Mycobacterium avium subsp. paratuberculosis Infected Cows Reveal Divergent Immune Response in Bovine Peripheral Blood Derived Lymphocyte Proteome
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Animals and Detection of MAP Infection Status
2.2. Preparation of PBMC and Co-Incubation of PBMC with Viable MAP In Vitro
2.3. Sample Digestion for Differential Proteome Analysis
2.4. Mass Spectrometric Analysis and Protein Identification
2.5. Data Processing
2.6. Flow Cytometric Analysis of PBMC
2.7. Immunocytology and Quantification of Signal Intensities
3. Results
3.1. The Proteome of Bovine Peripheral Blood Lymphocytes Consisted of 2631 Proteins and Showed Significant Differences between MAP-Resistant and Persistently MAP-Infected Cows after 48 h of Co-Incubation with MAP In Vitro
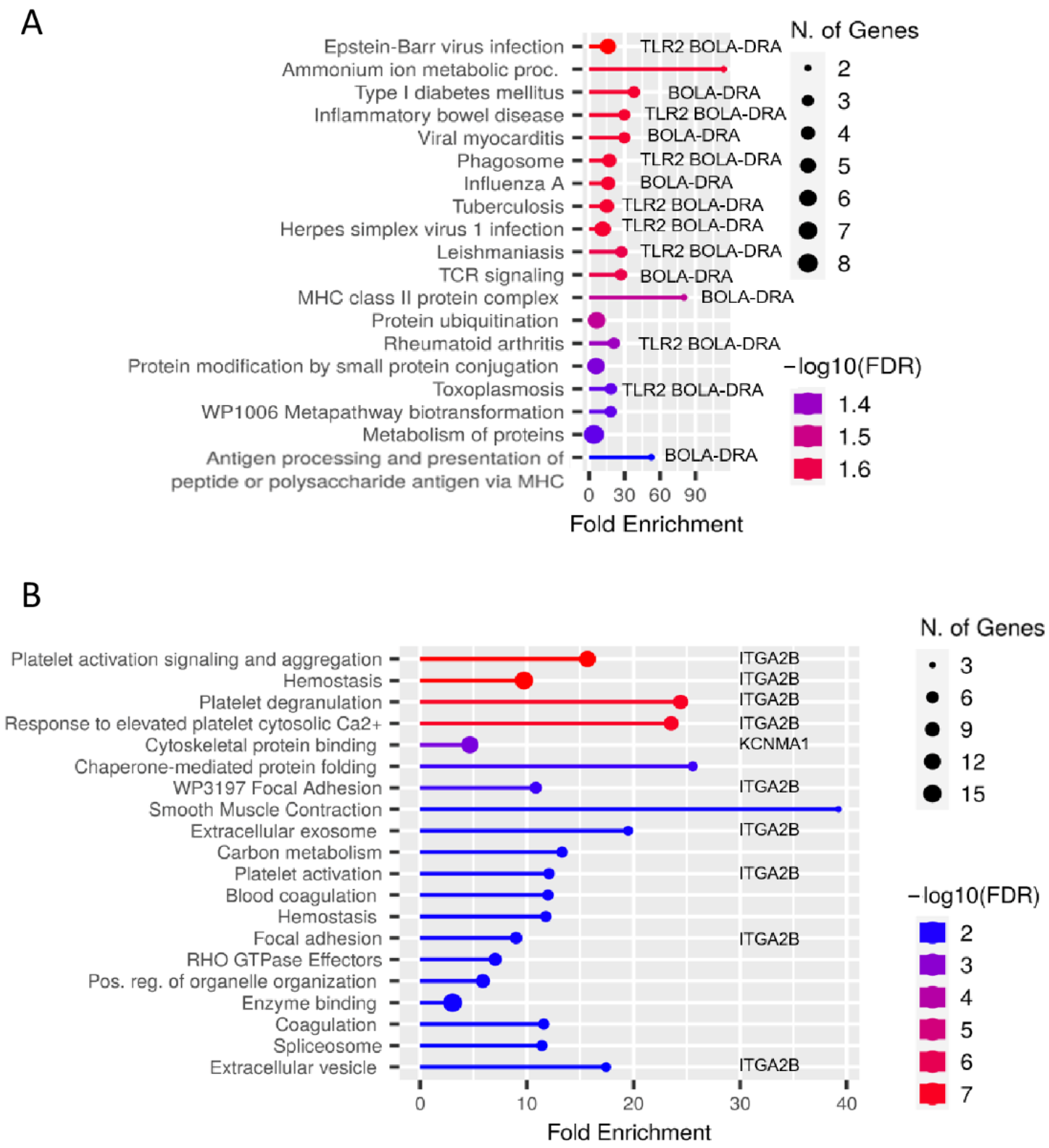
3.2. Analyses of Enriched Signaling Pathways Revealed Functional Differences between Lymphocytes from MAP-Resistant and Persistently MAP-Infected Cows after Co-Incubation with MAP In Vitro
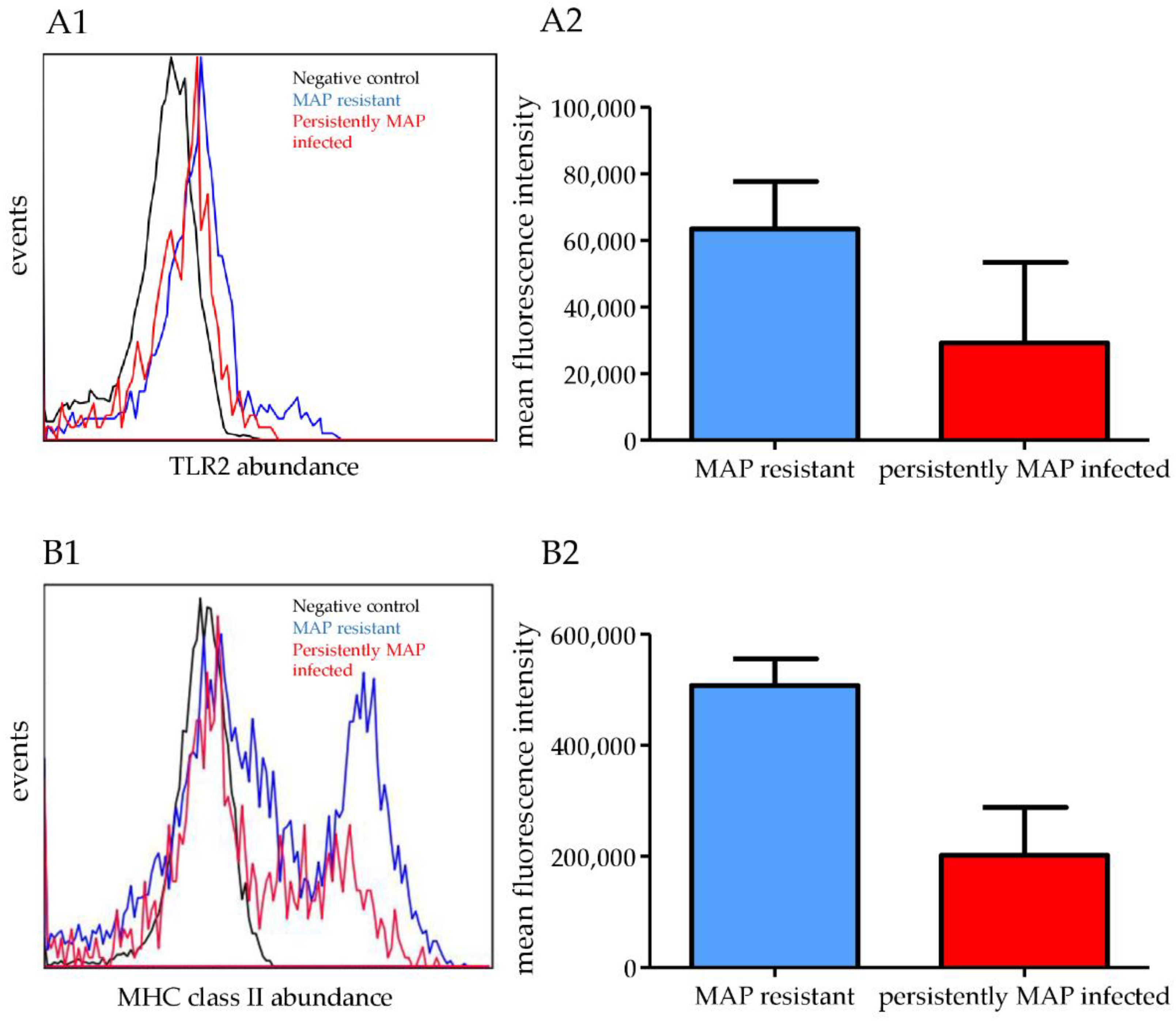
3.3. In Vitro Co-Incubation with MAP for 48 h Increased the Abundances of TLR2, BOLA-DRB3, and BOLA-DRA in MAP-Resistant Cows
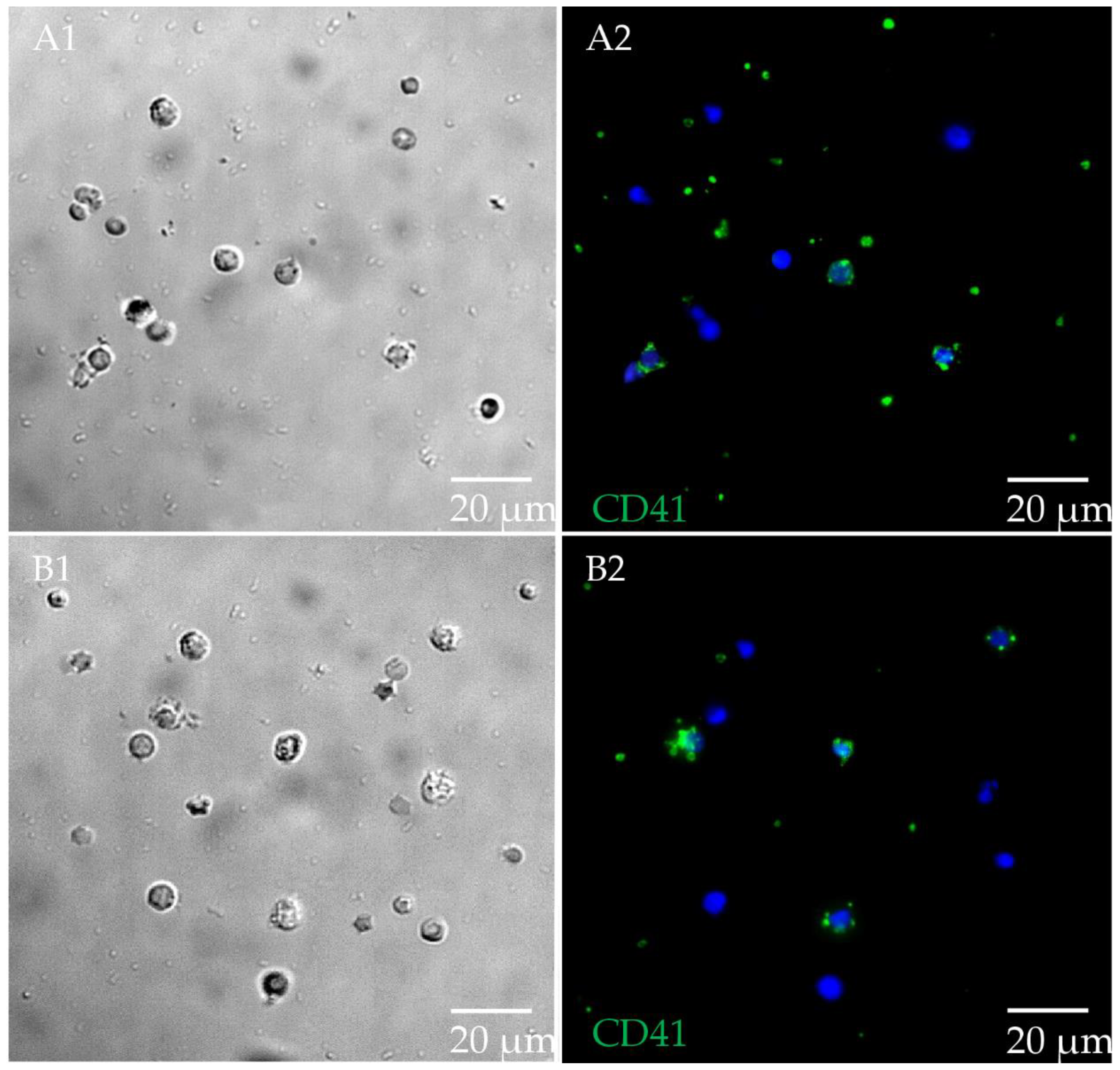
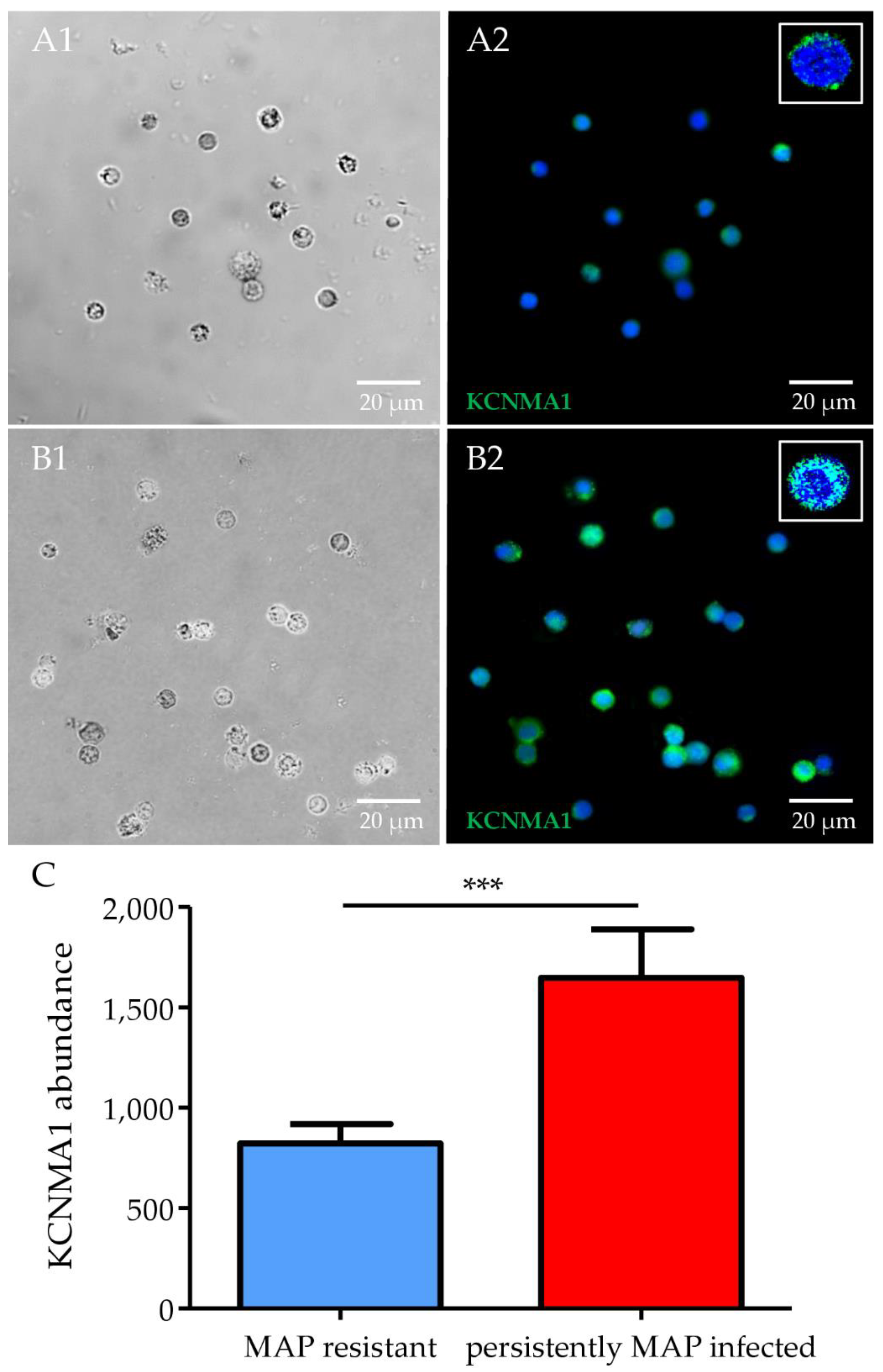
3.4. Significantly Higher Expression of ITGA2B and KCNMA1 in Persistently MAP-Infected Cows after Co-Incubation with MAP In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conde, C.; Branger, M.; Cochard, T.; Rossignol, M.-N.; Fourichon, C.; Delafosse, A.; Davergne, A.; Joly, A.; Ngwa-Mbot, D.; Journaux, L.; et al. Draft Genome Sequences of 142 Mycobacterium avium subsp. paratuberculosis Strains Isolated from Naturally Infected Dairy Cattle. Microbiol. Resour. Announc. 