Abstract
Assessment of dietary intake is challenging. Traditional methods suffer from both random and systematic errors; thus objective measures are important complements in monitoring dietary exposure. The study presented here aims to identify serum metabolites associated with reported food intake and to explore whether combinations of metabolites may improve predictive models. Fasting blood samples and a 4-day weighed food diary were collected from healthy Swedish subjects (n = 119) self-defined as having habitual vegan, vegetarian, vegetarian + fish, or omnivore diets. Serum was analyzed for metabolites by 1H-nuclear magnetic resonance spectroscopy. Associations between single and combined metabolites and 39 foods and food groups were explored. Area under the curve (AUC) was calculated for prediction models. In total, 24 foods or food groups associated with serum metabolites using the criteria of rho > 0.2, p < 0.01 and AUC ≥ 0.7 were identified. For the consumption of soybeans, citrus fruits and marmalade, nuts and almonds, green tea, red meat, poultry, total fish and shellfish, dairy, fermented dairy, cheese, eggs, and beer the final models included two or more metabolites. Our results indicate that a combination of metabolites improve the possibilities to use metabolites to identify several foods included in the current diet. Combined metabolite models should be confirmed in dose–response intervention studies.
1. Introduction
Poor diet is one of the leading risk factors for morbidity and mortality worldwide [1,2]. Nutritional epidemiology provides the main method for the assessment of long-term risk from diet in populations and accurate measurements of habitual diet is therefore crucial. However, measurement of true dietary intake is challenging. Traditionally used methods include food frequency questionnaires (FFQ), 24 h recalls and food records (weighed or unweighted). These methods are based on self-reported data and therefore prone to subjectivity and suffer from systematic and random errors such as recall bias and under-reporting [3]. As a result, epidemiological studies of diet/health relationships may suffer from inaccurate risk estimates and/or inconsistencies [3,4]. Consequently, objective measures, for example biomarkers, are of importance to reduce errors in the measure of dietary exposure and to correlate food intake to health outcomes.
Few validated dietary biomarkers exist and these include urinary sodium, nitrogen and sucrose/fructose for the estimation of salt, protein, and sugar intake, respectively, and the doubly labeled water technique for energy expenditure and alkylresorcinols for whole grain intake [4]. Nevertheless, putative dietary biomarkers have been proposed for several foods or food groups, including red meat, coffee, nuts, red wine, vegetables, legumes, fermented food products, citrus fruit, apples, milk, cheese, tea, and sugar-sweetened beverages [5,6,7,8,9,10,11,12,13] as well as for dietary patterns [14]. Metabolomics has been used to identify several of these dietary biomarkers and seems to be a promising approach for providing possible biomarkers of dietary intake [4]. Commonly, single biomarkers have been identified to estimate intake of a food, food groups, or nutrients. However, combining biomarkers may further improve the possibility to accurately predict dietary exposure. Using observational and intervention studies, multiple biomarkers as predictors of dietary exposure have been explored [15]. Even so, the food, food groups, and dietary patterns explored are to date limited.
Using 1H-nuclear magnetic resonance (1H-NMR) metabolomics analysis, we aimed to (1) identify serum metabolites associated with reported food intake and (2) explore the potential to use combinations of serum metabolites to improve predictive models of reported habitual food intake.
2. Results
2.1. Subject Characteristics
Subjects ranged in age from 19 to 57 years with a median age of 28 years, 63% of the subjects were female, and the median body mass index (BMI) (min-max) was 21.6 (18.2–28.9) kg/m2. The distribution of self-reported habitual dietary intake pattern was vegan (n = 43), lacto–ovo vegetarian (n = 25), lacto–ovo vegetarian + fish (n = 13), and omnivore (n = 38). For most food groups, consumers, as well as non-consumers, were identified; in short, 66% of subjects reported consumption of cruciferous vegetables, 26% of fish and shellfish, 31% of meat and poultry, 44% of eggs, 60% of dairy, and 23% reported consumption of beer during the 4 days. Relatively few subjects reported consuming soybeans (3.4%), white wine (14%), or spirits (5.9%).
2.2. Diet-Metabolite Associations
In total, 438 associations with absolute rho > 0.2 (rho = −0.506 to 0.628) and p < 0.01 were identified between the 39 foods/food groups and 237 1H-NMR -variables reflecting serum metabolites. These are presented in Supplemental Table S1.
Adding the criteria AUC ≥ 0.7, 95 of these associations, which included 24 of the food or food groups, remained of interest (Table 1). Reported consumption (non-consumers and consumers) of these 24 foods/food groups are shown in Supplemental Table S2.

