Changes of Plasma Tris(hydroxymethyl)aminomethane and 5-Guanidino-3-methyl-2-oxopentanoic Acid as Biomarkers of Heart Remodeling after Left Ventricular Assist Device Support
Abstract
1. Introduction
2. Materials and Methods
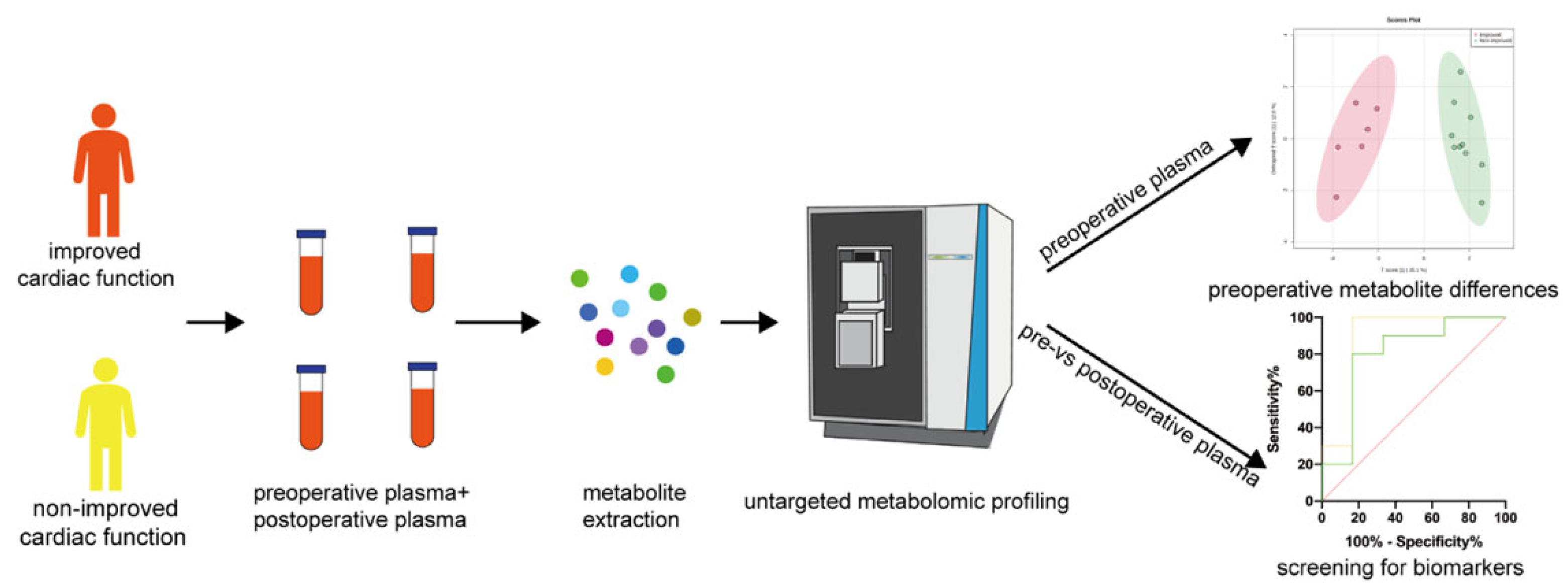
2.1. Study Design
2.2. Metabolite Extraction from Plasma Samples
2.3. Untargeted LC-MS/MS-Based Metabolomic Analysis
2.4. Data Processing
2.5. Statistical Analysis
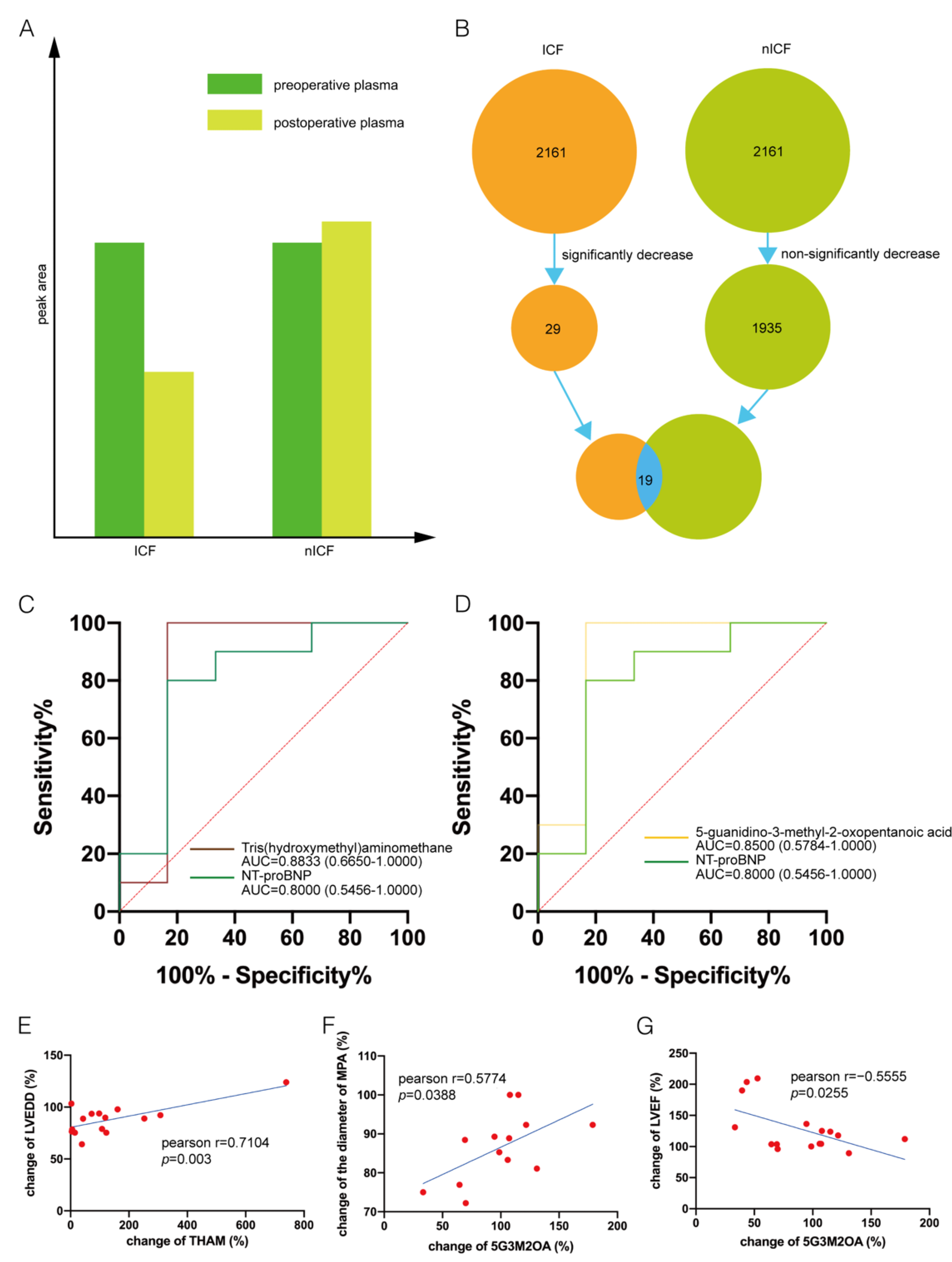
3. Results
3.1. Demographic Characteristics of the Patients
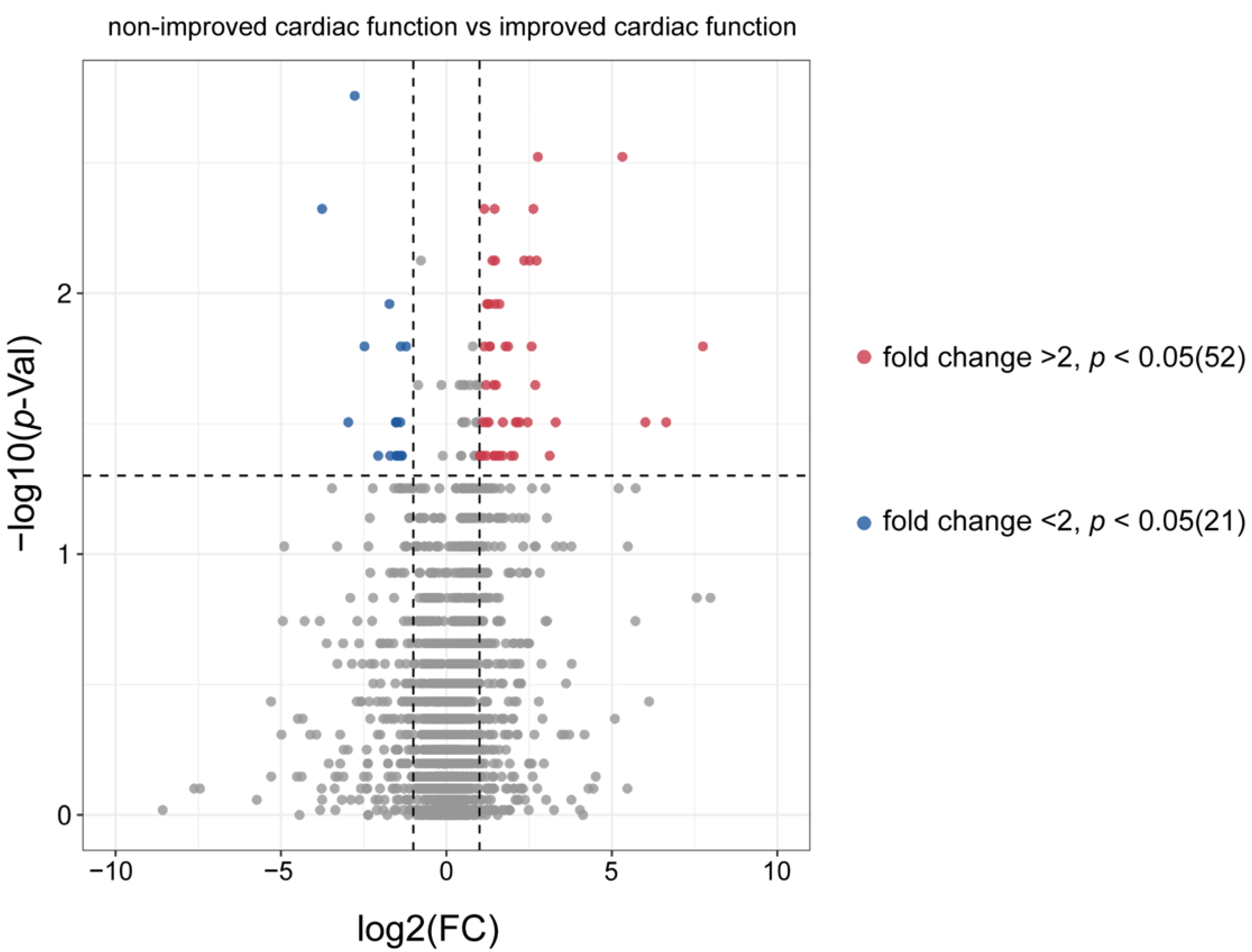
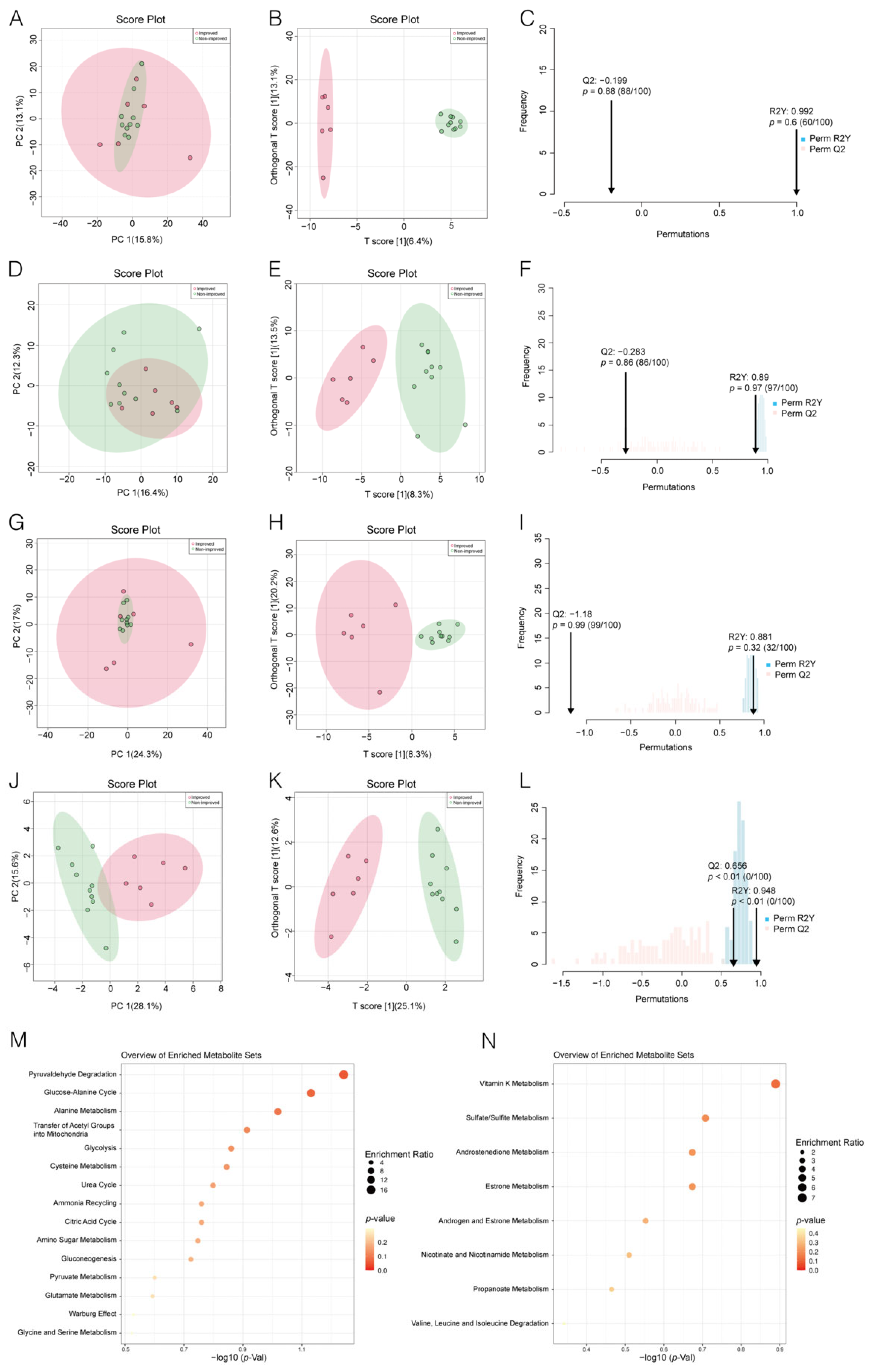
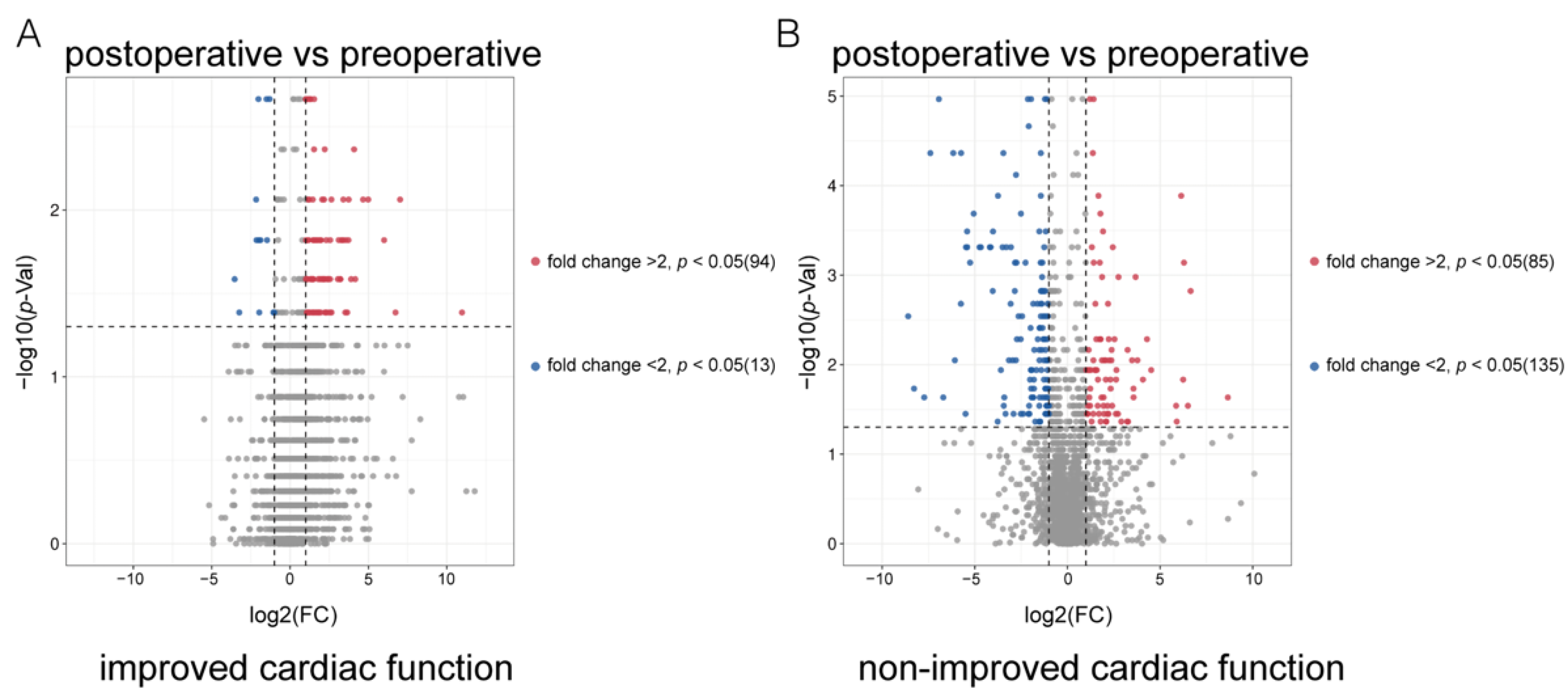
3.2. Distinguishing ICF Patients from nICF Patients Using Plasma Metabolomics
3.3. Identification of Potential Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slaughter, M.S.; Rogers, J.G.; Milano, C.A.; Russell, S.D.; Conte, J.V.; Feldman, D.; Sun, B.; Tatooles, A.J.; Delgado, R.M., III; Long, J.W.; et al. Advanced heart failure treated with continuous-flow left ventricular assist device. N. Engl. J. Med. 2009, 361, 2241–2251. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.A.; Bachman, T.N.; Watson, J.; Baldwin, J.T.; Wagner, W.R.; Borovetz, H.S. Current and Future Considerations in the Use of Mechanical Circulatory Support Devices: An Update, 2008–2018. Annu. Rev. Biomed. Eng. 2019, 21, 33–60. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.C.; Yamada, K.A.; Patel, A.Y.; Topkara, V.K.; George, I.; Cheema, F.H.; Ewald, G.A.; Mann, D.L.; Nerbonne, J.M. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014, 129, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Morley-Smith, A.C.; Mills, A.; Jacobs, S.; Meyns, B.; Rega, F.; Simon, A.R.; Pepper, J.R.; Lyon, A.R.; Thum, T. Circulating microRNAs for predicting and monitoring response to mechanical circulatory support from a left ventricular assist device. Eur. J. Heart Fail. 2014, 16, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Shahinian, J.H.; Rog-Zielinska, E.A.; Schlimpert, M.; Mayer, B.; Tholen, S.; Kammerer, B.; Biniossek, M.L.; Beyersdorf, F.; Schilling, O.; Siepe, M. Impact of left ventricular assist device therapy on the cardiac proteome and metabolome composition in ischemic cardiomyopathy. Artif. Organs 2020, 44, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Badolia, R.; Ramadurai, D.K.A.; Abel, E.D.; Ferrin, P.; Taleb, I.; Shankar, T.S.; Krokidi, A.T.; Navankasattusas, S.; McKellar, S.H.; Yin, M.; et al. The Role of Nonglycolytic Glucose Metabolism in Myocardial Recovery Upon Mechanical Unloading and Circulatory Support in Chronic Heart Failure. Circulation 2020, 142, 259–274. [Google Scholar] [CrossRef]