2021, 10, e0069721. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Reyes, A.L.; Chávez-Gris, G.; Maldonado-Castro, E.; Alcaraz-Sosa, E.L.; Díaz-Negrete, M.T. First identification of Mycobacterium avium subsp. paratuberculosis in wild ruminants in a zoo in Mexico. Vet. World 2022, 12, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, P.; Barkema, H.W.; Mason, S.; Beaulieu, E.; Hall, D.C. Economic losses due to Johne’s disease (paratuberculosis) in dairy cattle. J. Dairy Sci. 2021, 104, 3123–3143. [Google Scholar] [CrossRef] [PubMed]
- Kravitz, A.; Pelzer, K.; Sriranganathan, N. The Paratuberculosis Paradigm Examined: A Review of Host Genetic Resistance and Innate Immune Fitness in Mycobacterium avium subsp. Paratuberculosis Infection. Front. Vet. Sci. 2021, 8, 721706. [Google Scholar] [CrossRef]
- Alonso-Hearn, M.; Canive, M.; Blanco-Vazquez, C.; Torremocha, R.; Balseiro, A.; Amado, J.; Varela, E.; Ramos, R.; Jugo, B.M.; Casais, R. RNA-Seq analysis of ileocecal valve and peripheral blood from Holstein cattle infected with Mycobacterium avium subsp. paratuberculosis revealed dysregulation of the CXCL8/IL8 signaling pathway. Sci. Rep. 2019, 9, 14845. [Google Scholar] [CrossRef]
- Whittington, R.; Donat, K.; Weber, M.F.; Kelton, D.; Nielsen, S.S.; Eisenberg, S.; Arrigoni, N.; Juste, R.; Sáez, J.L.; Dhand, N.; et al. Control of paratuberculosis: Who, why and how. A review of 48 countries. BMC Vet. Res. 2019, 15, 198. [Google Scholar] [CrossRef]
- Taylor, E.N.; Beckmann, M.; Villarreal-Ramos, B.; Vordermeier, H.-M.; Hewinson, G.; Rooke, D.; Mur, L.A.J.; Koets, A.P. Metabolomic Changes in Naturally MAP-Infected Holstein-Friesian Heifers Indicate Immunologically Related Biochemical Reprogramming. Metabolites 2021, 11, 727. [Google Scholar] [CrossRef]