Table 1.
Serum metabolites associated with reported food intake among healthy Swedish subjects (n = 119).
2.2.1. Plant-Derived Foods
No associations with metabolites were identified for legume consumption, but consumption of soy beverages (rho = 0.32, AUC = 71%) and soy products (rho = 0.44, AUC = 78%) was associated with glycine (Table 1). In addition, soybean consumption was associated with asparagine and valine; however, among the study subjects, overall consumption of soybeans was low (3.4%). Diet-metabolite associations were also identified for cruciferous vegetables and citrus fruits. Cruciferous vegetable consumption was associated with glutamine + an unidentified metabolite. Citrus fruits and citrus products were associated with two unidentified variables, and this was also the case for the reported consumption of nuts and almonds (Table 1).
2.2.2. Animal-Derived Foods
Food-metabolite associations for meat, poultry, eggs, and dairy consumption were mainly characterized by amino acids (Table 1). Consumption of red meat, meat products/processed meat, poultry, eggs, and total dairy were predominantly associated with the branched-chain amino acids (BCAAs) valine, leucine, and isoleucine (rho = 0.25–0.44, AUC = 70–77%). These foods also were associated with 3-hydroxybutyrate (rho = 0.37–0.40, AUC = 75–82%) and creatine (rho = 0.27–0.54, AUC = 71–87%). In addition, consumption of red meat was associated with lipids, phosphocholine, acetylcholine, and phosphoethanolamine (rho = 0.27, AUC = 70%) and meat products/processed meat were associated with creatinine (rho = 0.303, AUC = 71%). Cheese and fermented dairy products were associated with 2-aminobutyrate (uncertain identification) but not with 3-hydroxybutyrate. Total fish and seafood consumption were also associated with creatine (rho = 0.41, AUC = 75%), creatine + lysine (rho = 0.44, AUC = 77%), and 3-hydroxybutyrate (rho = 0.30, AUC = 71%).
2.2.3. Alcoholic Beverages
Beer consumption was associated with several metabolites: isoleucine + unidentified; lipids/free fatty acids + methylguanidine; and glucose + lysine + unidentified and proline (rho = 0.24–0.38, AUC = 71–80%).
2.3. Combined Serum Metabolites for Prediction of Food Intake
To improve the prediction of reported food exposure by serum metabolites, combinations of two or more metabolites were created. Combinations of metabolites were selected for all 39 food/food groups using forward stepwise logistic regression. For soybeans, citrus fruits and marmalade, nuts and almonds, green tea, red meat, poultry, total fish and shellfish, dairy, fermented dairy, cheese, eggs, and beer the final regression model included two or more metabolites (Table 2).

Table 2.
Combined metabolite models to predict reported food intake among healthy Swedish subjects (n = 119).
AUC for soybean consumption was improved using combined metabolites (valine and asparagine). For citrus fruits and marmalade as well as for nuts and almonds, combined metabolites (all unidentified) only marginally improved the model (Table 2); note that unidentified variables may originate from the same metabolite. The predictive ability of consumption of meat products/processed meat and poultry consumption increased when combining metabolites (creatinine, creatine + lysine, and valine). For red meat consumption, the combined metabolite model (3-methylhistidine, leucine, and creatine + lysine) improved the AUC (92%) compared to each separate metabolite (70–82%) (Figure 1A).
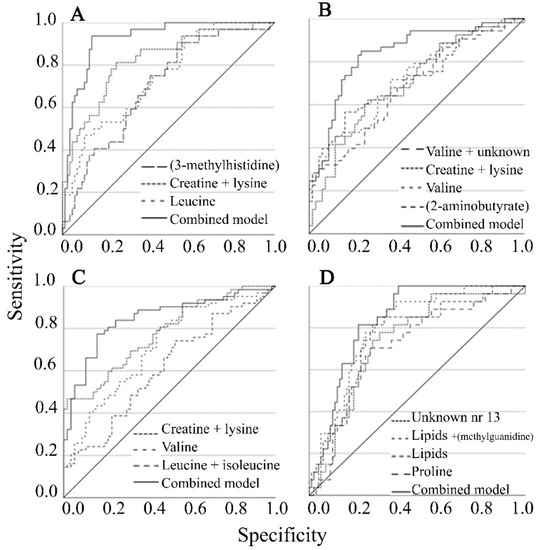
Figure 1.
Area under the curve for single metabolites and combined metabolite models for (A) red meat, (B) eggs, (C) fermented dairy, (D) beer. Diagonal line represents the reference line.