- Stanley, W.C.; Recchia, F.A.; Lopaschuk, G.D. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005, 85, 1093–1129. [Google Scholar] [CrossRef] [PubMed]
- Byrne, N.J.; Levasseur, J.; Sung, M.M.; Masson, G.; Boisvenue, J.; Young, M.E.; Dyck, J.R. Normalization of cardiac substrate utilization and left ventricular hypertrophy precede functional recovery in heart failure regression. Cardiovasc. Res. 2016, 110, 249–257. [Google Scholar] [CrossRef]
- Hu, J.R.; Coresh, J.; Inker, L.A.; Levey, A.S.; Zheng, Z.; Rebholz, C.M.; Tin, A.; Appel, L.J.; Chen, J.; Sarnak, M.J.; et al. Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int. 2018, 94, 381–389. [Google Scholar] [CrossRef]
- Barderas, M.G.; Laborde, C.M.; Posada, M.; de la Cuesta, F.; Zubiri, I.; Vivanco, F.; Alvarez-Llamas, G. Metabolomic profiling for identification of novel potential biomarkers in cardiovascular diseases. J. Biomed. Biotechnol. 2011, 2011, 790132. [Google Scholar] [CrossRef]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877 e822. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Shu, S.; Li, Y.; Yan, X.; Chen, X.; Chen, Z.; Hu, Y.; Chang, Y.; Hu, Z.; Wang, X.; et al. Plasma Metabolites-Based Prediction in Cardiac Surgery-Associated Acute Kidney Injury. J. Am. Heart Assoc. 2021, 10, e021825. [Google Scholar] [CrossRef] [PubMed]