- Kleinwort, K.J.; Hauck, S.M.; Degroote, R.; Scholz, A.M.; Hölzel, C.; Maertlbauer, E.P.; Deeg, C. Peripheral blood bovine lymphocytes and MAP show distinctly different proteome changes and immune pathways in host-pathogen interaction. PeerJ 2019, 7, e8130. [Google Scholar] [CrossRef]
- Hobmaier, B.F.; Lutterberg, K.; Kleinwort, K.J.; Mayer, R.; Hirmer, S.; Amann, B.; Hölzel, C.; Märtlbauer, E.P.; Deeg, C.A. Characterization of plant lectins for their ability to isolate Mycobacterium avium subsp. paratuberculosis from milk. Food Microbiol. 2019, 82, 231–239. [Google Scholar] [CrossRef]
- Canive, M.; Fernandez-Jimenez, N.; Casais, R.; Vázquez, P.; Lavín, J.L.; Bilbao, J.R.; Blanco-Vázquez, C.; Garrido, J.M.; Juste, R.A.; Alonso-Hearn, M. Identification of loci associated with susceptibility to bovine paratuberculosis and with the dysregulation of the MECOM, eEF1A2, and U1 spliceosomal RNA expression. Sci. Rep. 2021, 11, 313. [Google Scholar] [CrossRef]
- Juste, R.; Geijo, M.; Elguezabal, N.; Sevilla, I.; Alonso-Hearn, M.; Garrido, J. Paratuberculosis vaccination specific and non-specific effects on cattle lifespan. Vaccine 2021, 39, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Rosseels, V.; Huygen, K. Vaccination against paratuberculosis. Expert Rev. Vaccines 2008, 7, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Crociati, M.; Grispoldi, L.; Chalias, A.; Monaci, M.; Cenci-Goga, B.; Sylla, L. Effect of Culling Management Practices on the Seroprevalence of Johne’s Disease in Holstein Dairy Cattle in Central Italy. Vet. Sci. 2022, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Mitchell, R.; Smith, R.; Van Kessel, J.; Chapagain, P.; Schukken, Y.; Grohn, Y. The importance of culling in Johne’s disease control. J. Theor. Biol. 2008, 254, 135–146. [Google Scholar] [CrossRef]
- McAloon, C.G.; Roche, S.; Ritter, C.; Barkema, H.W.; Whyte, P.; More, S.J.; O’Grady, L.; Green, M.J.; Doherty, M.L. A review of paratuberculosis in dairy herds—Part 2: On-farm control. Vet. J. 2019, 246, 54–58. [Google Scholar] [CrossRef]
- Ritter, C.; Wolf, R.; Adams, C.; Kelton, D.; Pickel, C.; Mason, S.; Orsel, K.; De Buck, J.; Barkema, H. Short communication: Herd-level prevalence of Mycobacterium avium ssp. paratuberculosis is not associated with participation in a voluntary Alberta Johne’s disease control program. J. Dairy Sci. 2016, 99, 2157–2160. [Google Scholar] [CrossRef] [PubMed]
- Lutterberg, K.; Kleinwort, K.J.H.; Hobmaier, B.F.; Hauck, S.M.; Nüske, S.; Scholz, A.M.; Deeg, C.A. A Functionally Different Immune Phenotype in Cattle Is Associated With Higher Mastitis Incidence. Front. Immunol. 2018, 9, 2884. [Google Scholar] [CrossRef]
- Rodríguez-Gil, A.; Ritter, O.; Saul, V.V.; Wilhelm, J.; Yang, C.-Y.; Grosschedl, R.; Imai, Y.; Kuba, K.; Kracht, M.; Schmitz, M.L. The CCR4-NOT complex contributes to repression of Major Histocompatibility Complex class II transcription. Sci. Rep. 2017, 7, 3547. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The Proteome of Native Adult Müller Glial Cells from Murine Retina. Mol. Cell Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Ezz, M.A.; Marey, M.A.; Elweza, A.E.; Kawai, T.; Heppelmann, M.; Pfarrer, C.; Balboula, A.Z.; Montaser, A.; Imakawa, K.; Zaabel, S.M.; et al. TLR2/4 signaling pathway mediates sperm-induced inflammation in bovine endometrial epithelial cells in vitro. PLoS ONE 2019, 14, e0214516. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Houe, H.; Denwood, M.; Nielsen, L.R.; Forkman, B.; Otten, N.D.; Agger, J.F. Application of Methods to Assess Animal Welfare and Suffering Caused by Infectious Diseases in Cattle and Swine Populations. Animals 2021, 11, 3017. [Google Scholar] [CrossRef] [PubMed]
- Gondaira, S.; Nishi, K.; Iwano, H.; Fujiki, J.; Watanabe, R.; Eguchi, A.; Hirano, Y.; Higuchi, H.; Nagahata, H. Transcriptome analysis of Mycoplasma bovis stimulated bovine peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 2021, 232, 110166. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.S.; Parsons, R.; Patterson, R.; Coffey, T.J.; Thonur, L.; Chang, J.-S.; Russell, G.; Haig, D.; Werling, D.; Hope, J.C. Characterisation of antibodies to bovine Toll-like receptor (TLR)-2 and cross-reactivity with ovine TLR2. Vet. Immunol. Immunopathol. 2011, 139, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Ozinsky, A.; Smith, K.D.; Aderem, A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. USA 1999, 96, 14459–14463. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, S.; de Silva, K.; Whittington, R.J.; Plain, K.M. In vivo and in vitro expression pattern of Toll-like receptors in Mycobacterium avium subspecies paratuberculosis infection. Vet. Immunol. Immunopathol. 2013, 156, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Drennan, M.B.; Nicolle, D.; Quesniaux, V.J.; Jacobs, M.; Allie, N.; Mpagi, J.; Frémond, C.; Wagner, H.; Kirschning, C.; Ryffel, B. Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am. J. Pathol. 2004, 164, 49–57. [Google Scholar] [CrossRef]
- Park, H.-T.; Bin Park, W.; Kim, S.; Lim, J.-S.; Nah, G.; Yoo, H.S. Revealing immune responses in the Mycobacterium avium subsp. paratuberculosis-infected THP-1 cells using single cell RNA-sequencing. PLoS ONE 2021, 16, e0254194. [Google Scholar] [CrossRef]