For egg consumption, the combined metabolite model improved AUC (84%) in comparison to models with each separate metabolite (AUC = 68–72%) (Table 2, Figure 1B). For fermented dairy products, the combined metabolite model improved AUC (85%) in comparison to models with each separate metabolite (AUC = 61–77%) (Table 2, Figure 1C). Finally, for beer consumption, the combined metabolite model improved AUC (84%) in comparison to models with each separate metabolite (AUC = 71–80%) (Table 2, Figure 1D).
3. Discussion
The results from our explorative analysis show that serum metabolites can be associated to reported intake of different foods, when applying partial correlation and stepwise forward logistic regression analysis. In addition, the use of a combination of several serum metabolites improved predictive models for several foods and food groups, including soybeans, meat products/processed meat, poultry, red meat, eggs, fermented dairy, and beer.
Comparing the predictive power of our combined metabolite models with the prediction by one metabolite (AUC) from a paper by Wang et al. [16] shows that the predictability was higher in our combined metabolite models for soy beans, red meat, poultry, total meat, eggs, and beer.
Combining metabolites using stepwise logistic regression analyses have only been used in a few previous studies. In contrast to our study, these studies aimed at exploring associations between intake of specific foods (cocoa [17], bread [18], red wine [11], and walnuts [19]) and changes in urine metabolites. In addition, the dietary assessment methods used in these studies were FFQs to capture habitual intake, while a 4-day dietary record to capture recent intake was used in the current analyses. Even though both methods may capture habitual intake, dietary records also reflect the specific foods actually consumed the days before sampling, and this is an advantage when aiming to identify patterns of metabolites in serum associated with food consumption.
We did not identify some of the known food-metabolite associations such as, for example, genistein and daidzein for soy consumption [7]. This reflects that 1H-NMR metabolomics, the method used for our serum analysis, is a less sensitive method than for example mass spectrometry and therefore hampered the identification of low concentration metabolites.
Soy products are becoming more common in our diets due to the protein shift from animal-based products to plant-based products. In this study sample, with a large proportion of vegans and vegetarians, about 56% reported consuming soy products, and these reported intakes were associated with a high content of the amino acid glycine in serum. Glycine is a nonessential amino acid which can be found in many types of foods [20]. However, soy protein has a glycine content of 3.8 g/100 g, which is about twice the content of other foods also regarded to have a high glycine content [21]. Although many subjects consumed soy products, few subjects consumed soybeans. Surprisingly, in our study, soybeans did not associate with glycine but with valine and asparagine. However, the content of glycine in dried and cooked soybeans is only about 0.46 g/100 g, which probably explains our result. This highlights the complexity of using food groups when trying to identify markers for food intake. In our study, a combination of valine and asparagine improved the specificity to capture the intake of soybeans. Legumes and nuts, including soybeans, have a comparatively high content of valine and asparagine, but foods from animal sources also contribute to these amino acids, thus complicating the picture.
In a combined metabolite model to predict intake of green/herbal tea, glycine and asparagine jointly qualified. It is unlikely that these amino acids would increase after green/herbal tea intake, and we therefore tested if subjects who consumed soy products also were more prone to drink green/herbal tea. This was not the case, but intake of green/herbal tea may be correlated to consumption of some other foods that we were unable to capture or this could be a chance finding due to multiple testing.
Reported intake of cruciferous vegetables was associated with a variable including glutamine and an unidentified metabolite. This may be explained by the finding that subjects with a high intake of cruciferous vegetables concurrently had a low intake of protein. Similarly, serum glutamine has been reported to be inversely correlated to protein intake in other studies [22]. In line with this, glutamine has been associated with vegan diet in a previous report [23]. A higher glutamine-to-glutamate ratio has been associated with an improved cardiometabolic risk profile, making this marker interesting to explore further [24]. Nevertheless, a direct link between dietary intake pattern and serum glutamine/glutamate ratio has not yet been established.
In our study, foods from animal sources were associated with a wide range of amino acids, most notably the branched amino acids leucine, valine, and isoleucine and creatine. Interestingly, most animal protein sources were associated with 3-hydroxyisobutyrate, although this metabolite did not qualify in any of the combined metabolite models.
When comparing models among foods of different animal sources, creatinine was only included in the model for processed meat and 3-methylhistidine was only included in the model for red meat. Still, both metabolites were associated with not only meat intake but also with muscle mass in our data. When adjusting for lean body mass in the analyses, none of these markers remained significant, suggesting endogenous origin, and hence cannot be seen as reliable markers for meat intake. In contrast to our findings, 3-methylhistidine has previously been associated with poultry intake [25,26]. The difference in results may be explained by the adjustment for lean body mass in our study. To improve models further, the combination of serum metabolites with lipidomic data or with fatty acid concentrations may be applied since many amino acids are overlapping among animal products.