- Kaewkod, T.; Bovonsombut, S.; Tragoolpua, Y. Efficacy of Kombucha Obtained from Green, Oolong, and Black Teas on Inhibition of Pathogenic Bacteria, Antioxidation, and Toxicity on Colorectal Cancer Cell Line. Microorganisms 2019, 7, 700. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.L.; Fiehn, O.; Kaddurah-Daouk, R.; Lindon, J.C. Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Brief Bioinform 2006, 7, 159–165. [Google Scholar] [CrossRef]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 13. [Google Scholar] [CrossRef]
- Abraham, W.T.; Smith, S.A. Devices in the management of advanced, chronic heart failure. Nat. Rev. Cardiol. 2013, 10, 98–110. [Google Scholar] [CrossRef]
- Ammirati, E.; Oliva, F.G.; Colombo, T.; Russo, C.F.; Cipriani, M.G.; Garascia, A.; Guida, V.; Colombo, G.; Verde, A.; Perna, E.; et al. Mid-term survival after continuous-flow left ventricular assist device versus heart transplantation. Heart Vessel. 2016, 31, 722–733. [Google Scholar] [CrossRef]
- Bedi, K.C., Jr.; Snyder, N.W.; Brandimarto, J.; Aziz, M.; Mesaros, C.; Worth, A.J.; Wang, L.L.; Javaheri, A.; Blair, I.A.; Margulies, K.B.; et al. Evidence for Intramyocardial Disruption of Lipid Metabolism and Increased Myocardial Ketone Utilization in Advanced Human Heart Failure. Circulation 2016, 133, 706–716. [Google Scholar] [CrossRef]
- Chen, L.; Song, J.; Hu, S. Metabolic remodeling of substrate utilization during heart failure progression. Heart Fail Rev. 2019, 24, 143–154. [Google Scholar] [CrossRef]
