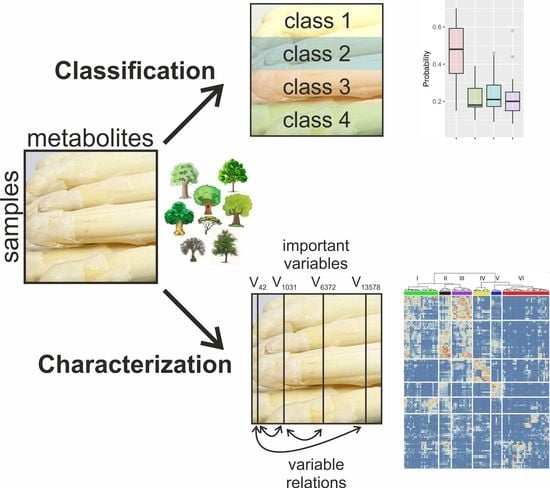
Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth
Abstract
:1. Introduction
2. Results and Discussion
2.1. Multi-Level Classification
2.2. Probability Machines
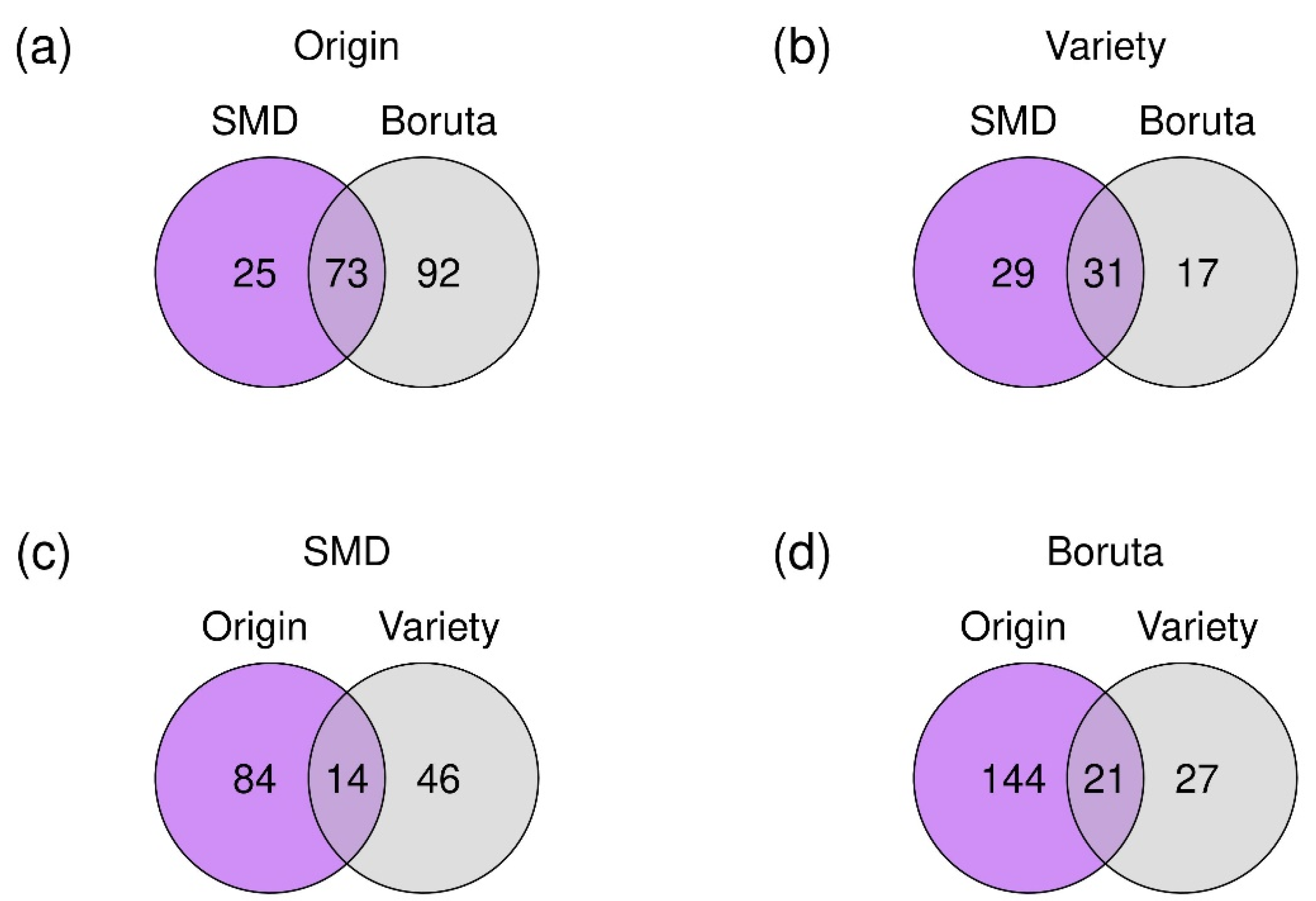
2.3. Feature Selection
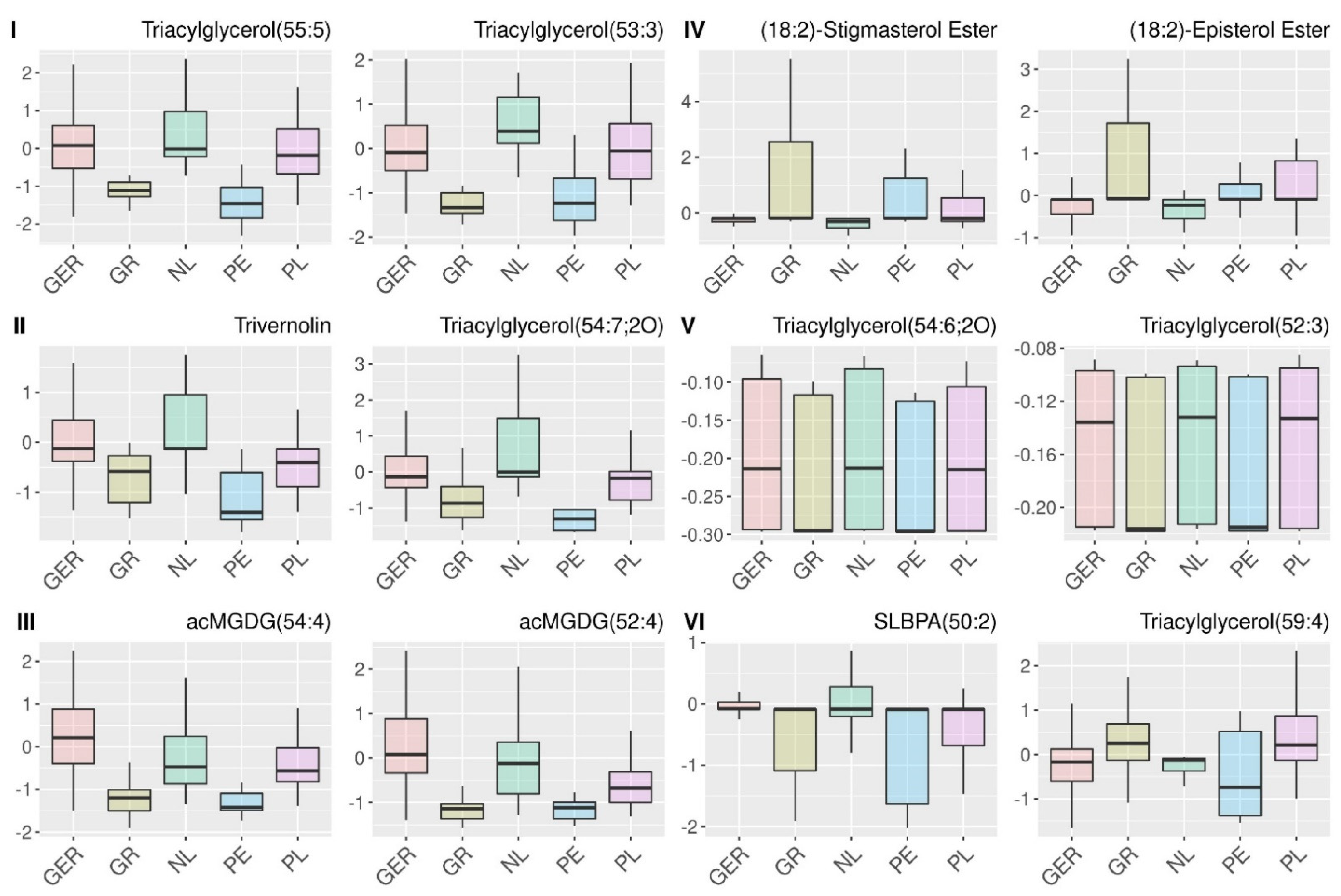
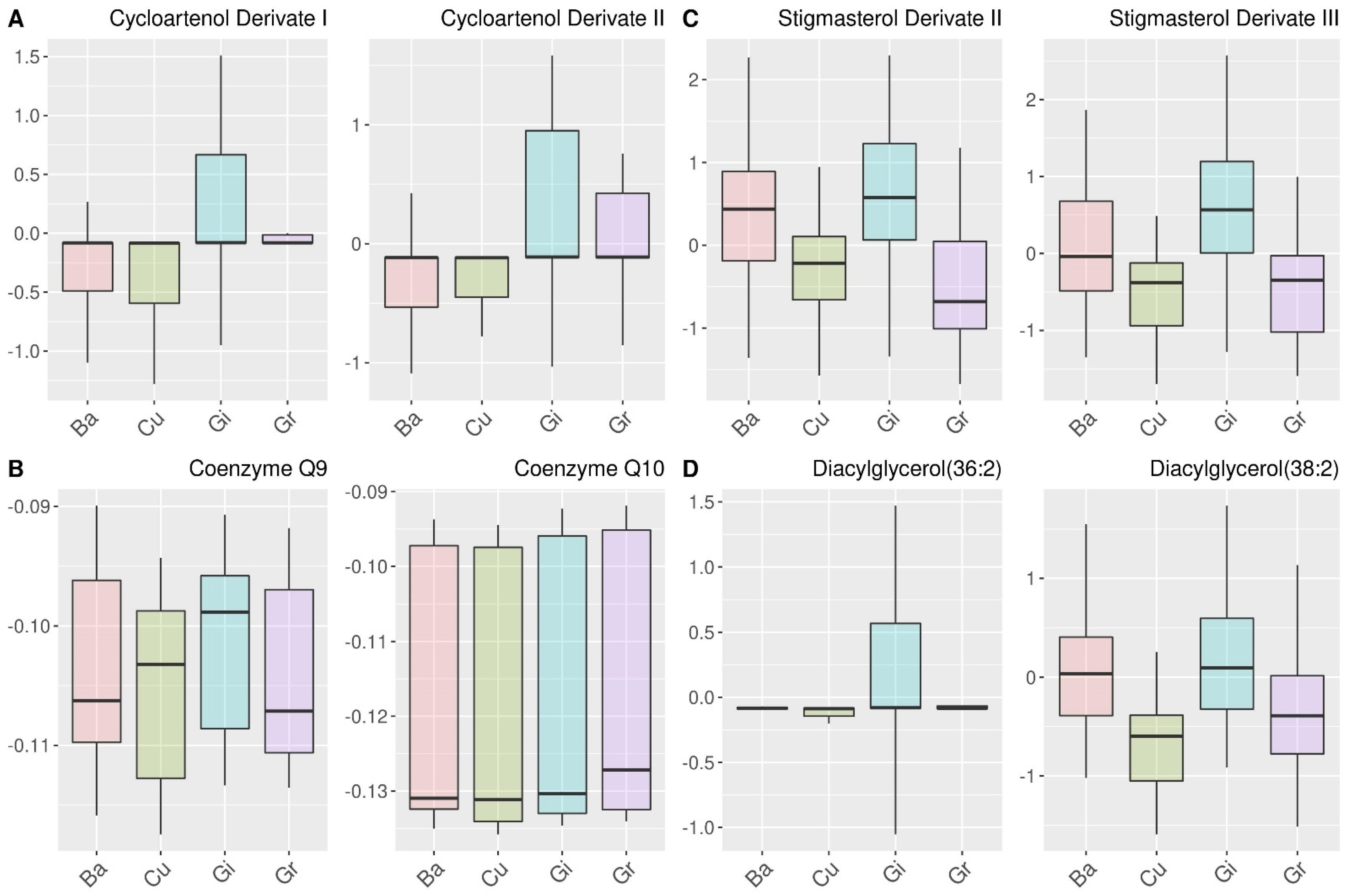
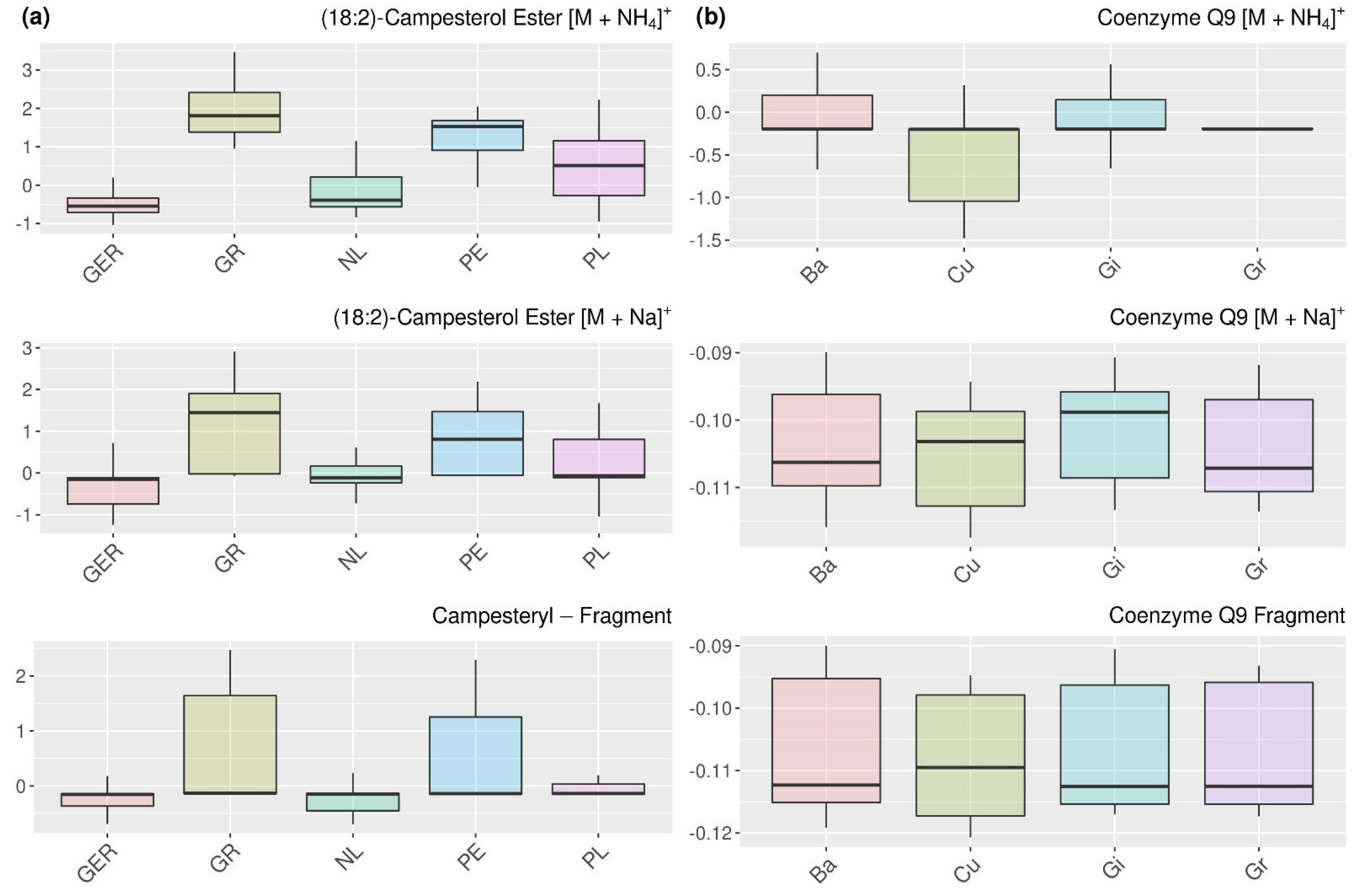
2.4. Analysis of the Relations of Selected Features
3. Materials and Methods
3.1. Data Acquisition and Preprocessing
3.2. Software and Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S. Current Progress in Computational Metabolomics. Brief. Bioinform. 2007, 8, 279–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O. Metabolomics—The Link between Genotypes and Phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Bachmann, R.; Klockmann, S.; Haerdter, J.; Fischer, M.; Hackl, T. H-NMR Spectroscopy for Determination of the Geographical Origin of Hazelnuts. J. Agric. Food Chem. 2018, 66, 11873–11879. [Google Scholar] [CrossRef]
- Ernst, M.; Silva, D.B.; Silva, R.R.; Vêncio, R.Z.N.; Lopes, N.P. Mass Spectrometry in Plant Metabolomics Strategies: From Analytical Platforms to Data Acquisition and Processing. Nat. Prod. Rep. 2014, 31, 784. [Google Scholar] [CrossRef]