In contrary to other animal-foods, the models for eggs and cheese contained the metabolite 2-aminobutyrate (L-alpha-aminobutyric acid), which is a metabolite from the amino acid metabolism. Few studies have reported 2-aminobutyrate concentrations in relation to diet. However, Gu et al. found increased concentrations of 2-aminobutyrate in relation to a diet high in eggs and other animal proteins [27].
Beer consumption was associated with several metabolites, among them lipids, glucose, and proline. In a combined metabolite model, lipids/free fatty acids, proline, and a variable including isoleucine + unidentified increased the precision. Since accurate reported intake of alcoholic beverages is notoriously difficult to obtain, this could be of special interest for future research. In our setting, beer consumption 1–3 days before sampling had a significant impact on blood lipids, a fact that might be of importance when studying blood lipids and for clinical sampling. In support of our results, previous studies have shown an association between alcohol intake and increased levels of high-density lipoprotein levels [28,29]. In our study, for other alcoholic beverages only single metabolites were associated, presumably xylose, for white wine and citrate and a combined variable with lactate, proline and 3-hydroxybuturate for spirits. Contrary to our results, in a meta-analysis of three cohorts the reported alcohol intake was inversely correlated with citrate in serum samples [30]. However, in our study alcohol could be consumed up to the day before sampling, whereas the number of days between consumption of alcohol and sample collection was unknown in the study by Würtz et al. [30].
Since metabolites from certain foods have different half-times in serum, a combination of several metabolites may jointly cover both short- and long-term metabolites, resulting in a more accurate picture of the dietary intake. In addition, the effect of endogenous metabolites may have less influence on the total concentration using a combination of several markers.
This study has several strengths. Fasting serum samples were used and these were rigorously handled following a strict protocol, resulting in high-quality 1H-NMR data. The wide range of consumption of food specifically from animal sources is another strength, as it makes it possible to find correlations between concentration of metabolites and specifically meat but also dairy and egg. In a normal population, few individuals exclude meat. However, this is also a weakness of the study as there can be other lifestyle factors coincident among vegans in addition to excluding meat, fish, egg, and dairy that might influence the results. Therefore, findings from this study should be confirmed in other settings.
To sum up, using models with combinations of metabolites quantified by 1H-NMR metabolomics analysis shows the potential to improve the precision of assessing certain food intake compared to today’s standard subjective methods, such as FFQ. It is important to notice that when a certain food represents a minor portion of all the carbohydrates, proteins, lipids, or overall calories, it is unlikely its individual fingerprint can be identified by 1H-NMR metabolomics analysis in serum since metabolites unique for individual foods are often found in low concentrations.
Findings in this work should be confirmed in intervention studies and evaluated in large epidemiological cohorts.
4. Materials and Methods
4.1. Subjects
The current work was based on data from a cross-sectional metabolomics study which included 124 healthy subjects living in the Gothenburg area, Sweden, registered at Clinicaltrials.gov as NCT02039609. Recruitment, study design, subject characteristics, and dietary intake have been described in detail elsewhere [31,32]. Briefly, healthy females and males complying with self-reported habitual vegan, (lacto-/ovo-) vegetarian, vegetarian adding fish, or omnivore diets were recruited during 2013 and 2015. Inclusion criteria were age between 18 and 65 y, no regular use of medications, and having a BMI between 18 and 30 kg/m2. Subjects who were pregnant, lactating, or used nicotine products regularly were excluded. Subjects were not allowed to drink alcohol the night before or to consume diet supplements one week before sampling.
Subjects with BMI < 18.0 kg/m2 (n = 2) or with food intake level (FIL; calculated by dividing total reported energy intake with estimated basal metabolic rate [33]) < 1.0 (n = 3) were excluded, leaving 119 subjects for the current analyses (Figure 2).
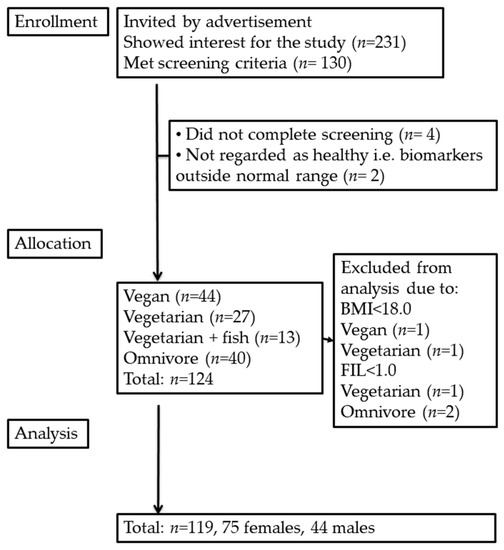
Figure 2.
