Bioengineering of Escherichia coli Nissle 1917 for Production and Excretion of Spermidine, a Key Metabolite in Human Health
Abstract
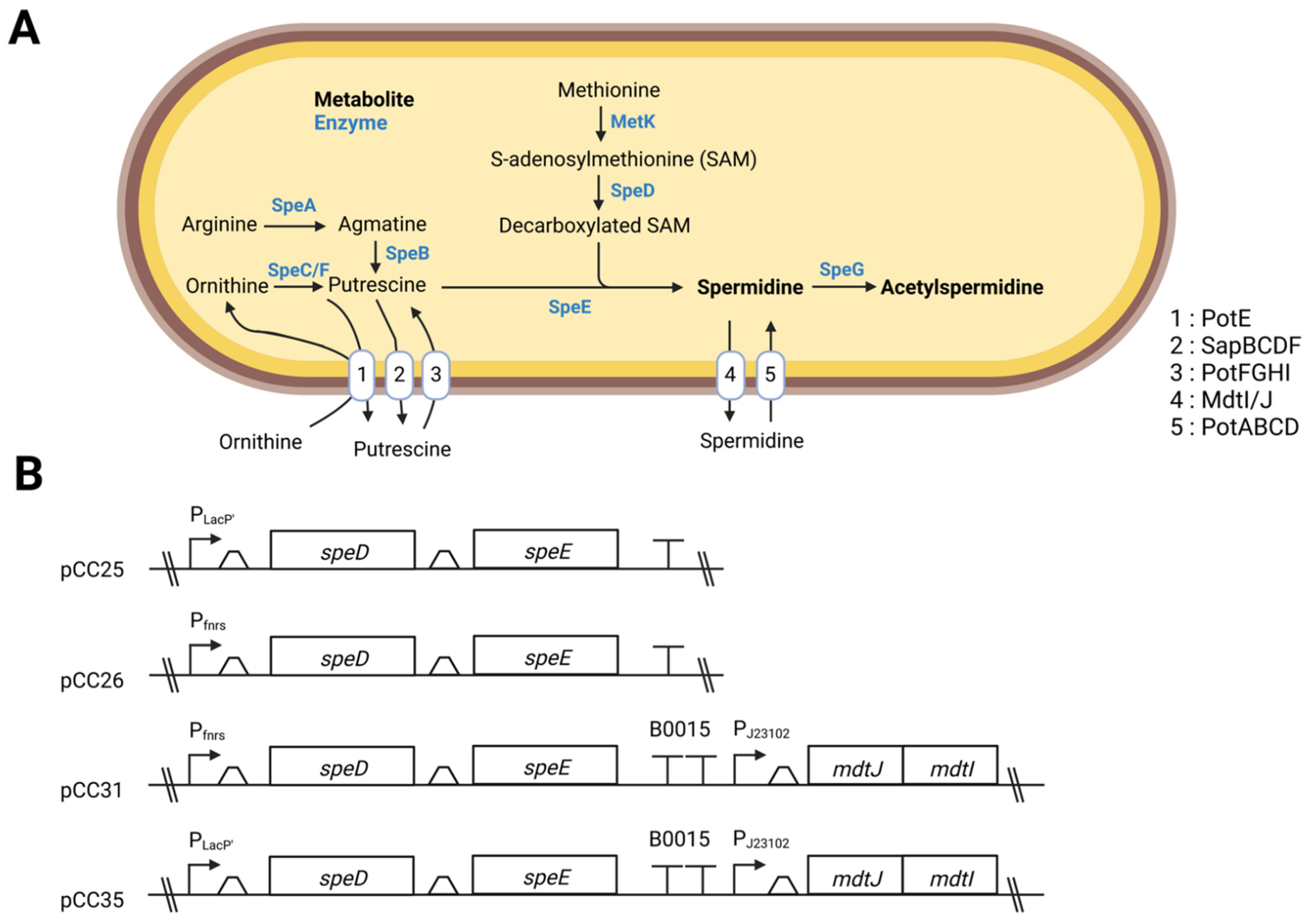
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Plasmids Construction
2.3. Sample Collection and Sample Preparation
2.4. Liquid Chromatography Coupled with High-Resolution Mass Spectrometry (LC-HRMS) Analysis
- The mobile phases consisted of A (100% water + 0.1% formic acid) and B (100% acetonitrile (ACN) + 0.1% formic acid). Elution started with an isocratic step of 3 min at 1% mobile phase B, followed by a linear gradient from 1% to 100% mobile phase B for the next 4 min. These proportions were kept constant for the next 3 min before returning to 1% B for 4 min.