- Johnstone, I.M.; Titterington, D.M. Statistical Challenges of High-Dimensional Data. Philos. Trans. Royal Soc. 2009, 367, 4237–4253. [Google Scholar] [CrossRef] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Cain, N.; Fischer, M. Food Targeting: Geographical Origin Determination of Hazelnuts ( Corylus Avellana ) by LC-QqQ-MS/MS-Based Targeted Metabolomics Application. J. Agric. Food Chem. 2017, 65, 1456–1465. [Google Scholar] [CrossRef]
- Long, N.P.; Lim, D.K.; Mo, C.; Kim, G.; Kwon, S.W. Development and Assessment of a Lysophospholipid-Based Deep Learning Model to Discriminate Geographical Origins of White Rice. Sci. Rep. 2017, 7, 8552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A Tutorial Review: Metabolomics and Partial Least Squares-Discriminant Analysis—a Marriage of Convenience or a Shotgun Wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Erban, A.; Fehrle, I.; Martinez-Seidel, F.; Brigante, F.; Más, A.L.; Baroni, V.; Wunderlin, D.; Kopka, J. Discovery of Food Identity Markers by Metabolomics and Machine Learning Technology. Sci. Rep. 2019, 9, 9697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y. Random Forest for Bioinformatics. In Ensemble Machine Learning; Zhang, C., Ma, Y., Eds.; Springer: Boston, MA, USA, 2012; pp. 307–323. ISBN 978-1-4419-9325-0. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Malley, J.D.; Kruppa, J.; Dasgupta, A.; Malley, K.G.; Ziegler, A. Probability Machines: Consistent Probability Estimation Using Nonparametric Learning Machines. Methods Inf. Med. 2012, 51, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Nembrini, S.; König, I.R.; Wright, M.N. The Revival of the Gini Importance? Bioinformatics 2018, 34, 3711–3718. [Google Scholar] [CrossRef] [Green Version]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of Variable Selection Methods for Random Forests and Omics Data Sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Seifert, S.; Gundlach, S.; Szymczak, S. Surrogate Minimal Depth as an Importance Measure for Variables in Random Forests. Bioinformatics 2019, 35, 3663–3671. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Routledge: Abingdon, UK, 2017; ISBN 978-1-315-13947-0. [Google Scholar]
- Shakiba, N.; Gerdes, A.; Holz, N.; Wenck, S.; Bachmann, R.; Schneider, T.; Seifert, S.; Fischer, M.; Hackl, T. Determination of the Geographical Origin of Hazelnuts (Corylus Avellana L.) by Near-Infrared Spectroscopy (NIR) and a Low-Level Fusion with Nuclear Magnetic Resonance (NMR). Microchem. J. 2022, 174, 107066. [Google Scholar] [CrossRef]
- Seifert, S. Application of Random Forest Based Approaches to Surface-Enhanced Raman Scattering Data. Sci. Rep. 2020, 10, 5436. [Google Scholar] [CrossRef]
- Živanović, V.; Seifert, S.; Drescher, D.; Schrade, P.; Werner, S.; Guttmann, P.; Szekeres, G.P.; Bachmann, S.; Schneider, G.; Arenz, C.; et al. Optical Nanosensing of Lipid Accumulation Due to Enzyme Inhibition in Live Cells. ACS Nano 2019, 13, 9363–9375. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.; Gurk, S.; Wagner, D.; Bockmayr, M.; Fischer, M. Food Authentication: Multi-Elemental Analysis of White Asparagus for Provenance Discrimination. Food Chem. 2019, 286, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.; Rurik, M.; Gurk, S.; Kohlbacher, O.; Fischer, M. Food Monitoring: Screening of the Geographical Origin of White Asparagus Using FT-NIR and Machine Learning. Food Control 2019, 104, 318–325. [Google Scholar] [CrossRef]
- Klare, J.; Rurik, M.; Rottmann, E.; Bollen, A.; Kohlbacher, O.; Fischer, M.; Hackl, T. Determination of the Geographical Origin of Asparagus Officinalis L. by 1 H-NMR Spectroscopy. J. Agric. Food Chem. 2020, 68, 14353–14363. [Google Scholar] [CrossRef] [PubMed]
- Creydt, M.; Hudzik, D.; Rurik, M.; Kohlbacher, O.; Fischer, M. Food Authentication: Small-Molecule Profiling as a Tool for the Geographic Discrimination of German White Asparagus. J. Agric. Food Chem. 2018, 66, 13328–13339. [Google Scholar] [CrossRef]
- Creydt, M.; Fischer, M. Metabolic Imaging: Analysis of Different Sections of White Asparagus Officinalis Shoots Using High-Resolution Mass Spectrometry. J. Plant Physiol. 2020, 250, 153179. [Google Scholar] [CrossRef] [PubMed]
- Creydt, M.; Arndt, M.; Hudzik, D.; Fischer, M. Plant Metabolomics: Evaluation of Different Extraction Parameters for Nontargeted UPLC-ESI-QTOF-Mass Spectrometry at the Example of White Asparagus Officinalis. J. Agric. Food Chem. 2018, 66, 12876–12887. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, Z.; Fang, Y.; Qi, F.; Liu, H.; Miao, L.; Du, P.; Shi, L.; Gao, W.; Han, S.; et al. Genetic Diversity, Population Structure, and Botanical Variety of 320 Global Peanut Accessions Revealed through Tunable Genotyping-by-Sequencing. Sci. Rep. 2018, 8, 14500. [Google Scholar] [CrossRef]
- Scharf, A.; Lang, C.; Fischer, M. Genetic Authentication: Differentiation of Fine and Bulk Cocoa (Theobroma Cacao L.) by a New CRISPR/Cas9-Based in Vitro Method. Food Control 2020, 114, 107219. [Google Scholar] [CrossRef]
- Torres-Moreno, M.; Torrescasana, E.; Salas-Salvadó, J.; Blanch, C. Nutritional Composition and Fatty Acids Profile in Cocoa Beans and Chocolates with Different Geographical Origin and Processing Conditions. Food Chem. 2015, 166, 125–132. [Google Scholar] [CrossRef]
- Arena, E.; Campisi, S.; Fallico, B.; Maccarone, E. Distribution of Fatty Acids and Phytosterols as a Criterion to Discriminate Geographic Origin of Pistachio Seeds. Food Chem. 2007, 104, 403–408. [Google Scholar] [CrossRef]
- Cossignani, L.; Blasi, F.; Simonetti, M.S.; Montesano, D. Fatty Acids and Phytosterols to Discriminate Geographic Origin of Lycium Barbarum Berry. Food Anal. Methods 2018, 11, 1180–1188. [Google Scholar] [CrossRef]
- He, M.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef]
- Sauveplane, V.; Kandel, S.; Kastner, P.-E.; Ehlting, J.; Compagnon, V.; Werck-Reichhart, D.; Pinot, F. Arabidopsis Thaliana CYP77A4 Is the First Cytochrome P450 Able to Catalyze the Epoxidation of Free Fatty Acids in Plants: CYP77A4, an Epoxy Fatty Acid-Forming Enzyme. FEBS J. 2009, 276, 719–735. [Google Scholar] [CrossRef]
- Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- Creydt, M.; Fischer, M. Mass-Spectrometry-Based Food Metabolomics in Routine Applications: A Basic Standardization Approach Using Housekeeping Metabolites for the Authentication of Asparagus. J. Agric. Food Chem. 2020, 68, 14343–14352. [Google Scholar] [CrossRef]
- Rezzonico, E.; Moire, L.; Delessert, S.; Poirier, Y. Level of Accumulation of Epoxy Fatty Acid in Arabidopsis Thaliana Expressing a Linoleic Acid ?12-Epoxygenase Is Influenced by the Availability of the Substrate Linoleic Acid. Theor. Appl. Genet. 2004, 109, 1077–1082. [Google Scholar] [CrossRef]
- Ferrer, A.; Altabella, T.; Arró, M.; Boronat, A. Emerging Roles for Conjugated Sterols in Plants. Prog. Lipid Res. 2017, 67, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F.V. Plant Sterols: Diversity, Biosynthesis, and Physiological Functions. Biochemistry 2016, 81, 819–834. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Korbozova, N.K.; Kudrina, N.O.; Kobylina, T.N.; Kurmanbayeva, M.S.; Meduntseva, N.D.; Tolstikova, T.G. The Influence of Abiotic Stress Factors on the Morphophysiological and Phytochemical Aspects of the Acclimation of the Plant Rhodiola Semenowii Boriss. Plants 2021, 10, 1196. [Google Scholar] [CrossRef]
- Swiezewska, E. Ubiquinone and Plastoquinone Metabolism in Plants. Methods Enzymol. 2004, 378, 124–131, ISBN 978-0-12-182782-3. [Google Scholar] [PubMed]
- Liu, M.; Lu, S. Plastoquinone and Ubiquinone in Plants: Biosynthesis, Physiological Function and Metabolic Engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, S.; Gundlach, S.; Junge, O.; Szymczak, S. Integrating Biological Knowledge and Gene Expression Data Using Pathway-Guided Random Forests: A Benchmarking Study. Bioinformatics 2020, 36, 4301–4308. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Buhlmann, P. MissForest-Non-Parametric Missing Value Imputation for Mixed-Type Data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Soft. 2017, 77. [Google Scholar] [CrossRef] [Green Version]
- Kucheryavskiy, S. Mdatools—R Package for Chemometrics. Chemometrics Intell. Lab. Sys. 2020, 198, 103937. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Chen, X.; Minn, A.J. Random Survival Forests for High-Dimensional Data: Random Survival Forests for High-Dimensional Data. Stat. Anal. Data Min. 2011, 4, 115–132. [Google Scholar] [CrossRef]
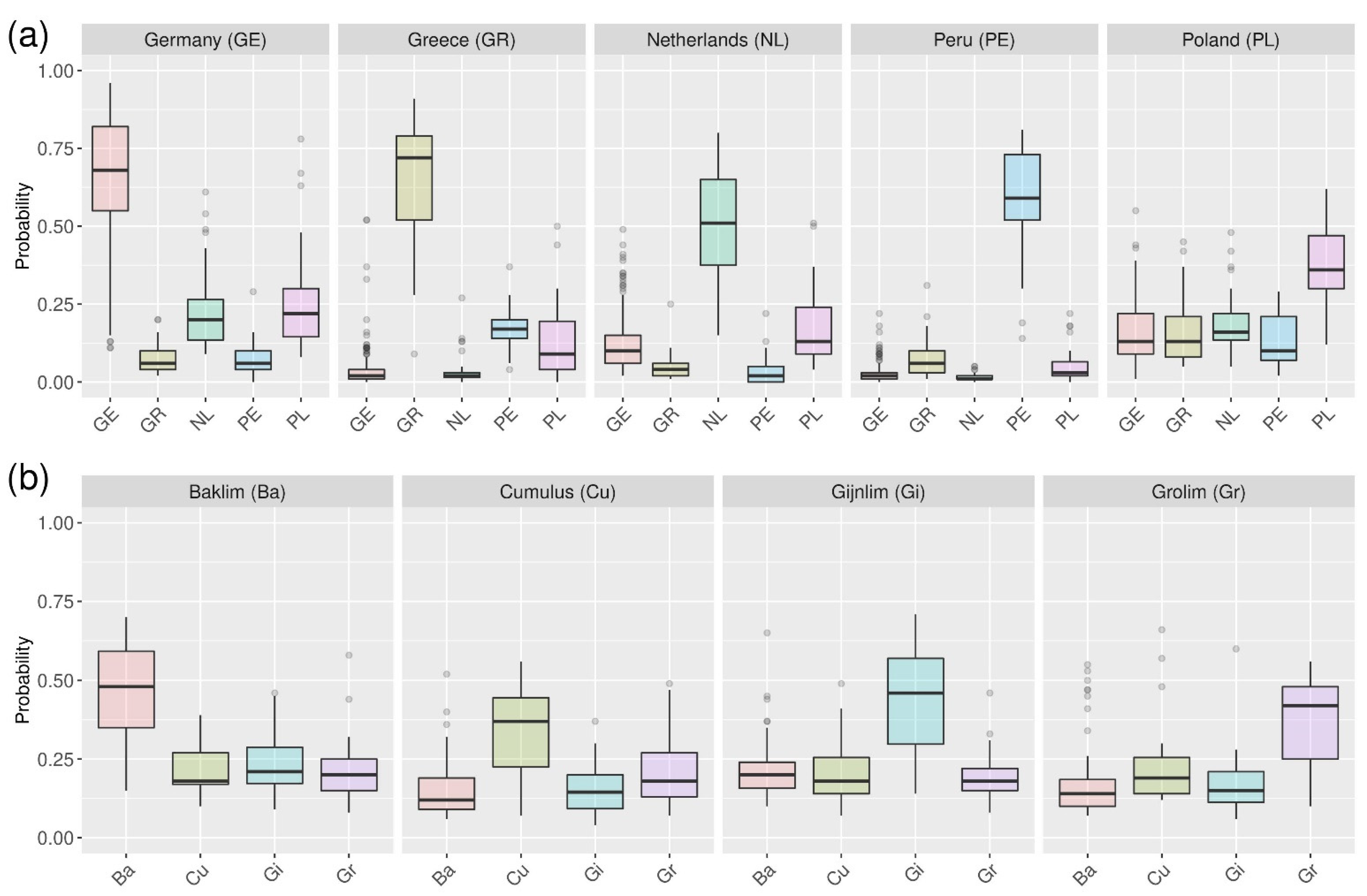

| Germany | Greece | Netherlands | Peru | Poland | Sensitivity [%] | |
|---|---|---|---|---|---|---|
| Germany | 196 | 4 | 5 | 0 | 8 | 92.0 |
| Greece | 0 | 23 | 0 | 0 | 2 | 92.0 |
| Netherlands | 5 | 0 | 23 | 0 | 3 | 74.2 |
| Peru | 1 | 0 | 0 | 11 | 1 | 84.6 |
| Poland | 7 | 2 | 3 | 0 | 23 | 65.7 |
| Specificity [%] | 93.8 | 79.3 | 74.2 | 100 | 62.2 |
| Backlim | Cumulus | Gijnlim | Grolim | Sensitivity [%] | |
|---|---|---|---|---|---|
| