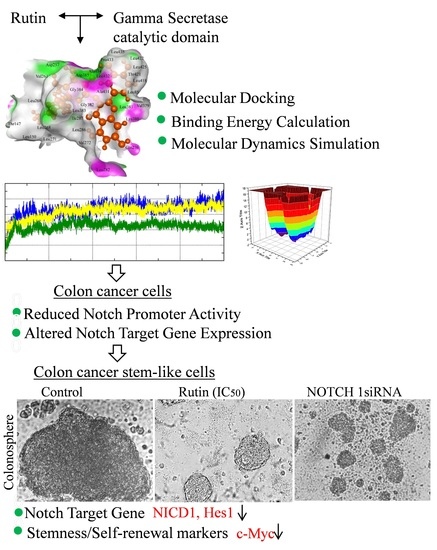
Rutin Potentially Binds the Gamma Secretase Catalytic Site, Down Regulates the Notch Signaling Pathway and Reduces Sphere Formation in Colonospheres
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein, Ligand Retrieval and Preparation
2.2. Receptor Grid Generation and Molecular Docking
2.3. MM-GBSA Calculation
2.4. Molecular Dynamics Simulation
2.5. Trajectory Analysis
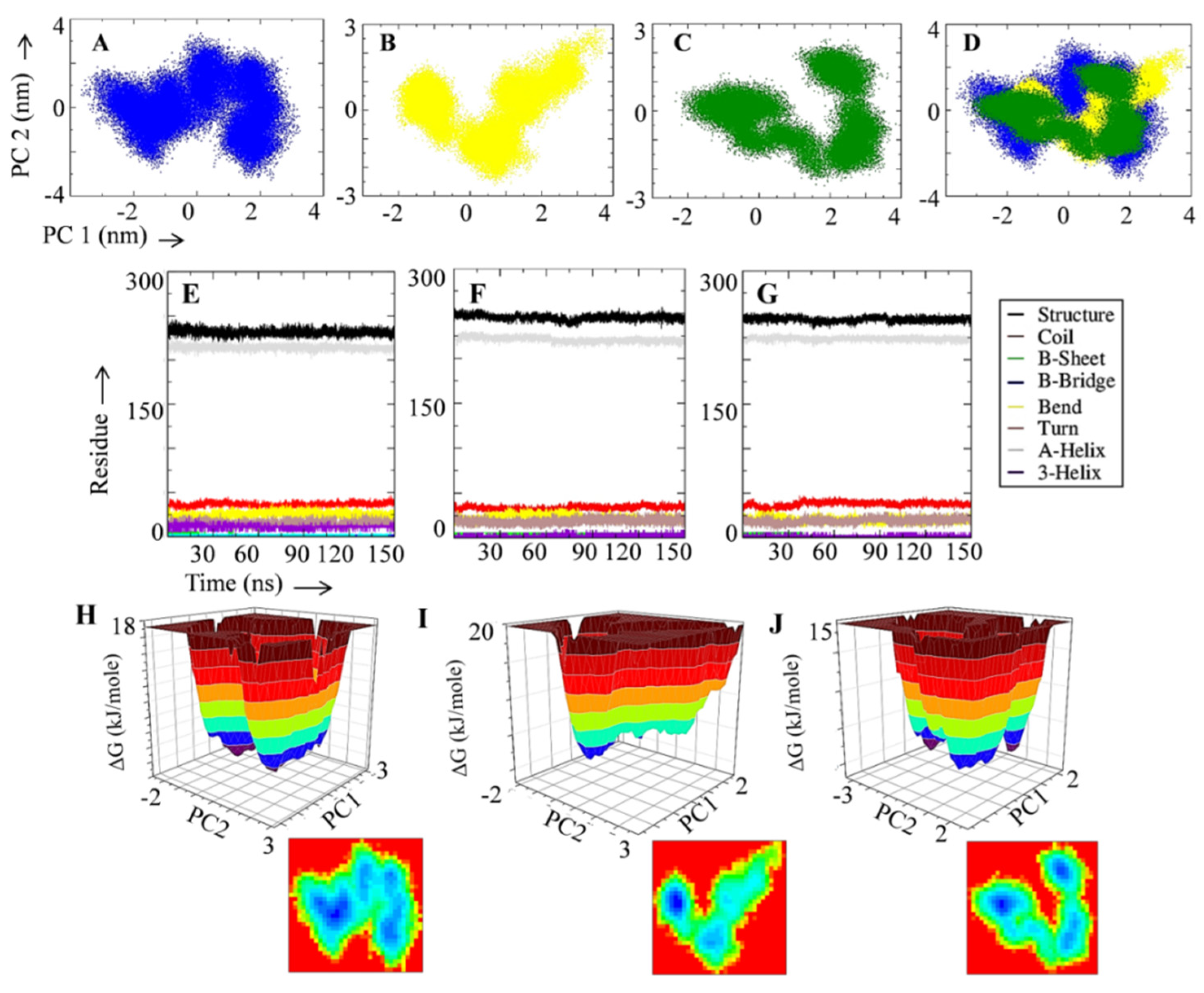
2.6. Principal Component Analysis and Free Energy Landscape
2.7. Reagents
2.8. Cell Culture and MTT Assay
2.9. Silencing of Notch-1 by siRNA
2.10. Colonosphere Formation and Phytochemical Treatment
2.11. Notch Promoter Activity
2.12. RNA Isolation, cDNA Synthesis, and Quantitative RT-PCR Analysis
2.13. Western Blotting
2.14. Statistical Analysis
3. Results and Discussion
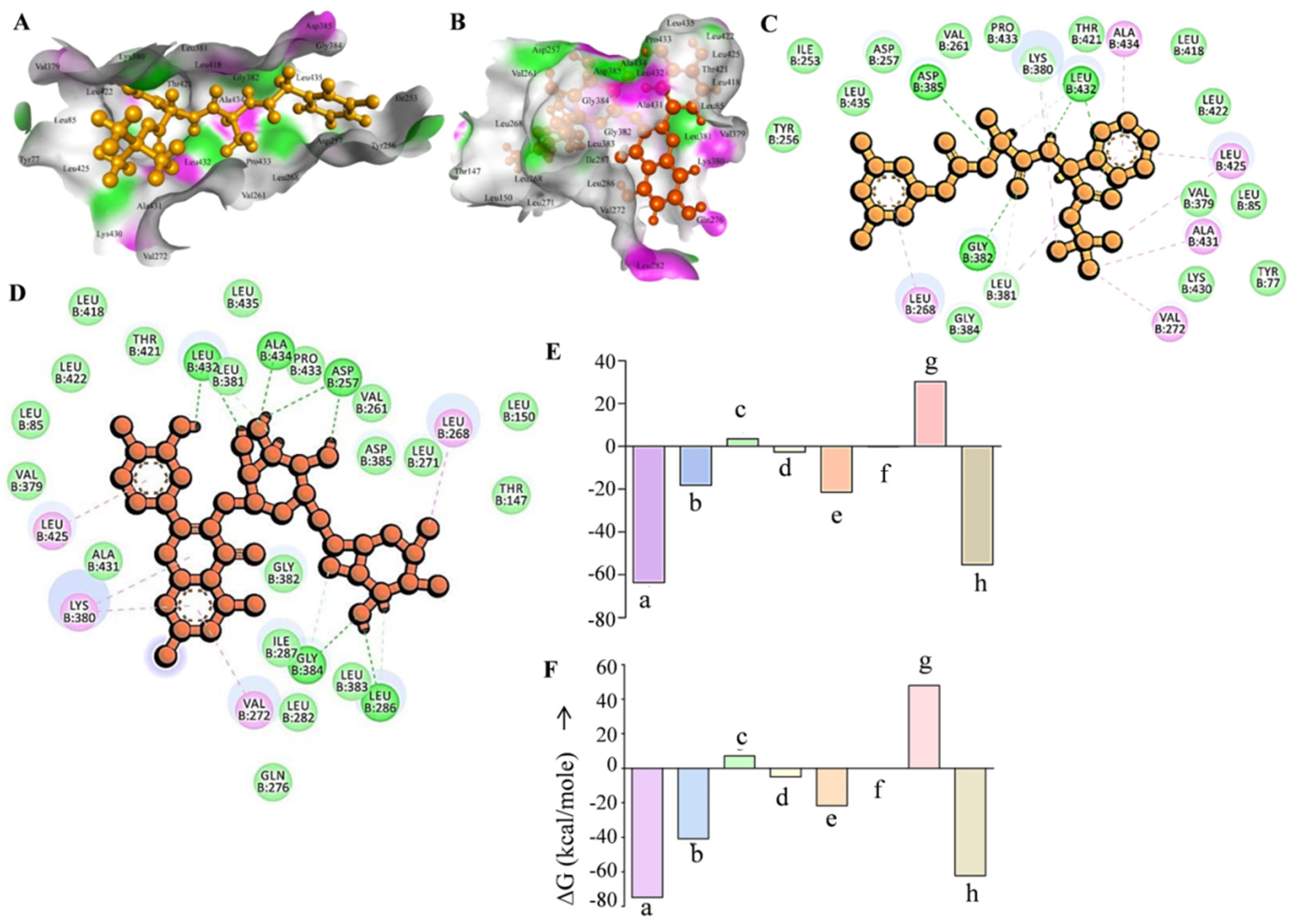
3.1. Rutin Binds Efficiently to the Gamma Secretase Catalytic Site
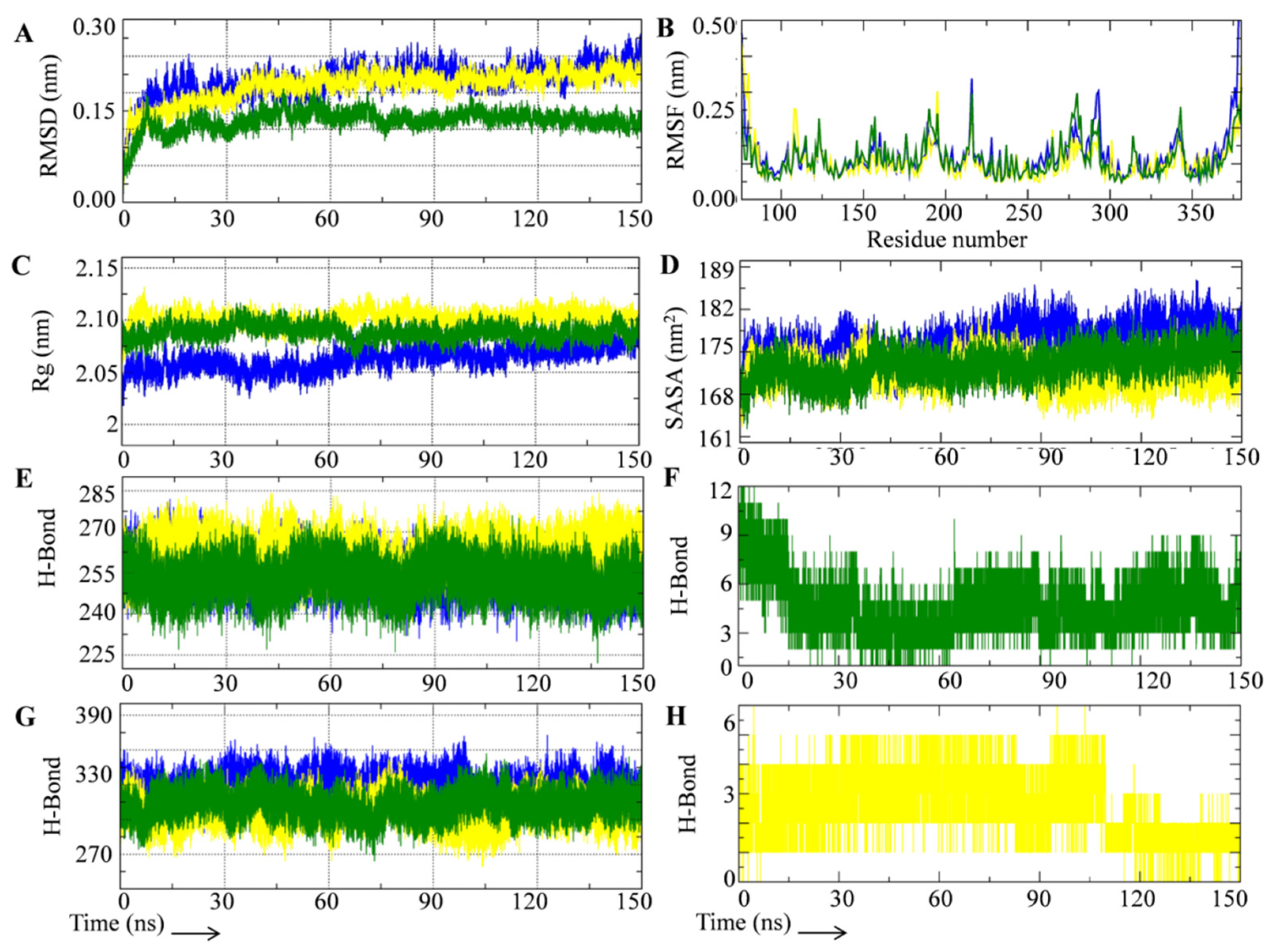
3.2. Rutin Forms Stable and an Energetically Favorable Complex with a Gamma Secretase Catalytic Subunit
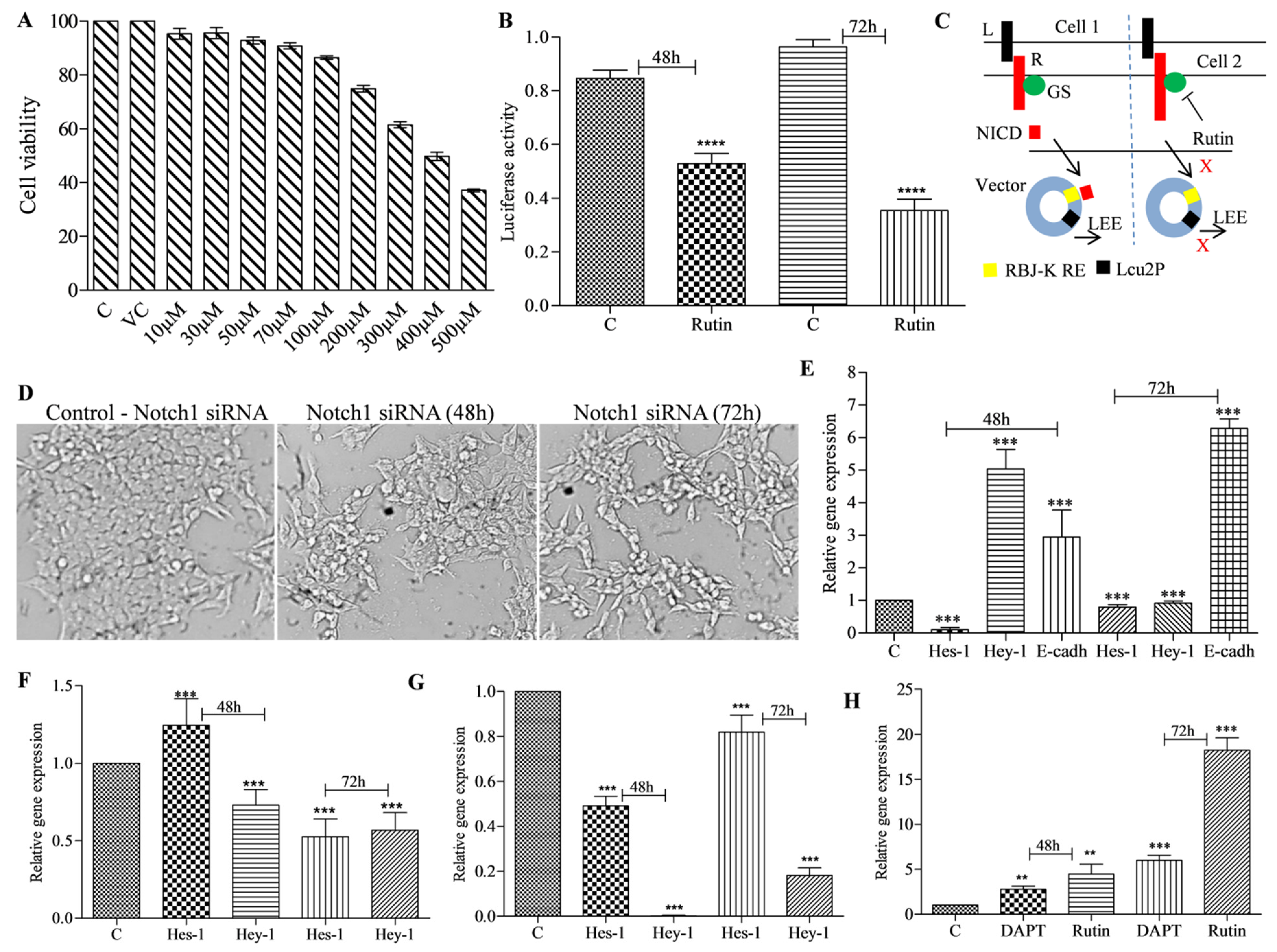
3.3. Rutin Treatment Decreases Notch Promoter Activity and Alters Notch Target Gene Expression in HCT-116 Cells
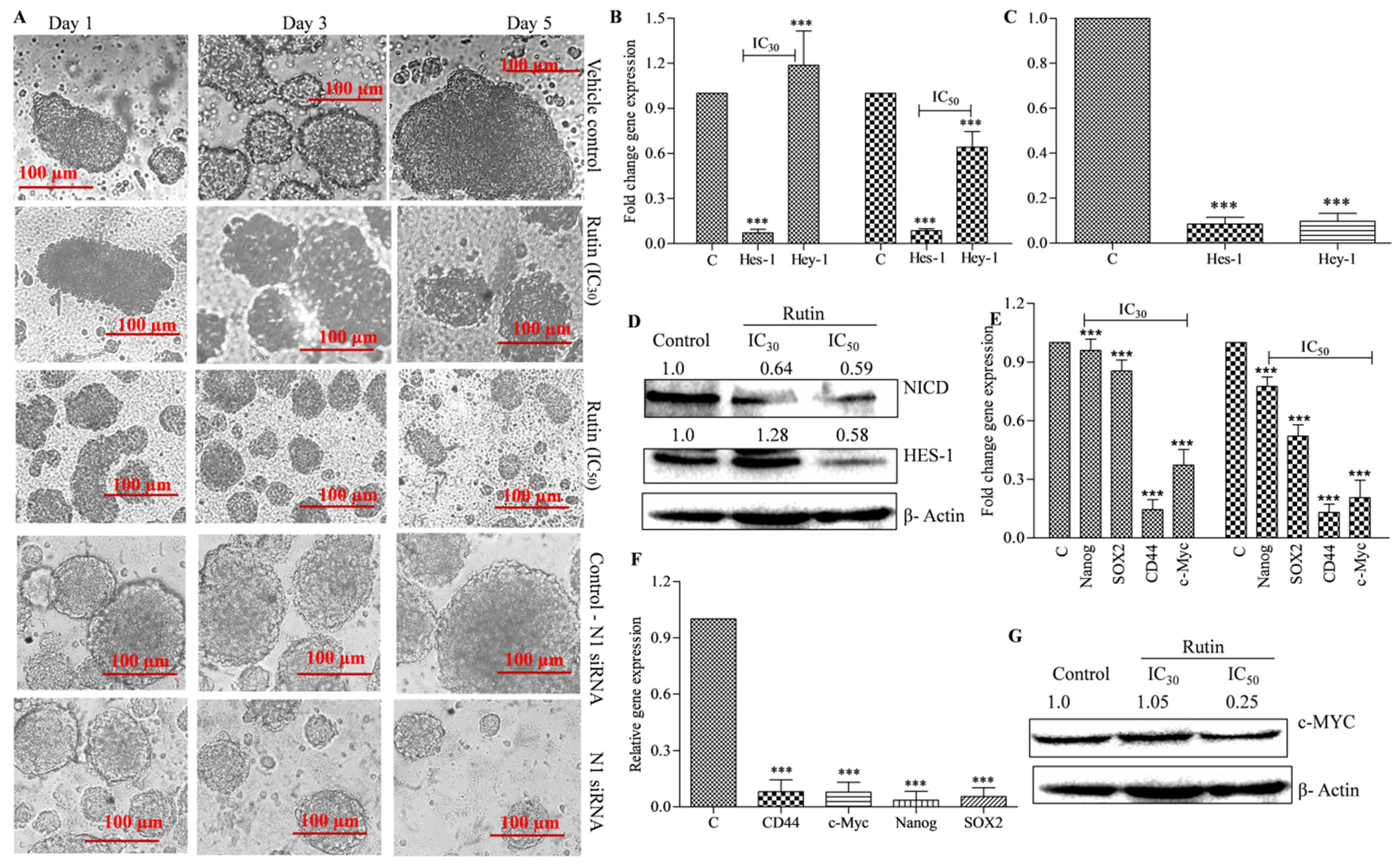
3.4. Rutin Decreases Colonosphere Formation by Downregulating Notch Signaling Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cutsem, E.V.; Köhne, C.H.; Hitre, J.; Zaluski, E.; Chang Chien, C.R.; Makhson, A.; D’Haens, G.; Pintér, T.; Lim, R.; Bodoky, G.; et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, P.J.; Sundaresan, R.S. Surgical management of colorectal lung metastasis. Clin. Colon Rectal Surg. 2009, 22, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Sethi, N.; Dai, X.; Winter, C.G.; Kang, Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 2011, 19, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ye, X.; Fan, F.; Xia, L.; Bhattacharya, R.; Bellister, S.; Tozzi, F.; Sceusi, E.; Zhou, Y.; Tachibana, I.; et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 2013, 23, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Arcaroli, J.J.; Tai, W.M.; McWilliams, R.; Bagby, S.; Blatchford, P.J.; Varella-Garcia, M.; Purkey, A.; Quackenbush, K.S.; Song, E.K.; Pitts, T.M.; et al. A NOTCH1 gene copy number gain is a prognostic indicator of worse survival and a predictive biomarker to a Notch1 targeting antibody in colorectal cancer. Int. J. Cancer 2016, 138, 195–205. [Google Scholar] [CrossRef]
- Jackstadt, R.; Van Hooff, S.R.; Leach, J.D.; Cortes-Lavaud, X.; Lohuis, J.O.; Ridgway, R.A.; Wouters, V.M.; Roper, J.; Kendall, T.J.; Roxburgh, C.S.; et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell 2019, 36, 319–336. [Google Scholar] [CrossRef]
- Fender, A.W.; Nutter, J.M.; Fitzgerald, T.L.; Bertrand, F.E.; Sigounas, G. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J. Cell. Biochem. 2015, 116, 2517–2527. [Google Scholar] [CrossRef]
- Tyagi, A.; Sharma, A.K.; Damodaran, C. A review on notch signaling and colorectal cancer. Cells 2020, 9, 1549. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Vardhan, P.S.; Kapewangolo, P.; Shuaib, M.; Prajapati, S.K.; Singh, A.K.; Kumar, S. Bulbine frutescens phytochemical inhibits notch signaling pathway and induces apoptosis in triple negative and luminal breast cancer cells. Life Sci. 2019, 234, 116783. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Prajapati, K.S.; Shuaib, M.; Kushwaha, P.P.; Tuli, H.S.; Singh, A.K. Five-Decade Update on Chemopreventive and Other Pharmacological Potential of Kurarinone: A Natural Flavanone. Front. Pharmacol. 2021, 12, 737137. [Google Scholar] [CrossRef]