Study flowchart. Body mass index, BMI; food intake level, FIL.
4.2. Dietary Assessment
Food intake was estimated from a 4-day weighed food diary recorded by subjects at one time point before blood sampling. Subjects were instructed to weigh all food and drinks consumed during 3 weekdays and 1 weekend day using a household scale. Records were registered in the nutritional calculation program DietistNet version 18.12.16, (Kost och Näringsdata AB, Bromma, Sweden). For nutritional calculations the databases from Sweden (National Food Agency, Uppsala, Sweden, version 17.12.15) and Finland (Fineli, National Institute for Health and Welfare, Helsinki, Finland, version 18.02.28) were used. Micro- and macro-nutrient intakes have been reported in detail elsewhere [32].
Dietary intake was categorized into 32 foods/food groups and 7 combined groups such as total meat (Supplemental Table S1). The foods/food groups have all previously been associated with different metabolites in serum in metabolomic studies [6,7,8,9,10,26,34,35].
4.3. Covariate Data
Weight and height were measured, and BMI was calculated by weight (kg)/height2 (m). Physical activity was self-reported and estimated by two questions capturing physical exercise (six scores) and everyday physical activity (seven scores) [36]. A physical activity score was calculated by multiplying physical exercise with a factor of two (to consider higher intensity) and adding everyday physical activity resulting in an individual score of 3–19. Physical activity scores were divided into tertiles.
4.4. Data Acquisition
4.4.1. Sampling, Sample Handling, and Preprocessing
Fasting venous blood was collected at one time point. Sample handling and preprocessing have been reported in detail elsewhere [31]. Briefly, blood was drawn into a 5 mL BD vacutainer glass tube, allowed to clot for 30 min, and centrifuged (2600× g, 10 min). Serum aliquots were placed at −20 °C within 1 h and stored at −80 °C within 2 h until analysis. Before 1H-NMR analysis, serum samples were thawed and mixed with phosphate buffer whereafter 180 µL of the sample mix was transferred to 3.0-mm NMR tubes (Bruker BioSpin, Billerica, MA, USA, 96 sample racks for SampleJet) using a SamplePro liquid handling robot (Bruker BioSpin). Samples were kept at 6 °C until analysis.
4.4.2. NMR Spectroscopy
1H-NMR analysis has been described in detail previously [31]. In short, spectra were recorded at 800 MHz with a Bruker Advance III HD spectrometer with a 3-mm TCI cryoprobe. NMR data were recoded using the Bruker pulse sequence “zgespe”. A total of 128 scans were collected into 64 k data points. Data processing was performed with TopSpin 3.2p16 (Bruker BioSpin) and MatLab (MathWorks Inc., Natick, MA, USA), using TSP-d4 for referencing. In total 237 peaks were manually aligned and integrated peak-by-peak, and these variables represent ∼70 metabolites. A variable could also reflect more than one metabolite. Only variables of interest were identified.
For annotation Chenomx NMR suite 8.31 (Chenomx Inc., Edmonton, AB, Canada), the Human Metabolome Database [37] and an in-house implementation of the statistical total correlation spectroscopy (STOCSY) routine [38] were used.
4.5. Statistics
Subject characteristics and reported dietary intake are presented as median (min-max), mean (SD), or proportions (n, %). Partial correlation (Spearman’s rank correlation) was used to determine associations between foods and metabolites controlling for age (y, continuous), sex, physical activity score (categorical), BMI (kg/m2, continuous), and reported energy intake (kcal/d, continuous). Correlations were considered statistically significant if p values were <0.01. We further set absolute values of the correlation coefficients (rho) >0.2 to be considered relevant. Furthermore, to evaluate the predictive accuracy of dietary biomarkers to discriminate consumers from non-consumers, the area under the curve (AUC) was calculated from the receiver operating characteristics (ROC) curve. AUC <0.7 was deemed as low, 0.7 to <0.8 as moderate and ≥0.8 as high predictive accuracy. Because selected variables had to meet these three statistical requirements, no other correction for multiple hypothesis tests were done.
To assess if combined biomarkers would increase predictive models, a stepwise forward logistic regression was applied. All metabolites with an absolute correlation coefficient > 0.2 and p < 0.01 were included in the regression model. If two or more metabolites were included in the final model of stepwise forward logistic regression, these were further evaluated in combination using predictive probabilities. Diet–metabolite associations for combined biomarkers were evaluated using Spearman’s partial correlation. Further, AUC was calculated for the combined model and compared with the AUC for each single metabolite and ROC presented.