- Taylor, B.C.; Choi, K.Y.; Scibienski, R.J.; Moore, P.F.; Stott, J.L. Differential expression of bovine MHC class II antigens identified by monoclonal antibodies. J. Leukoc. Biol. 1993, 53, 479–489. [Google Scholar] [CrossRef]
- Miyasaka, T.; Takeshima, S.-N.; Sentsui, H.; Aida, Y. Identification and diversity of bovine major histocompatibility complex class II haplotypes in Japanese Black and Holstein cattle in Japan. J. Dairy Sci. 2012, 95, 420–431. [Google Scholar] [CrossRef]
- Weiss, D.J.; Evanson, O.A.; McClenahan, D.J.; Abrahamsen, M.S.; Walcheck, B.K. Regulation of expression of major histocompatibility antigens by bovine macrophages infected with Mycobacterium avium subsp. paratuberculosis or Mycobacterium avium subsp. avium. Infect. Immun. 2001, 69, 1002–1008. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khan, A.; Bakhru, P.; Saikolappan, S.; Das, K.; Soudani, E.; Singh, C.R.; Estrella, J.L.; Zhang, D.; Pasare, C.; Ma, Y.; et al. An autophagy-inducing and TLR-2 activating BCG vaccine induces a robust protection against tuberculosis in mice. NPJ Vaccines 2019, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, L.; Noss, E.; Boom, W.H.; Harding, C.V. Processing of Mycobacterium tuberculosis antigen 85B involves intraphagosomal formation of peptide-major histocompatibility complex II complexes and is inhibited by live bacilli that decrease phagosome maturation. J. Exp. Med. 2001, 194, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Dusinsky, R.; Horovska, L.; Bilka, F.; Hluchy, S. Immunohistochemical reactivity of anti-platelet monoclonal antibodies. Vet. Immunol. Immunopathol. 1996, 52, 377–382. [Google Scholar] [CrossRef]
- Ammari, M.; McCarthy, F.M.; Nanduri, B.; Pinchuk, L.M. Analysis of Bovine Viral Diarrhea Viruses-infected monocytes: Identification of cytopathic and non-cytopathic biotype differences. BMC Bioinform. 2010, 11 (Suppl. S6), S9. [Google Scholar] [CrossRef]
- Fox, K.A.; Kirwan, D.E.; Whittington, A.M.; Krishnan, N.; Robertson, B.D.; Gilman, R.H.; López, J.W.; Singh, S.; Porter, J.C.; Friedland, J.S. Platelets Regulate Pulmonary Inflammation and Tissue Destruction in Tuberculosis. Am. J. Respir. Crit. Care Med. 2018, 198, 245–255. [Google Scholar] [CrossRef]
- Forde, N.; Carter, F.; Spencer, T.; Bazer, F.; Sandra, O.; Mansouri-Attia, N.; Okumu, L.; McGettigan, P.; Mehta, J.; McBride, R.; et al. Conceptus-induced changes in the endometrial transcriptome: How soon does the cow know she is pregnant? Biol. Reprod. 2011, 85, 144–156. [Google Scholar] [CrossRef]
- Salkoff, L.; Butler, A.; Ferreira, G.; Santi, C.; Wei, A. High-conductance potassium channels of the SLO family. Nat. Rev. Neurosci. 2006, 7, 921–931. [Google Scholar] [CrossRef]
- Augustino, S.M.A.; Xu, Q.; Liu, X.; Liu, L.; Zhang, Q.; Yu, Y. Transcriptomic Study of Porcine Small Intestine Epithelial Cells Reveals Important Genes and Pathways Associated With Susceptibility to Escherichia coli F4ac Diarrhea. Front. Genet. 2020, 11, 68. [Google Scholar] [CrossRef]
- Santos, A.S.; Cunha-Neto, E.; Gonfinetti, N.V.; Bertonha, F.B.; Brochet, P.; Bergon, A.; Moreira-Filho, C.A.; Chevillard, C.; da Silva, M.E.R. Prevalence of Inflammatory Pathways Over Immuno-Tolerance in Peripheral Blood Mononuclear Cells of Recent-Onset Type 1 Diabetes. Front. Immunol. 2021, 12, 765264. [Google Scholar] [CrossRef]