- Lukšič, L.; Bonafaccia, G.; Timoracka, M.; Vollmannova, A.; Trček, J.; Nyambe, T.K.; Melini, V.; Acquistucci, R.; Germ, M.; Kreft, I. Rutin and quercetin transformation during preparation of buckwheat sourdough bread. J. Cereal Sci. 2016, 69, 71–76. [Google Scholar] [CrossRef]
- Satari, A.; Ghasemi, S.; Habtemariam, S.; Asgharian, S.; Lorigooini, Z. Rutin: A flavonoid as an effective sensitizer for anticancer therapy; insights into multifaceted mechanisms and applicability for combination therapy. Evid. Based Complement. Altern. Med. 2021, 2021, 9913179. [Google Scholar] [CrossRef] [PubMed]
- Nasri Nasrabadi, P.; Zareian, S.; Nayeri, Z.; Salmanipour, R.; Parsafar, S.; Gharib, E.; Asadzadeh Aghdaei, H.; Zali, M.R. A detailed image of rutin underlying intracellular signaling pathways in human SW480 colorectal cancer cells based on miRNAs-lncRNAs-mRNAs-TFs interactions. J. Cell. Physiol 2019, 234, 15570–15580. [Google Scholar] [CrossRef] [PubMed]
- Ben Sghaier, M.; Pagano, A.; Mousslim, M.; Ammari, Y.; Kovacic, H.; Luis, J. Rutin inhibits proliferation, attenuates superoxide production and decreases adhesion and migration of human cancerous cells. Biomed. Pharmacother. 2016, 84, 1972–1978. [Google Scholar] [CrossRef]
- Vijay, M.; Sivagami, G.; Thayalan, K.; Nalini, N. Radiosensitizing potential of rutin against human colon adenocarcinoma HT-29 cells. Bratisl. Lek. 2016, 117, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Khan, F.; Qari, H.A.; Oves, M. Rutin (Bioflavonoid) as Cell Signaling Pathway Modulator: Prospects in Treatment and Chemoprevention. Pharmaceuticals 2021, 14, 1069. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F.; Alzahrani, F.A.; Qari, H.A.; Oves, M. A Novel Approach to Unraveling the Apoptotic Potential of Rutin (Bioflavonoid) via Targeting Jab1 in Cervical Cancer Cells. Molecules 2021, 26, 5529. [Google Scholar] [CrossRef]
- Khan, F.; Pandey, P.; Jha, N.K.; Khalid, M.; Ojha, S. Rutin Mediated Apoptotic Cell Death in Caski Cervical Cancer Cells via Notch-1 and Hes-1 Downregulation. Life 2021, 11, 761. [Google Scholar] [CrossRef]
- Miyake, T.; Kuge, M.; Matsumoto, Y.; Shimada, M. α-glucosyl-rutin activates immediate early genes in human induced pluripotent stem cells. Stem Cell Res. 2021, 56, 102511. [Google Scholar] [CrossRef]
- Kalita, B.; Ranjan, R.; Gupta, M.L. Combination treatment of podophyllotoxin and rutin promotes mouse Lgr5+ ve intestinal stem cells survival against lethal radiation injury through Wnt signaling. Apoptosis 2019, 24, 326–340. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, R.; Guo, X.; Yan, C.; Lei, J.; Shi, Y. Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell 2021, 184, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modelling. Proteins 2011, 79, 2794–2812. [Google Scholar] [CrossRef]
- Dhanjal, J.K.; Kumar, V.; Garg, S.; Subramani, C.; Agarwal, S.; Wang, J.; Zhang, H.; Kaul, A.; Kalra, R.S.; Kaul, S.; et al. Molecular mechanism of anti-SARS-CoV2 activity of Ashwagandha-derived withanolides. Int. J. Biol. Macromol. 2021, 184, 297–312. [Google Scholar] [CrossRef]
- Zhang, W.; Qiu, K.X.; Yu, F.; Xie, X.G.; Zhang, S.Q.; Chen, Y.J.; Xie, H.D. Virtual screening of B-Raf kinase inhibitors: A combination of pharmacophore modelling, molecular docking, 3D-QSAR model and binding free energy calculation studies. Comput. Biol. Chem. 2017, 70, 186–190. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, A.K.; Kushwaha, P.P.; Prajapati, K.S.; Shuaib, M.; Senapati, S.; Kumar, S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2021, 39, 4334–4345. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, 370–376. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Haug, E.J.; Arora, J.S.; Matsui, K. A steepest-descent method for optimization of mechanical systems. J. Optim. Theory Appl. 1976, 19, 401–424. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; Van Gunsteren, W.F.; DiNola, A.R.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Evans, D.J.; Holian, B.L. The nose–hoover thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Rana, N.; Singh, A.K.; Shuaib, M.; Gupta, S.; Habiballah, M.M.; Alkhanani, M.F.; Haque, S.; Reshi, M.S.; Kumar, S. Drug Resistance Mechanism of M46I-Mutation-Induced Saquinavir Resistance in HIV-1 Protease Using Molecular Dynamics Simulation and Binding Energy Calculation. Viruses 2022, 14, 697. [Google Scholar] [CrossRef]
- Wolf, A.; Kirschner, K.N. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11· 23S subdomain. J. Mol. Model. 2013, 19, 539–549. [Google Scholar] [CrossRef]
- Huang, B.; Fan, S.; Liu, Y.; Zhao, Y.; Lin, D.; Liao, X. Solution structure and backbone dynamics for S1 domain of ribosomal protein S1 from Mycobacterium tuberculosis. Eur. Biophys. J. 2019, 48, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.P.; Singh, A.K.; Bansal, T.; Yadav, A.; Prajapati, K.S.; Shuaib, M.; Kumar, S. Identification of natural inhibitors against SARS-CoV-2 drugable targets using molecular docking, molecular dynamics simulation, and MM-PBSA approach. Front. Cell. Infect. Microbiol. 2021, 11, 730288. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.P.; Singh, A.K.; Prajapati, K.S.; Shuaib, M.; Fayez, S.; Bringmann, G.; Kumar, S. Induction of apoptosis in breast cancer cells by naphthylisoquinoline alkaloids. Toxicol. Appl. Pharmacol. 2020, 409, 115297. [Google Scholar] [CrossRef]