- Zhabyeyev, P.; Gandhi, M.; Mori, J.; Basu, R.; Kassiri, Z.; Clanachan, A.; Lopaschuk, G.D.; Oudit, G.Y. Pressure-overload-induced heart failure induces a selective reduction in glucose oxidation at physiological afterload. Cardiovasc. Res. 2013, 97, 676–685. [Google Scholar] [CrossRef]
- Amorim, P.A.; Nguyen, T.D.; Shingu, Y.; Schwarzer, M.; Mohr, F.W.; Schrepper, A.; Doenst, T. Myocardial infarction in rats causes partial impairment in insulin response associated with reduced fatty acid oxidation and mitochondrial gene expression. J. Thorac. Cardiovasc. Surg. 2010, 140, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Doenst, T.; Pytel, G.; Schrepper, A.; Amorim, P.; Farber, G.; Shingu, Y.; Mohr, F.W.; Schwarzer, M. Decreased rates of substrate oxidation ex vivo predict the onset of heart failure and contractile dysfunction in rats with pressure overload. Cardiovasc. Res. 2010, 86, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Bayes-Genis, A.; Liu, P.P.; Lanfear, D.E.; de Boer, R.A.; Gonzalez, A.; Thum, T.; Emdin, M.; Januzzi, J.L. Omics phenotyping in heart failure: The next frontier. Eur. Heart J. 2020, 41, 3477–3484. [Google Scholar] [CrossRef] [PubMed]
- Sirieix, D.; Delayance, S.; Paris, M.; Massonnet-Castel, S.; Carpentier, A.; Baron, J.F. Tris-hydroxymethyl aminomethane and sodium bicarbonate to buffer metabolic acidosis in an isolated heart model. Am. J. Respir. Crit. Care Med. 1997, 155, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Mattiazzi, A.; Vittone, L.; Mundina-Weilenmann, C. Ca2+/calmodulin-dependent protein kinase: A key component in the contractile recovery from acidosis. Cardiovasc. Res. 2007, 73, 648–656. [Google Scholar] [CrossRef]
- Feng, J.; Wu, J.; Gao, J.; Xia, Z.; Deng, Z.; He, X. Biosynthesis of the beta-methylarginine residue of peptidyl nucleoside arginomycin in Streptomyces arginensis NRRL 15941. Appl. Environ. Microbiol. 2014, 80, 5021–5027. [Google Scholar] [CrossRef]
- Guridi, J.; Borgatello, C.; Scremin, O.U. Arginine NO-dependent and NO-independent effects on hemodynamics. Eur. J. Pharmacol. 2014, 729, 138–143. [Google Scholar] [CrossRef]
| Variable | ICF (n = 6) | nICF (n = 10) | p-Value |
|---|---|---|---|
| Age at implantation | 39.2 (30.50–46.25) | 40.5 (30.75–47.75) | 0.8125 |
| Male sex | 6 (100%) | 9 (90%) | 0.4577 |
| HF etiology | 0.2769 | ||
| ICM | 2 (33%) | 1 (10%) | |
| DCM | 4 (67%) | 6 (60%) | |
| VHD | 0 | 2 (20%) | |
| ACM | 0 | 1 (10%) | |
| BMI (kg/m2) | 25.5 ± 4.27 | 23.3 ± 5.01 | 0.4167 |
| NYHA functional class | 0.4250 | ||
| I | 0 | 0 | |
| II | 1 (16.7%) | 0 | |
| III | 0 | 1 (10%) | |
| IV | 5 (83.3%) | 9 (90%) | |
| Diabetes | 0 | 1(10%) | 0.4577 |
| Creatinine (μmol/L) | 87.4 ± 28.47 | 99.4 ± 19.48 | 0.3324 |