The computer software package SPSS for Windows, version 28 (IBM, New York, NY, USA) was used for statistical analyses.
5. Conclusions
Our results show that few serum metabolites are unique for a certain food item, but they can possibly be used in combinations to predict intake of some foods or food groups ingested in a habitual diet. The overall protein intake seems to be crucial for many of the metabolites found by 1H-NMR -analysis to associate with different foods. However, since many metabolites from animal products are both provided by the diet and are endogenously produced, it is important to adjust for factors such as lean body mass, which could contribute to the metabolite concentration in serum. Combinations of metabolites associated to food intake identified here might be of interest to evaluate further in dose–response intervention studies as potential combinatorial biomarkers. Finally, as the data analysis was performed without correction for multiple testing, we view the results presented here as an exploratory report on a potential method to combine metabolites from 1H-NMR -analysis to predict the intake of foods from serum samples.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo12100908/s1, Table S1: Serum metabolites associated with reported food intake; Table S2: Reported consumption of 24 foods/food groups.
Author Contributions
Conceptualization, A.W., L.E. and H.M.L.; data curation, M.R. and A.P.; formal analysis, T.K.; writing—original draft preparation, T.K. and H.M.L.; writing—review and editing, A.W., T.K., M.R. and H.M.L.; funding acquisition, A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Swedish Research Council grants numbers 521-2011-2664, 2016-01216 and 2021-00954.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Regional Ethical Review Board in Gothenburg (reference number 561-12).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on reasonable request and should be made to Helen M. Lindqvist, helen.lindqvist@gu.se. The data are not publicly available due to Swedish law.
Acknowledgments
We thank Sofia Isaksson for her help with recruiting study subjects and collecting the data.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Collaborators GBDD. Health effects of dietary risks in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 393, 1958–1972. [Google Scholar] [CrossRef]
- Collaborators GBDRF. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef]
- Kipnis, V.; Midthune, D.; Freedman, L.; Bingham, S.; Day, N.E.; Riboli, E.; Ferrari, P.; Carroll, R.J. Bias in dietary-report instruments and its implications for nutritional epidemiology. Public Health Nutr. 2002, 5, 915–923. [Google Scholar] [CrossRef]
- Brennan, L.; Hu, F.B. Metabolomics-Based Dietary Biomarkers in Nutritional Epidemiology-Current Status and Future Opportunities. Mol. Nutr. Food Res. 2019, 63, e1701064. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef]
- Garcia-Aloy, M.; Hulshof, P.J.M.; Estruel-Amades, S.; Oste, M.C.J.; Lankinen, M.; Geleijnse, J.M.; de Goede, J.; Ulaszewska, M.; Mattivi, F.; Bakker, S.J.L.; et al. Biomarkers of food intake for nuts and vegetable oils: An extensive literature search. Genes Nutr. 2019, 14, 7. [Google Scholar] [CrossRef]
- Sri Harsha, P.S.C.; Wahab, R.A.; Garcia-Aloy, M.; Madrid-Gambin, F.; Estruel-Amades, S.; Watzl, B.; Andres-Lacueva, C.; Brennan, L. Biomarkers of legume intake in human intervention and observational studies: A systematic review. Genes Nutr. 2018, 13, 25. [Google Scholar] [CrossRef]
- Cuparencu, C.; Pratico, G.; Hemeryck, L.Y.; Sri Harsha, P.S.C.; Noerman, S.; Rombouts, C.; Xi, M.; Vanhaecke, L.; Hanhineva, K.; Brennan, L.; et al. Biomarkers of meat and seafood intake: An extensive literature review. Genes Nutr. 2019, 14, 35. [Google Scholar] [CrossRef]
- Li, K.J.; Brouwer-Brolsma, E.M.; Burton-Pimentel, K.J.; Vergeres, G.; Feskens, E.J.M. A systematic review to identify biomarkers of intake for fermented food products. Genes Nutr. 2021, 16, 5. [Google Scholar] [CrossRef]
- Rothwell, J.A.; Madrid-Gambin, F.; Garcia-Aloy, M.; Andres-Lacueva, C.; Logue, C.; Gallagher, A.M.; Mack, C.; Kulling, S.E.; Gao, Q.; Pratico, G.; et al. Biomarkers of intake for coffee, tea, and sweetened beverages. Genes Nutr. 2018, 13, 15. [Google Scholar] [CrossRef]
- Vazquez-Fresno, R.; Llorach, R.; Urpi-Sarda, M.; Khymenets, O.; Bullo, M.; Corella, D.; Fito, M.; Martinez-Gonzalez, M.A.; Estruch, R.; Andres Lacueva, C. An NMR metabolomics spproach reveals a combined-biomarkers model in a wine interventional trial with validation in free-living individuals of the PREDIMED study. Metabolomics 2015, 11, 797–806. [Google Scholar] [CrossRef]
- Trimigno, A.; Münger, L.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Vionnet, N.; Pralong, F.; Capozzi, F.; Badertscher, R.; Vergères, G. GC-MS Based Metabolomics and NMR Spectroscopy Investigation of Food Intake Biomarkers for Milk and Cheese in Serum of Healthy Humans. Metabolites 2018, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Mcnamara, A.E.; Collins, C.; Harsha, P.S.C.S.; González-Peña, D.; Gibbons, H.; Mcnulty, B.A.; Nugent, A.P.; Walton, J.; Flynn, A.; Brennan, L. Metabolomic-Based Approach to Identify Biomarkers of Apple Intake. Mol. Nutr. Food Res. 2020, 64, 1901158. [Google Scholar] [CrossRef]
- Liang, S.; Nasir, R.F.; Bell-Anderson, K.S.; Toniutti, C.A.; O’Leary, F.M.; Skilton, M.R. Biomarkers of dietary patterns: A systematic review of randomized controlled trials. Nutr. Rev. 2022, 80, 1856–1895. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aloy, M.; Rabassa, M.; Casas-Agustench, P.; Hidalgo-Liberona, N.; Llorach, R.; Andres-Lacueva, C. Novel strategies for improving dietary exposure assessment: Multiple-data fusion is a more accurate measure than the traditional single-biomarker approach. Trends Food Sci. Technol. 2017, 69, 220–229. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Hodge, R.A.; Stevens, V.L.; Hartman, T.J.; McCullough, M.L. Identification and Reproducibility of Plasma Metabolomic Biomarkers of Habitual Food Intake in a US Diet Validation Study. Metabolites 2020, 10, 382. [Google Scholar] [CrossRef]
- Garcia-Aloy, M.; Llorach, R.; Urpi-Sarda, M.; Jauregui, O.; Corella, D.; Ruiz-Canela, M.; Salas-Salvado, J.; Fito, M.; Ros, E.; Estruch, R.; et al. A metabolomics-driven approach to predict cocoa product consumption by designing a multimetabolite biomarker model in free-living subjects from the PREDIMED study. Mol. Nutr. Food Res. 2015, 59, 212–220. [Google Scholar] [CrossRef]
- Garcia-Aloy, M.; Llorach, R.; Urpi-Sarda, M.; Tulipani, S.; Salas-Salvado, J.; Martinez-Gonzalez, M.A.; Corella, D.; Fito, M.; Estruch, R.; Serra-Majam, L.; et al. Nutrimetabolomics fingerprinting to identify biomarkers of bread exposure in a free-living population from the PREDIMED study cohort. Metabolomics 2015, 11, 155–165. [Google Scholar] [CrossRef]
- Garcia-Aloy, M.; Llorach, R.; Urpi-Sarda, M.; Tulipani, S.; Estruch, R.; Martinez-Gonzalez, M.A.; Corella, D.; Fito, M.; Ros, E.; Salas-Salvado, J.; et al. Novel multimetabolite prediction of walnut consumption by a urinary biomarker model in a free-living population: The PREDIMED study. J. Proteome Res. 2014, 13, 3476–3483. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Livsmedelsverk, S. Livsmedelstabeller: Energi Och Näringsämnen; Statens Livsmedelsverk: Uppsala, Sweden, 1986. [Google Scholar]
- Matthews, D.E.; Campbell, R.G. The effect of dietary protein intake on glutamine and glutamate nitrogen metabolism in humans. Am. J. Clin. Nutr. 1992, 55, 963–970. [Google Scholar] [CrossRef]