- Xu, Y.; Shu, B.; Tian, Y.; Wang, G.; Wang, Y.; Wang, J.; Dong, Y. Oleanolic acid induces osteosarcoma cell apoptosis by inhibition of Notch signaling. Mol. Carcinog. 2018, 57, 896–902. [Google Scholar] [CrossRef]
- Jaggi, M.; Chauhan, S.C.; Du, C.; Balaji, K.C. Bryostatin 1 modulates β-catenin subcellular localization and transcription activity through protein kinase D1 activation. Mol. Cancer Ther. 2008, 7, 2703–2712. [Google Scholar] [CrossRef]
- Shaheen, S.; Ahmed, M.; Lorenzi, F.; Nateri, A.S. Spheroid-formation (colonosphere) assay for in vitro assessment and expansion of stem cells in colon cancer. Stem Cell Rev. Rep. 2016, 12492–12499. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Singh, A.K.; Shuaib, M.; Prajapati, K.S.; Vardhan, P.S.; Gupta, S.; Kumar, S. 3-O-(E)-p-Coumaroyl betulinic acid possess anticancer activity and inhibit Notch signaling pathway in breast cancer cells and mammosphere. Chem. Biol. Interact. 2020, 328, 109200. [Google Scholar] [CrossRef] [PubMed]
- Chien, N.Q.; Hung, N.V.; Santarsiero, B.D.; Mesecar, A.D.; Cuong, N.M.; Soejarto, D.D.; Pezzuto, J.M.; Fong, H.H.; Tan, G.T. New 3-O-acyl betulinic acids from Strychnos vanprukii Craib. J. Nat. Prod. 2004, 67, 994–998. [Google Scholar] [CrossRef]
- Sahoo, B.M.; Ravi Kumar, B.V.; Sruti, J.; Mahapatra, M.K.; Banik, B.K.; Borah, P. Drug repurposing strategy (DRS): Emerging approach to identify potential therapeutics for treatment of novel coronavirus infection. Front. Mol. Biosci. 2021, 8, 628144. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kushwaha, P.P.; Prajapati, K.S.; Shuaib, M.; Gupta, S.; Kumar, S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1pp domain inhibitor: An in silico study. Comput. Biol. Med. 2021, 130, 104185. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided. Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, A.K.; Kushwaha, P.P.; Prajapati, K.S.; Senapati, S.; Shuaib, M.; Gupta, S. Identification of Compounds from Curcuma longa with In Silico Binding Potential against SARS-CoV-2 and Human Host Proteins Involve in Virus Entry and Pathogenesis. Indian J. Pharm. Sci. 2021, 83, 1181–1195. [Google Scholar] [CrossRef]
- Tandon, A.; Fraser, P. The presenilins. Genome Biol. 2002, 3, 1–9. [Google Scholar] [CrossRef]
- Wrigley, J.D.; Nunn, E.J.; Nyabi, O.; Clarke, E.E.; Hunt, P.; Nadin, A.; De Strooper, B.; Shearman, M.S.; Beher, D. Conserved residues within the putative active site of γ-secretase differentially influence enzyme activity and inhibitor binding. J. Neurochem. 2004, 90, 1312–1320. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Maurya, S.K.; Singh, A.; Prajapati, K.S.; Singh, A.K.; Shuaib, M.; Kumar, S. Bulbine frutescens phytochemicals as novel ABC-transporter inhibitor: A molecular docking and molecular dynamics simulation study. J. Cancer Metastasis Treat. 2021, 7, 2. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Singh, A.K.; Prajapati, K.S.; Shuaib, M.; Gupta, S.; Kumar, S. Phytochemicals present in Indian ginseng possess potential to inhibit SARS-CoV-2 virulence: A molecular docking and MD simulation study. Microb. Pathog. 2021, 157, 104954. [Google Scholar] [CrossRef]
