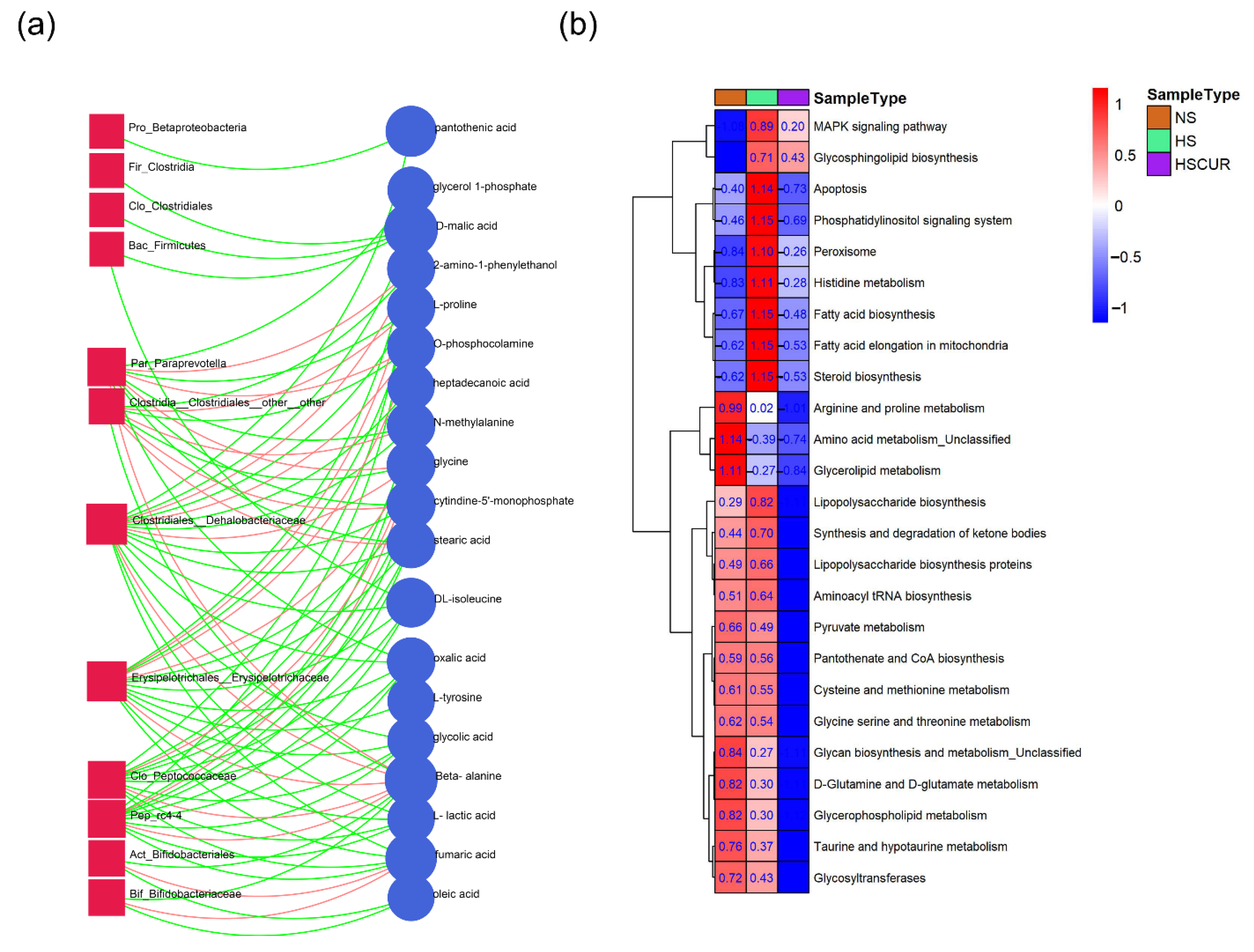
3.5. HSD and Curcumin Administration Influences Hepatic Metabolism by Changing the Gut Microbial Composition
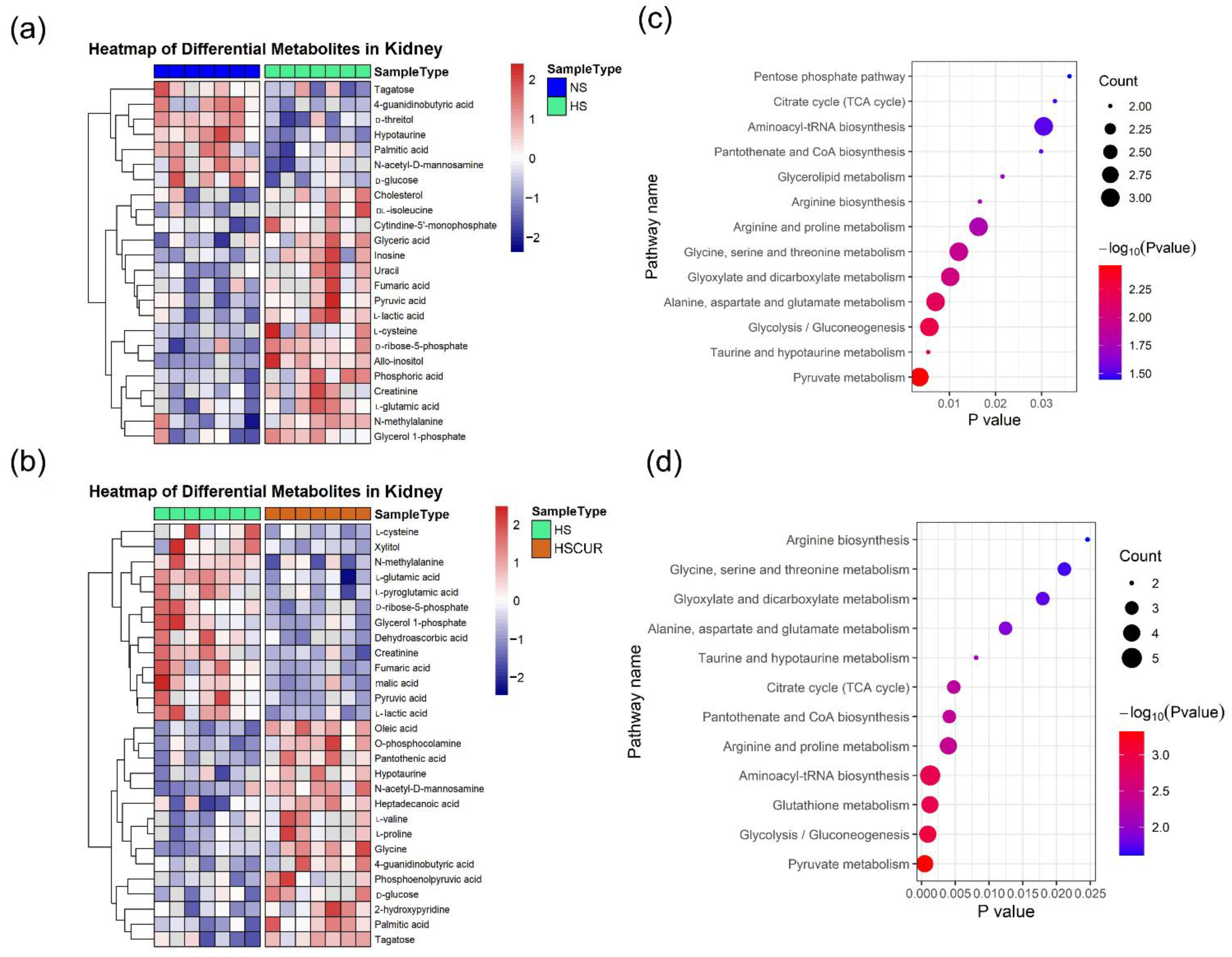
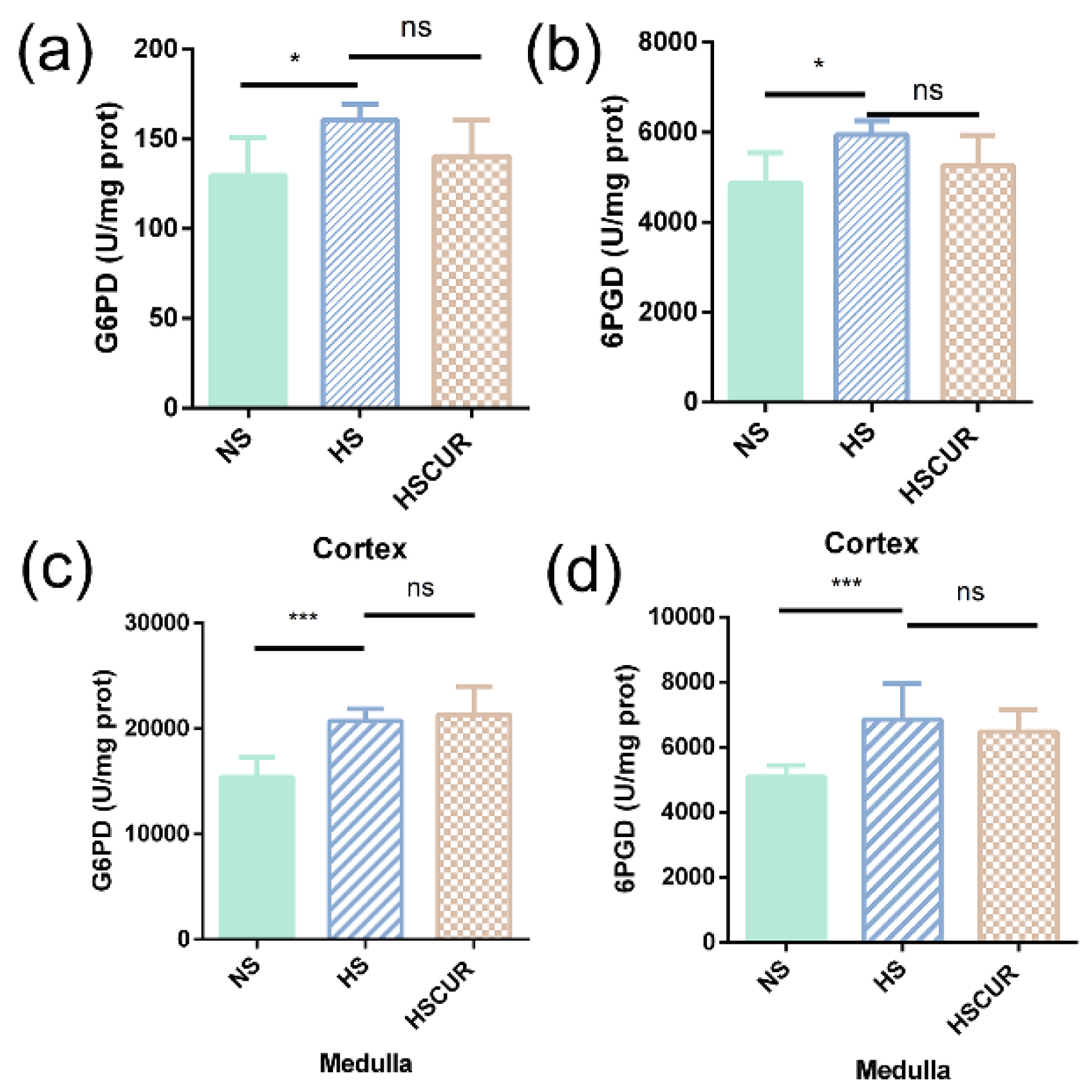
3.6. Curcumin Administration Reversed the Salt-Induced Changes in Renal Metabolism
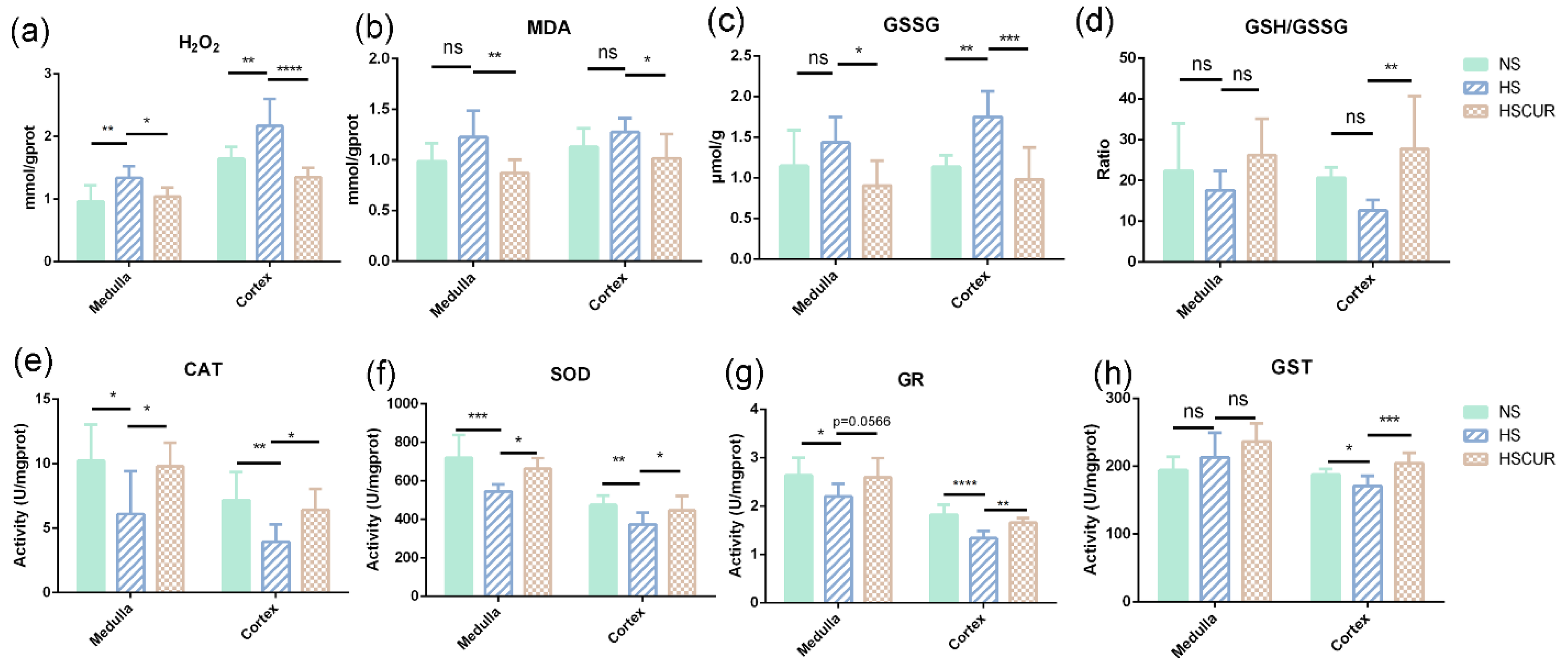
3.7. Effect of Curcumin on REDOX Balance in SS Rats after HSD Feeding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Elijovich, F.; Laffer, C.L.; Sahinoz, M.; Pitzer, A.; Ferguson, J.F.; Kirabo, A. The gut microbiome, inflammation, and salt-sensitive hypertension. Curr. Hypertens. Rep. 2020, 22, 79. [Google Scholar] [CrossRef]
- Härtl, G. WHO issues new guidance on dietary salt and potassium. Cent. Eur. J. Public Health 2013, 21, 16. [Google Scholar]
- Appel, L.J.; Frohlich, E.D.; Hall, J.E.; Pearson, T.A.; Sacco, R.L.; Seals, D.R.; Sacks, F.M.; Smith, S.C., Jr.; Vafiadis, D.K.; Van Horn, L.V. The importance of population-wide sodium reduction as a means to prevent cardiovascular disease and stroke: A call to action from the American Heart Association. Circulation 2011, 123, 1138–1143. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, X.; Jin, Y.; Zhou, L.; Yang, P.; Ahmad, H.; Tian, Z. High salt diet contributes to hypertension by weakening the medullary tricarboxylic acid cycle and antioxidant system in Dahl salt-sensitive rats. Biochimie 2021, 181, 154–161. [Google Scholar] [CrossRef]
- Frame, A.A.; Wainford, R.D. Renal sodium handling and sodium sensitivity. Kidney Res. Clin. Pract. 2017, 36, 117–131. [Google Scholar] [CrossRef]