- The mobile phases consisted of A (100% water + 0.1% formic acid) and B (100% acetonitrile (ACN) + 0.1% formic acid). Elution started with an isocratic step of 2 min at 1% mobile phase B, followed by a linear gradient from 1% to 100% mobile phase B for the next 12 min. These proportions were kept constant for the next 4 min before returning to 1% B for 5 min.
2.5. LC-HRMS Data Processing
2.6. Metabolite Annotation
2.7. Statistical Analysis
3. Results
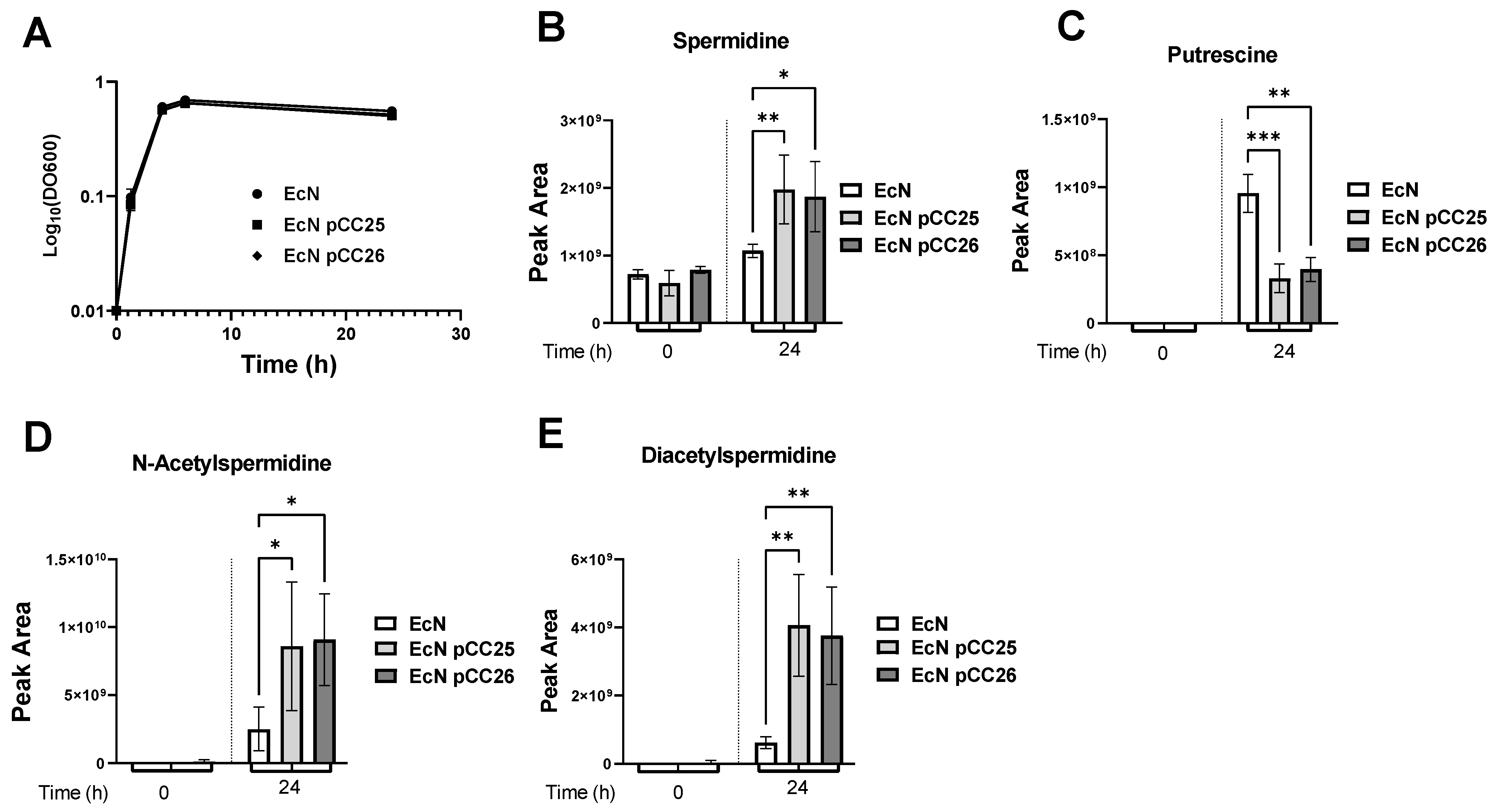
3.1. Overexpression of the Genes speD and speE Increases the Amount of Spermidine
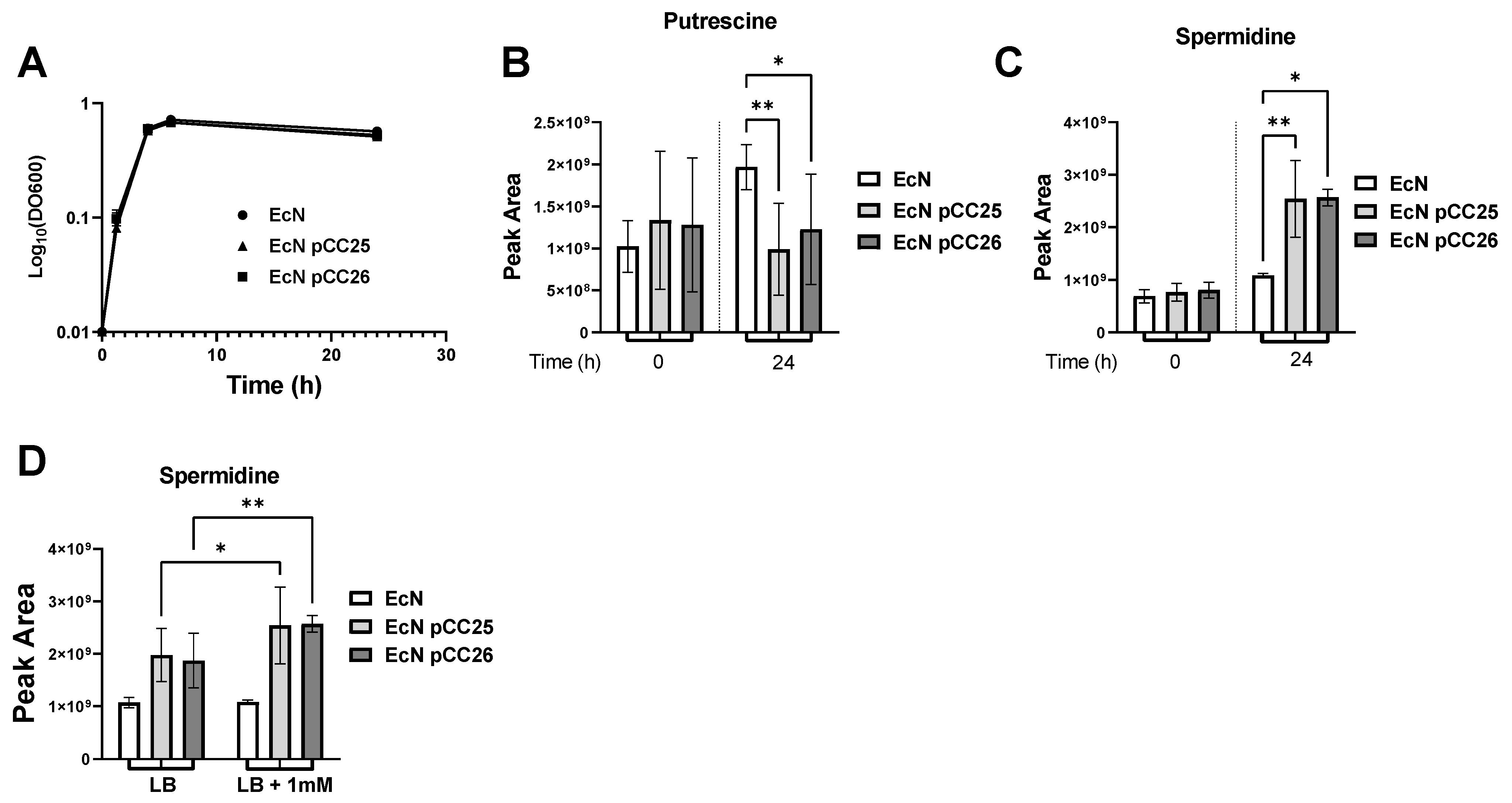
3.2. Putrescine Supplementation of the Medium Increases Spermidine Production
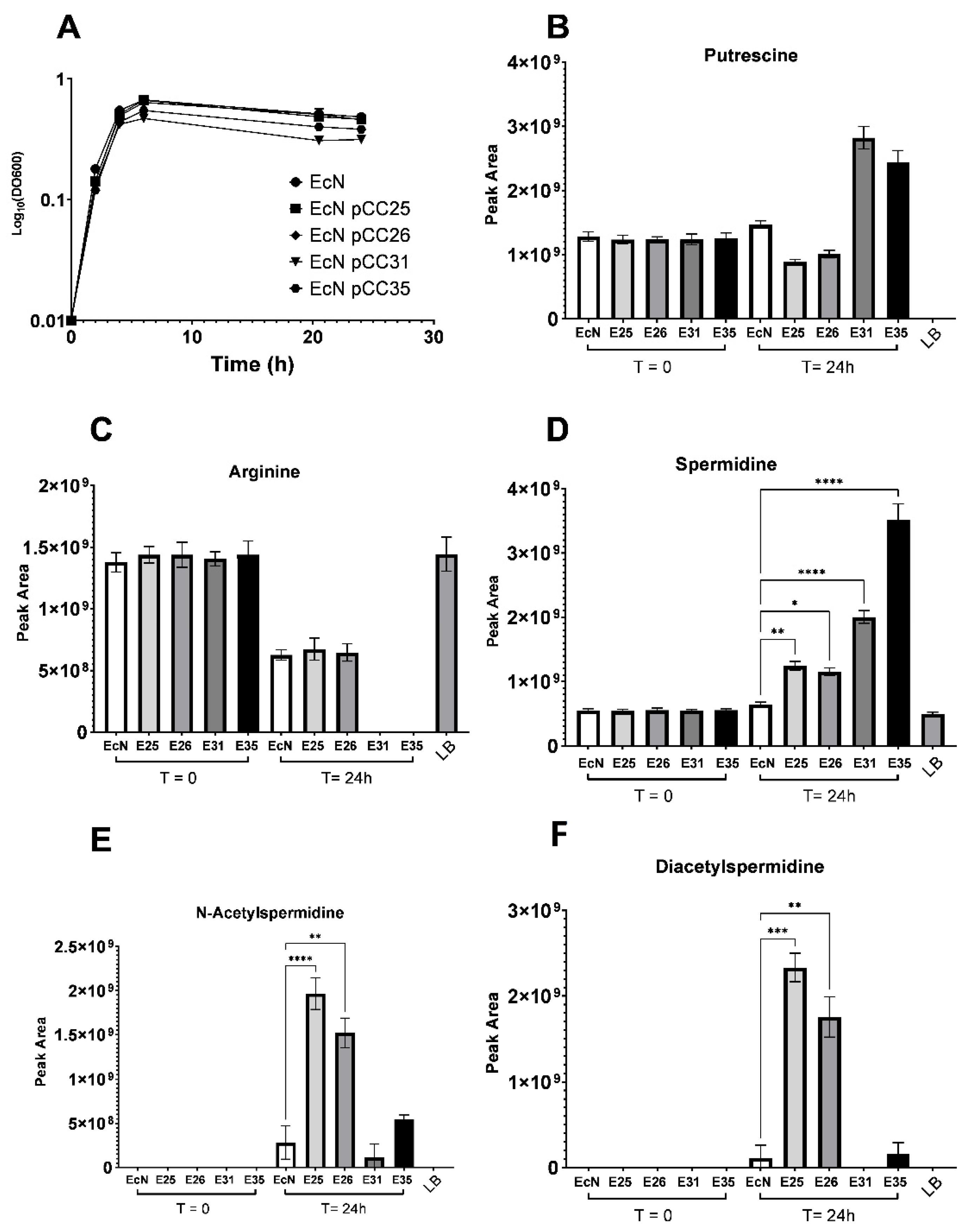
3.3. Excretion and Production of Acetylated Forms of Spermidine Can Be Limited by Overexpressing the Spermidine Exporter MdtI/MdtJ
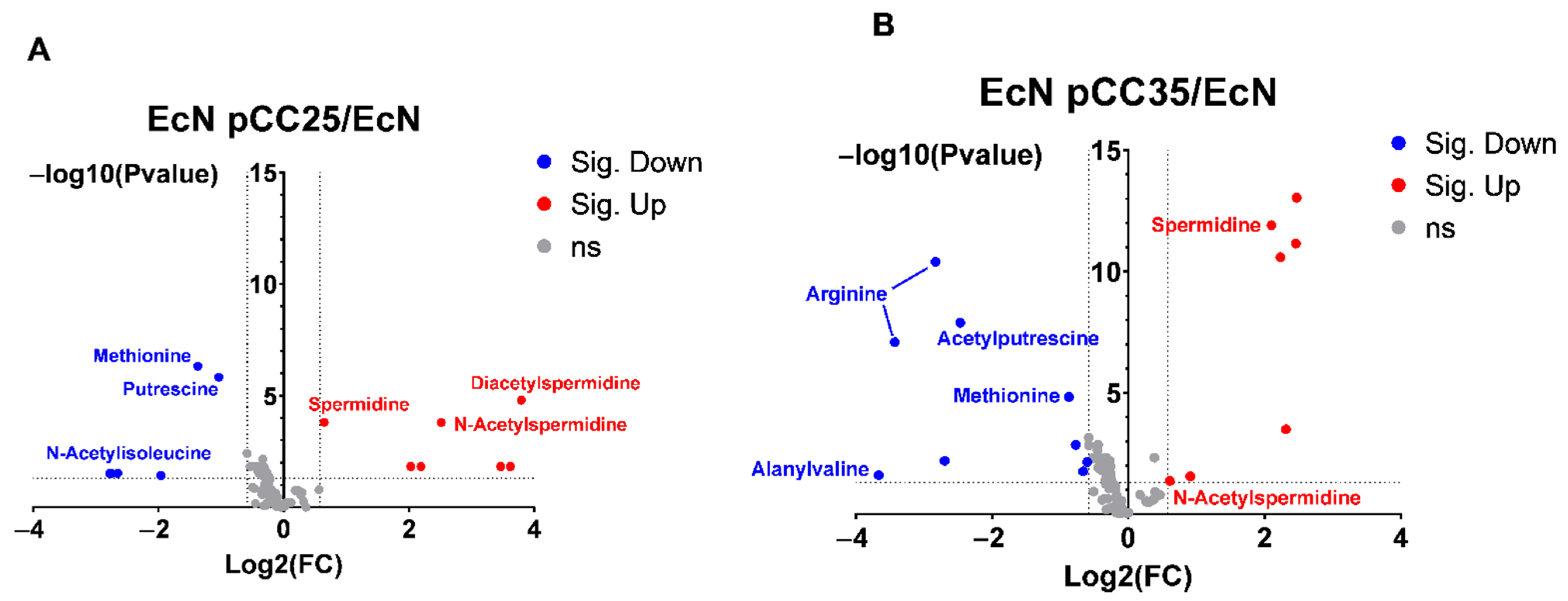
3.4. EcN Engineering Seems to Have a Limited Impact on Its Overall Metabolism
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caffaratti, C.; Plazy, C.; Mery, G.; Tidjani, A.R.; Fiorini, F.; Thiroux, S.; Toussaint, B.; Hannani, D.; Le Gouellec, A. What we know so far about the metabolite-mediated microbiota-intestinal immunity dialogue and how to hear the sound of this crosstalk. Metabolites 2021, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- Sorbara, M.T.; Pamer, E.G. Microbiome-based therapeutics. Nat. Rev. Microbiol. 2022, 20, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Isabella, V.M.; Ha, B.N.; Castillo, M.J.; Lubkowicz, D.J.; Rowe, S.E.; Millet, Y.A.; Anderson, C.L.; Li, N.; Fisher, A.B.; West, K.A.; et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018, 36, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, C.; Yu, J.; Qi, Q.; Wang, Q. Bioengineered Escherichia coli Nissle 1917 for tumour-targeting therapy. Microb. Biotechnol. 2020, 13, 629–636. [Google Scholar] [CrossRef]
- Charbonneau, M.R.; Isabella, V.M.; Li, N.; Kurtz, C.B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 2020, 11, 1738. [Google Scholar] [CrossRef]
- Rodríguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Utrilla, M.P.; Chueca, N.; Fernández-Caballero, J.A.; García, F.; Rodríguez-Cabezas, M.E.; Gálvez, J. The administration of Escherichia coli Nissle 1917 ameliorates development of DSS-induced colitis in mice. Front. Pharmacol. 2018, 9, 468. [Google Scholar] [CrossRef]
- Souza, É.L.; Elian, S.D.; Paula, L.M.; Garcia, C.C.; Vieira, A.T.; Teixeira, M.M.; Arantes, R.M.; Nicoli, J.R.; Martins, F.S. Escherichia coli strain Nissle 1917 ameliorates experimental colitis by modulating intestinal permeability, the inflammatory response and clinical signs in a faecal transplantation model. J. Med. Microbiol. 2016, 65, 201–210. [Google Scholar] [CrossRef]
- Ukena, S.N.; Singh, A.; Dringenberg, U.; Engelhardt, R.; Seidler, U.; Hansen, W.; Bleich, A.; Bruder, D.; Franzke, A.; Rogler, G.; et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2007, 2, e1308. [Google Scholar] [CrossRef]
- Rembacken, B.J.; Snelling, A.M.; Hawkey, P.M.; Chalmers, D.M.; Axon, A.T.R. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: A randomised trial. Lancet 1999, 354, 635–639. [Google Scholar] [CrossRef]
- Kruis, W.; Frič, P.; Pokrotnieks, J.; Lukáš, M.; Fixa, B.; Kaščák, M.; Kamm, M.A.; Weismueller, J.; Beglinger, C.; Stolte, M.; et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 2004, 53, 1617–1623. [Google Scholar] [CrossRef]
- Forbes, A.; Escher, J.; Hébuterne, X.; Kłęk, S.; Krznaric, Z.; Schneider, S.; Shamir, R.; Stardelova, K.; Wierdsma, N.; Wiskin, A.E.; et al. ESPEN guideline: Clinical nutrition in inflammatory bowel disease. Clin. Nutr. 2017, 36, 321–347. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kibe, R.; Ooga, T.; Aiba, Y.; Kurihara, S.; Sawaki, E.; Koga, Y.; Benno, Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2012, 2, 233. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Ooga, T.; Matsumoto, M. Intestinal luminal putrescine is produced by collective biosynthetic pathways of the commensal microbiome. Gut Microbes 2019, 10, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, 4548. [Google Scholar] [CrossRef]