- El Khatabi, K.; Aanouz, I.; El-Mernissi, R.; Singh, A.K.; Ajana, M.A.; Lakhlifi, T.; Kumar, S.; Bouachrine, M. Integrated 3D-QSAR, molecular docking, and molecular dynamics simulation studies on 1, 2, 3-triazole based derivatives for designing new acetylcholinesterase inhibitors. Turk. J. Chem. 2021, 45, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kumar, S. Naringin dihydrochalcone potentially binds to catalytic domain of matrix metalloproteinase-2: Molecular docking, MM-GBSA, and molecular dynamics simulation approach. Nat. Prod. Res. 2022, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Bhatt, L.K.; Johnston, T.P.; Prabhavalkar, K.S. Colon cancer stem cells: Potential target for the treatment of colorectal cancer. Cancer Biol. Ther. 2019, 20, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.K.; Shuaib, M.; Prajapati, K.S.; Kumar, S. Rutin Potentially Binds the Gamma Secretase Catalytic Site, Down Regulates the Notch Signaling Pathway and Reduces Sphere Formation in Colonospheres. Metabolites 2022, 12, 926. https://doi.org/10.3390/metabo12100926
Singh AK, Shuaib M, Prajapati KS, Kumar S. Rutin Potentially Binds the Gamma Secretase Catalytic Site, Down Regulates the Notch Signaling Pathway and Reduces Sphere Formation in Colonospheres. Metabolites. 2022; 12(10):926. https://doi.org/10.3390/metabo12100926
Chicago/Turabian StyleSingh, Atul Kumar, Mohd Shuaib, Kumari Sunita Prajapati, and Shashank Kumar. 2022. "Rutin Potentially Binds the Gamma Secretase Catalytic Site, Down Regulates the Notch Signaling Pathway and Reduces Sphere Formation in Colonospheres" Metabolites 12, no. 10: 926. https://doi.org/10.3390/metabo12100926
APA StyleSingh, A. K., Shuaib, M., Prajapati, K. S., & Kumar, S. (2022). Rutin Potentially Binds the Gamma Secretase Catalytic Site, Down Regulates the Notch Signaling Pathway and Reduces Sphere Formation in Colonospheres. Metabolites, 12(10), 926. https://doi.org/10.3390/metabo12100926