- Tian, Z.; Liang, M. Renal metabolism and hypertension. Nat. Commun. 2021, 12, 963. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Schreiner, J.; Weber, M.; Loeschke, K. Sodium chloride transport of normal and dietary enlarged rat cecum in vitro. Digestion 1998, 59, 676–682. [Google Scholar] [CrossRef]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, M.; Cui, B.; Chen, X. Antibiotic disruption of the gut microbiota enhances the murine hepatic dysfunction associated with a high-salt diet. Front. Pharmacol. 2022, 13, 829686. [Google Scholar] [CrossRef]
- Gao, B.; Jeong, W.I.; Tian, Z. Liver: An organ with predominant innate immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.H.; Lee, K.J.; Lim, J.S.; Lee, M.Y.; Park, H.J.; Kim, M.Y.; Kim, J.W.; Chung, C.H.; Shin, J.Y.; Kim, H.S.; et al. High dietary sodium intake assessed by estimated 24-h urinary sodium excretion is associated with NAFLD and hepatic fibrosis. PLoS ONE 2015, 10, e0143222. [Google Scholar] [CrossRef] [PubMed]
- Tavares, G.; Venturini, G.; Padilha, K.; Zatz, R.; Pereira, A.C.; Thadhani, R.I.; Rhee, E.P.; Titan, S.M.O. 1,5-Anhydroglucitol predicts CKD progression in macroalbuminuric diabetic kidney disease: Results from non-targeted metabolomics. Metabolomics 2018, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Abais-Battad, J.M.; Alsheikh, A.J.; Pan, X.; Fehrenbach, D.J.; Dasinger, J.H.; Lund, H.; Roberts, M.L.; Kriegel, A.J.; Cowley, A.W., Jr.; Kidambi, S.; et al. Dietary effects on Dahl salt-sensitive hypertension, renal damage, and the T lymphocyte transcriptome. Hypertension 2019, 74, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhou, L.; Chen, M.; Zeng, L.; Ouyang, Y.; Zheng, X.; Chen, X.; Yang, Z.; Tian, Z. Supplementation of amino acids and organic acids prevents the increase in blood pressure induced by high salt in Dahl salt-sensitive rats. Food Funct. 2022, 13, 891–903. [Google Scholar] [CrossRef]