- Nakamura, A.; Kurihara, S.; Takahashi, D.; Ohashi, W.; Nakamura, Y.; Kimura, S.; Onuki, M.; Kume, A.; Sasazawa, Y.; Furusawa, Y.; et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun. 2021, 12, 2105. [Google Scholar] [CrossRef]
- Morón, B.; Spalinger, M.; Kasper, S.; Atrott, K.; Frey-Wagner, I.; Fried, M.; McCole, D.F.; Rogler, G.; Scharl, M. Activation of Protein Tyrosine Phosphatase Non-Receptor Type 2 by Spermidine Exerts Anti-Inflammatory Effects in Human THP-1 Monocytes and in a Mouse Model of Acute Colitis. PLoS ONE 2013, 8, e73703. [Google Scholar] [CrossRef]
- Carriche, G.M.; Almeida, L.; Stüve, P.; Velasquez, L.; Dhillon-LaBrooy, A.; Roy, U.; Lindenberg, M.; Strowig, T.; Plaza-Sirvent, C.; Schmitz, I.; et al. Regulating T-cell differentiation through the polyamine spermidine. J. Allergy Clin. Immunol. 2020, 147, 335–348. [Google Scholar] [CrossRef]
- Vick, J.E.; Johnson, E.T.; Choudhary, S.; Bloch, S.E.; Lopez-Gallego, F.; Srivastava, P.; Tikh, I.B.; Wawrzyn, G.T.; Schmidt-Dannert, C. Optimized compatible set of BioBrickTM vectors for metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2011, 92, 1275–1286. [Google Scholar] [CrossRef]
- Seo, S.W.; Yang, J.S.; Kim, I.; Yang, J.; Min, B.E.; Kim, S.; Jung, G.Y. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013, 15, 67–74. [Google Scholar] [CrossRef]
- Boudah, S.; Olivier, M.F.; Aros-Calt, S.; Oliveira, L.; Fenaille, F.; Tabet, J.C.; Junot, C. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 966, 34–47. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Aros-Calt, S.; Muller, B.H.; Boudah, S.; Ducruix, C.; Gervasi, G.; Junot, C.; Fenaille, F. Annotation of the Staphylococcus aureus metabolome using liquid chromatography coupled to high-resolution mass spectrometry and application to the study of methicillin resistance. J. Proteome Res. 2015, 14, 4863–4875. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Buch, J.K.; Boyle, S.M. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J. Bacteriol. 1985, 163, 522–527. [Google Scholar] [CrossRef]
- Boyle, S.M.; Markham, G.D.; Hafner, E.W.; Wright, J.M.; Tabor, H.; Tabor, C.W. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (speA, speB, speC and metK). Gene 1984, 30, 129–136. [Google Scholar] [CrossRef]
- Markham, G.D.; Tabor, C.W.; Tabor, H. S-adenosylmethionine decarboxylase of Escherichia coli. Studies on the covalently linked pyruvate required for activity. J. Biol. Chem. 1982, 257, 12063–12068. [Google Scholar] [CrossRef]
- White Tabor, C.; Tabor, H.; Xie, Q.W. Spermidine synthase of Escherichia coli: Localization of the speE gene. Proc. Natl. Acad. Sci. USA 1986, 83, 6040–6044. [Google Scholar] [CrossRef]
- Boysen, A.; Møller-Jensen, J.; Kallipolitis, B.; Valentin-Hansen, P.; Overgaard, M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J. Biol. Chem. 2010, 285, 10690–10702. [Google Scholar] [CrossRef]
- Samarra, I.; Ramos-Molina, B.; Queipo-Ortuño, M.I.; Tinahones, F.J.; Arola, L.; Delpino-Rius, A.; Herrero, P.; Canela, N. Gender-related differences on polyamine metabolome in liquid biopsies by a simple and sensitive two-step liquid-liquid extraction and LC-MS/MS. Biomolecules 2019, 9, 779. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Benno, Y. The Relationship between Microbiota and Polyamine Concentration in the Human Intestine: A Pilot Study. Microbiol. Immunol. 2007, 51, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, J.I.; Kashiwagi, K.; Yamagishi, M.; Ishihama, A.; Igarashi, K. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 1995, 270, 18831–18835. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Ishigure, H.; Demizu, R.; Uemura, T.; Nishino, K.; Yamaguchi, A.; Kashiwagi, K.; Igarashi, K. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008, 190, 872–878. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science 2018, 359, eaan2788. [Google Scholar] [CrossRef]
| Name | Primer Sequence (5′-3′) |
|---|---|
| Pfnrs-F | 5′ ATTGAATTCATCTAGAAAAAACGCCGCAAAGTT 3′ |
| Pfnrs-R | 5′ ATTCATATGTTTATTCTTCCCTCCTCTATC 3′ |
| speD-F | 5′ TAATTTAAGGGGGTAACATAATGTTGAAAAAACTGAAACTGC 3′ |
| speD-R | 5′ TCCGTCCTTACGTCCCCACTTTAAACAGCGGGCATATTGC 3′ |
| RBS-speE-F | 5′ AGTGGGGACGTAAGGACGGATTTGGATGGCCGAAAAAAAACAGT |
| RBS-speE-R | 5′ TTAGGACGGCTGTGAAGC 3′ |
| speE-F | 5′ GCAATATGCCCGCTGTTTAAAGTGGGGACGTAAGGACGGA 3′ |
| speE-R | 5′ TTATTTGATGCCTGGATGCATTAGGACGGCTGTGAAGCCA 3′ |
| B0015-F | 5′ ATTATGCATCCAGGCATCAAATAAAACG 3′ |
| B0015-R | 5′ ATTACTAGTTATAAACGCAGAAAGGCCC 3′ |
| mdtJ-mdtI-F | 5′ ATTCATATGTATATTTATTGGATTTTATTAGGTCTG 3′ |
| mdtJ-mdtI-R | 5′ ATTACTAGTTCAGGCAAGTTTCACCAT 3′ |
| oSeq1 | 5′ CTGACGTCTAAGAAACCATT 3′ |
| oSeq2 | 5′ GCAGGTCCTGAAGTTAACTAG 3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caffaratti, C.; Plazy, C.; Cunin, V.; Toussaint, B.; Le Gouellec, A. Bioengineering of Escherichia coli Nissle 1917 for Production and Excretion of Spermidine, a Key Metabolite in Human Health. Metabolites 2022, 12, 1061. https://doi.org/10.3390/metabo12111061
Caffaratti C, Plazy C, Cunin V, Toussaint B, Le Gouellec A. Bioengineering of Escherichia coli Nissle 1917 for Production and Excretion of Spermidine, a Key Metabolite in Human Health. Metabolites. 2022; 12(11):1061. https://doi.org/10.3390/metabo12111061
Chicago/Turabian StyleCaffaratti, Clément, Caroline Plazy, Valérie Cunin, Bertrand Toussaint, and Audrey Le Gouellec. 2022. "Bioengineering of Escherichia coli Nissle 1917 for Production and Excretion of Spermidine, a Key Metabolite in Human Health" Metabolites 12, no. 11: 1061. https://doi.org/10.3390/metabo12111061
APA StyleCaffaratti, C., Plazy, C., Cunin, V., Toussaint, B., & Le Gouellec, A. (2022). Bioengineering of Escherichia coli Nissle 1917 for Production and Excretion of Spermidine, a Key Metabolite in Human Health. Metabolites, 12(11), 1061. https://doi.org/10.3390/metabo12111061