| Hemoglobin (g/L) | 115.3 ± 28.65 | 135.9 ± 15.47 | 0.0805 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Cui, H.; Chen, X.; Hua, X.; Song, J.; Hu, S. Changes of Plasma Tris(hydroxymethyl)aminomethane and 5-Guanidino-3-methyl-2-oxopentanoic Acid as Biomarkers of Heart Remodeling after Left Ventricular Assist Device Support. Metabolites 2022, 12, 1068. https://doi.org/10.3390/metabo12111068
Xu M, Cui H, Chen X, Hua X, Song J, Hu S. Changes of Plasma Tris(hydroxymethyl)aminomethane and 5-Guanidino-3-methyl-2-oxopentanoic Acid as Biomarkers of Heart Remodeling after Left Ventricular Assist Device Support. Metabolites. 2022; 12(11):1068. https://doi.org/10.3390/metabo12111068
Chicago/Turabian StyleXu, Mengda, Hao Cui, Xiao Chen, Xiumeng Hua, Jiangping Song, and Shengshou Hu. 2022. "Changes of Plasma Tris(hydroxymethyl)aminomethane and 5-Guanidino-3-methyl-2-oxopentanoic Acid as Biomarkers of Heart Remodeling after Left Ventricular Assist Device Support" Metabolites 12, no. 11: 1068. https://doi.org/10.3390/metabo12111068
APA StyleXu, M., Cui, H., Chen, X., Hua, X., Song, J., & Hu, S. (2022). Changes of Plasma Tris(hydroxymethyl)aminomethane and 5-Guanidino-3-methyl-2-oxopentanoic Acid as Biomarkers of Heart Remodeling after Left Ventricular Assist Device Support. Metabolites, 12(11), 1068. https://doi.org/10.3390/metabo12111068