- Daily, J.W.; Yang, M.; Park, S. Efficacy of turmeric extracts and curcumin for alleviating the symptoms of joint arthritis: A systematic review and meta-analysis of randomized clinical trials. J. Med. Food 2016, 19, 717–729. [Google Scholar] [CrossRef]
- Nouri-Vaskeh, M.; Malek Mahdavi, A.; Afshan, H.; Alizadeh, L.; Zarei, M. Effect of curcumin supplementation on disease severity in patients with liver cirrhosis: A randomized controlled trial. Phytother. Res. 2020, 34, 1446–1454. [Google Scholar] [CrossRef]
- Mantzorou, M.; Pavlidou, E.; Vasios, G.; Tsagalioti, E.; Giaginis, C. Effects of curcumin consumption on human chronic diseases: A narrative review of the most recent clinical data. Phytother. Res. 2018, 32, 957–975. [Google Scholar] [CrossRef]
- Zia, A.; Farkhondeh, T.; Pourbagher-Shahri, A.M.; Samarghandian, S. The role of curcumin in aging and senescence: Molecular mechanisms. Biomed. Pharmacother. 2021, 134, 111119. [Google Scholar] [CrossRef]
- Wang, L.; Hou, E.; Wang, L.; Wang, Y.; Yang, L.; Zheng, X.; Xie, G.; Sun, Q.; Liang, M.; Tian, Z. Reconstruction and analysis of correlation networks based on GC-MS metabolomics data for young hypertensive men. Anal. Chim. Acta 2015, 854, 95–105. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Jia, H.; Zhou, C.; Yang, Y.; Zhao, Y.; Yang, M.; Zou, Z. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J. Pharm. Biomed. Anal. 2017, 10, 231–239. [Google Scholar] [CrossRef]
- Hu, S.; Li, A.; Huang, T.; Lai, J.; Li, J.; Sublette, M.E.; Lu, H.; Lu, Q.; Du, Y.; Hu, Z.; et al. Gut microbiota changes in patients with bipolar depression. Adv. Sci. 2019, 6, 1900752. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Jamilian, M.; Foroozanfard, F.; Kavossian, E.; Aghadavod, E.; Shafabakhsh, R.; Hoseini, A.; Asemi, Z. Effects of curcumin on body weight, glycemic control and serum lipids in women with polycystic ovary syndrome: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. ESPEN 2020, 36, 128–133. [Google Scholar] [CrossRef]
- Meng, Z.; Yu, X.H.; Chen, J.; Li, L.; Li, S. Curcumin attenuates cardiac fibrosis in spontaneously hypertensive rats through PPAR-γ activation. Acta Pharmacol. Sin. 2014, 35, 1247–1256. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Cui, B.; Jiang, A.; Tao, H.; Cheng, S.; Liu, Y. Combination of chronic alcohol consumption and high-salt intake elicits gut microbial alterations and liver steatosis in mice. J. Agric. Food Chem. 2020, 68, 1750–1759. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, J.E.; Chang, Y.; Kim, M.K.; Sung, E.; Shin, H.; Ryu, S. Dietary sodium and potassium intake in relation to non-alcoholic fatty liver disease. Br. J. Nutr. 2016, 116, 1447–1456. [Google Scholar] [CrossRef]
- Daneberga, Z.; Nakazawa-Miklasevica, M.; Berga-Svitina, E.; Murmane, D.; Isarova, D.; Cupane, L.; Masinska, M.; Nartisa, I.; Lazdane, A.; Miklasevics, E. Urinary organic acids spectra in children with altered gut microbiota composition and autistic spectrum disorder. Nord. J. Psychiatry 2021, 76, 523–529. [Google Scholar] [CrossRef]
- Golonka, R.M.; Vijay-Kumar, M. Atypical immunometabolism and metabolic reprogramming in liver cancer: Deciphering the role of gut microbiome. Adv. Cancer Res. 2021, 149, 171–255. [Google Scholar] [CrossRef] [PubMed]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 2019, 8, e00678. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Rius-Pérez, S.; Torres-Cuevas, I.; Millán, I.; Ortega, Á.L.; Pérez, S. PGC-1α, Inflammation, and oxidative stress: An integrative view in metabolism. Oxid. Med. Cell. Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.; Fanti, P.; Rossi, R. Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc. 2013, 8, 1660–1669. [Google Scholar] [CrossRef]
- Habashy, W.S.; Milfort, M.C.; Rekaya, R.; Aggrey, S.E. Cellular antioxidant enzyme activity and biomarkers for oxidative stress are affected by heat stress. Int. J. Biometeorol. 2019, 63, 1569–1584. [Google Scholar] [CrossRef]
- Gao, P.; You, M.; Li, L.; Zhang, Q.; Fang, X.; Wei, X.; Zhou, Q.; Zhang, H.; Wang, M.; Lu, Z.; et al. Salt-induced hepatic inflammatory memory contributes to cardiovascular damage through epigenetic modulation of SIRT3. Circulation 2022, 145, 375–391. [Google Scholar] [CrossRef]
- Wang, G.; Yeung, C.K.; Wong, W.Y.; Zhang, N.; Wei, Y.F.; Zhang, J.L.; Yan, Y.; Wong, C.Y.; Tang, J.J.; Chuai, M.; et al. Liver fibrosis can be induced by high salt intake through excess reactive oxygen species (ROS) production. J. Agric. Food Chem. 2016, 64, 1610–1617. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Revisiting the Warburg effect: Historical dogma versus current understanding. J. Physiol. 2021, 599, 1745–1757. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Zhang, C.; Wang, Z. High salt diet induces metabolic alterations in multiple biological processes of Dahl salt-sensitive rats. J. Nutr. Biochem. 