- Gry, M.; Rimini, R.; Strömberg, S.; Asplund, A.; Pontén, F.; Uhlén, M.; Nilsson, P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 2009, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Reimegård, J.; Tarbier, M.; Danielsson, M.; Schuster, J.; Baskaran, S.; Panagiotou, S.; Dahl, N.; Friedländer, M.R.; Gallant, C.J. A combined approach for single-cell mRNA and intracellular protein expression analysis. Commun. Biol. 2021, 4, 624. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korbonits, L.; Kleinwort, K.J.H.; Amann, B.; Didier, A.; Märtlbauer, E.; Hauck, S.M.; Deeg, C.A. Mycobacterium avium subsp. paratuberculosis Infected Cows Reveal Divergent Immune Response in Bovine Peripheral Blood Derived Lymphocyte Proteome. Metabolites 2022, 12, 924. https://doi.org/10.3390/metabo12100924
Korbonits L, Kleinwort KJH, Amann B, Didier A, Märtlbauer E, Hauck SM, Deeg CA. Mycobacterium avium subsp. paratuberculosis Infected Cows Reveal Divergent Immune Response in Bovine Peripheral Blood Derived Lymphocyte Proteome. Metabolites. 2022; 12(10):924. https://doi.org/10.3390/metabo12100924
Chicago/Turabian StyleKorbonits, Lucia, Kristina J. H. Kleinwort, Barbara Amann, Andrea Didier, Erwin Märtlbauer, Stefanie M. Hauck, and Cornelia A. Deeg. 2022. "Mycobacterium avium subsp. paratuberculosis Infected Cows Reveal Divergent Immune Response in Bovine Peripheral Blood Derived Lymphocyte Proteome" Metabolites 12, no. 10: 924. https://doi.org/10.3390/metabo12100924
APA StyleKorbonits, L., Kleinwort, K. J. H., Amann, B., Didier, A., Märtlbauer, E., Hauck, S. M., & Deeg, C. A. (2022). Mycobacterium avium subsp. paratuberculosis Infected Cows Reveal Divergent Immune Response in Bovine Peripheral Blood Derived Lymphocyte Proteome. Metabolites, 12(10), 924. https://doi.org/10.3390/metabo12100924