- Schmidt, J.A.; Fensom, G.K.; Rinaldi, S.; Scalbert, A.; Gunter, M.J.; Holmes, M.V.; Key, T.J.; Travis, R.C. NMR Metabolite Profiles in Male Meat-Eaters, Fish-Eaters, Vegetarians and Vegans, and Comparison with MS Metabolite Profiles. Metabolites 2021, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, C.; Hernandez-Alonso, P.; Bullo, M.; Ruiz-Canela, M.; Li, J.; Guasch-Ferre, M.; Toledo, E.; Clish, C.; Corella, D.; Estruch, R.; et al. High Plasma Glutamate and a Low Glutamine-to-Glutamate Ratio Are Associated with Increased Risk of Heart Failure but Not Atrial Fibrillation in the Prevencion con Dieta Mediterranea (PREDIMED) Study. J. Nutr. 2020, 150, 2882–2889. [Google Scholar] [CrossRef]
- Langenau, J.; Oluwagbemigun, K.; Brachem, C.; Lieb, W.; Giuseppe, R.D.; Artati, A.; Kastenmuller, G.; Weinhold, L.; Schmid, M.; Nothlings, U. Blood Metabolomic Profiling Confirms and Identifies Biomarkers of Food Intake. Metabolites 2020, 10, 468. [Google Scholar] [CrossRef]
- Wang, Y.; Gapstur, S.M.; Carter, B.D.; Hartman, T.J.; Stevens, V.L.; Gaudet, M.M.; McCullough, M.L. Untargeted Metabolomics Identifies Novel Potential Biomarkers of Habitual Food Intake in a Cross-Sectional Study of Postmenopausal Women. J. Nutr. 2018, 148, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhao, A.; Huang, F.; Zhang, Y.; Liu, J.; Wang, C.; Jia, W.; Xie, G.; Jia, W. Very low carbohydrate diet significantly alters the serum metabolic profiles in obese subjects. J. Proteome Res. 2013, 12, 5801–5811. [Google Scholar] [CrossRef]
- Brien, S.E.; Ronksley, P.E.; Turner, B.J.; Mukamal, K.J.; Ghali, W.A. Effect of alcohol consumption on biological markers associated with risk of coronary heart disease: Systematic review and meta-analysis of interventional studies. BMJ 2011, 342, d636. [Google Scholar] [CrossRef]
- van Tol, A.; S van der Gaag, M.; MScheek, L.; van Gent, T.; Hendriks, H.F. Changes in postprandial lipoproteins of low and high density caused by moderate alcohol consumption with dinner. Atherosclerosis. 1998, 141 (Suppl. S1), S101-3. [Google Scholar] [CrossRef]
- Wurtz, P.; Cook, S.; Wang, Q.; Tiainen, M.; Tynkkynen, T.; Kangas, A.J.; Soininen, P.; Laitinen, J.; Viikari, J.; Kahonen, M.; et al. Metabolic profiling of alcohol consumption in 9778 young adults. Int. J. Epidemiol. 2016, 45, 1493–1506. [Google Scholar] [CrossRef]
- Lindqvist, H.M.; Radjursoga, M.; Malmodin, D.; Winkvist, A.; Ellegard, L. Serum metabolite profiles of habitual diet: Evaluation by 1H-nuclear magnetic resonance analysis. Am. J. Clin. Nutr. 2019, 110, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, H.M.; Radjursoga, M.; Torstensson, T.; Jansson, L.; Ellegard, L.; Winkvist, A. Urine Metabolite Profiles and Nutrient Intake Based on 4-Day Weighed Food Diary in Habitual Vegans, Vegetarians, and Omnivores. J. Nutr. 2021, 151, 30–39. [Google Scholar] [CrossRef] [PubMed]
- NNR Working Group. Nordic Nutrition Recommendations 2012. Integrating Nutrition and Physical Activity, 5th ed.; Nordic Council of Ministers: Copenhagen, Denmark, 2014. [Google Scholar]
- Pallister, T.; Jennings, A.; Mohney, R.P.; Yarand, D.; Mangino, M.; Cassidy, A.; MacGregor, A.; Spector, T.D.; Menni, C. Characterizing Blood Metabolomics Profiles Associated with Self-Reported Food Intakes in Female Twins. PLoS ONE 2016, 11, e0158568. [Google Scholar] [CrossRef]
- Cheung, W.; Keski-Rahkonen, P.; Assi, N.; Ferrari, P.; Freisling, H.; Rinaldi, S.; Slimani, N.; Zamora-Ros, R.; Rundle, M.; Frost, G.; et al. A metabolomic study of biomarkers of meat and fish intake. Am. J. Clin. Nutr. 2017, 105, 600–608. [Google Scholar] [CrossRef]
- Ek, A.; Kallings, L.V.; Ekstrom, M.; Borjesson, M.; Ekblom, O. Subjective reports of physical activity levels and sedentary time prior to hospital admission can predict utilization of hospital care and all-cause mortality among patients with cardiovascular disease. Eur. J. Cardiovasc. Nurs. 2020, 19, 691–701. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res/ 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson J: Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005, 77, 1282–1289. [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).