2018, 56, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, K.R.; Milton, J.E.; Packard, D.P.; Shuler, L.A.; Short, R.A. Dietary amino acids and blood pressure: A cohort study of patients with cardiovascular disease. Am. J. Kidney Dis. 2012, 59, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Huang, C.; Su, J.; Pan, C.W.; Ke, C. Identification of biomarkers for essential hypertension based on metabolomics. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 382–395. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.A.; do Carmo, J.M.; Li, X.; Wang, Z.; Mouton, A.J.; Hall, J.E. Role of hyperinsulinemia and insulin resistance in hypertension: Metabolic syndrome revisited. Can. J. Cardiol. 2020, 36, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Mehrmohamadi, M.; Liu, X.; Shestov, A.A.; Locasale, J.W. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014, 9, 1507–1519. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, Y.; Usa, K.; Mladinov, D.; Fang, Y.; Ding, X.; Greene, A.S.; Cowley, A.W., Jr.; Liang, M. Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension 2009, 54, 255–260. [Google Scholar] [CrossRef]
- Kovarik, J.J.; Morisawa, N.; Wild, J.; Marton, A.; Takase-Minegishi, K.; Minegishi, S.; Daub, S.; Sands, J.M.; Klein, J.D.; Bailey, J.L.; et al. Adaptive physiological water conservation explains hypertension and muscle catabolism in experimental chronic renal failure. Acta Physiol. 2021, 232, e13629. [Google Scholar] [CrossRef]
- Do, M.H.; Lee, H.B.; Lee, E.; Park, H.Y. The effects of gelatinized wheat starch and high salt diet on gut microbiota and metabolic disorder. Nutrients 2020, 12, 301. [Google Scholar] [CrossRef]
- Gutierrez-Calabres, E.; Ortega-Hernandez, A.; Modrego, J.; Gomez-Gordo, R.; Caro-Vadillo, A.; Rodriguez-Bobada, C.; Gonzalez, P.; Gomez-Garre, D. Gut microbiota profile identifies transition from compensated cardiac hypertrophy to heart failure in hypertensive rats. Hypertension 2020, 76, 1545–1554. [Google Scholar] [CrossRef]
- Kwan, S.Y.; Jiao, J.; Joon, A.; Wei, P.; Petty, L.E.; Below, J.E.; Daniel, C.R.; Wu, X.; Zhang, J.; Jenq, R.R.; et al. Gut microbiome features associated with liver fibrosis in Hispanics, a population at high risk for fatty liver disease. Hepatology 2022, 75, 955–967. [Google Scholar] [CrossRef]
- Adams, L.A.; Wang, Z.; Liddle, C.; Melton, P.E.; Ariff, A.; Chandraratna, H.; Tan, J.; Ching, H.; Coulter, S.; de Boer, B.; et al. Bile acids associate with specific gut microbiota, low-level alcohol consumption and liver fibrosis in patients with non-alcoholic fatty liver disease. Liver Int. 2020, 40, 1356–1365. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Norris, K.; Vaziri, N.D. Dysregulation of hepatic fatty acid metabolism in chronic kidney disease. Nephrol. Dial. Transplant. 2013, 28, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Feng, Y.F.; Liu, X.T.; Li, Y.C.; Zhu, H.M.; Sun, M.R.; Li, P.; Liu, B.; Yang, H. Songorine promotes cardiac mitochondrial biogenesis via Nrf2 induction during sepsis. Redox Biol. 2021, 38, 101771. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Billi, A.M.; Mongiorgi, S.; Ratti, S.; McCubrey, J.A.; Suh, P.G.; Cocco, L.; Ramazzotti, G. Nuclear phosphatidylinositol signaling: Focus on phosphatidylinositol phosphate kinases and phospholipases C. J. Cell. Physiol. 2016, 231, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Song, Z.; Weng, J.; Fantus, I.G. Curcumin and other dietary polyphenols: Potential mechanisms of metabolic actions and therapy for diabetes and obesity. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E201–E205. [Google Scholar] [CrossRef] [PubMed]
- Au, A.; Cheng, K.K.; Wei, L.K. Metabolomics, lipidomics and pharmacometabolomics of human hypertension. Adv. Exp. Med. Biol. 2017, 956, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the role of the gut microbiome and microbial metabolites in Non-Alcoholic Fatty Liver Disease: Current evidence and perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef] [PubMed]
| Parameters | NS | HS | HSCUR |
|---|---|---|---|
| Body weight (g) | 284.7 ± 26.3 | 275.1 ± 17.8 | 257.0 ± 11.3 |
| Kidney weight (g) | 1.29 ± 0.107 | 1.23 ± 0.138 | 1.36 ± 0.151 |
| Heart weight (g) | 1.11 ± 0.034 | 1.11 ± 0.04 | 0.986 ± 0.046 * |
| Liver weight (g) | 7.99 ± 0.33 | 7.34 ± 0.16 | 7.60 ± 0.24 |
| Daily food intake (g/day) | 17.4 ± 0.525 | 23.1 ± 0.609 ## | 21.8 ± 1.98 |
| Daily water intake (mL/day) | 27.6 ± 1.30 | 77.3 ± 2.30 #### | 64.8 ± 3.43 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou-Yang, Y.-n.; Yuan, M.-d.; Yang, Z.-m.; Min, Z.; Jin, Y.-x.; Tian, Z.-m. Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis. Metabolites 2022, 12, 1076. https://doi.org/10.3390/metabo12111076
Ou-Yang Y-n, Yuan M-d, Yang Z-m, Min Z, Jin Y-x, Tian Z-m. Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis. Metabolites. 2022; 12(11):1076. https://doi.org/10.3390/metabo12111076
Chicago/Turabian StyleOu-Yang, Ya-nan, Meng-di Yuan, Zheng-mao Yang, Zhuo Min, Yue-xin Jin, and Zhong-min Tian. 2022. "Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis" Metabolites 12, no. 11: 1076. https://doi.org/10.3390/metabo12111076
APA StyleOu-Yang, Y.-n., Yuan, M.-d., Yang, Z.-m., Min, Z., Jin, Y.-x., & Tian, Z.-m. (2022). Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis. Metabolites, 12(11), 1076. https://doi.org/10.3390/metabo12111076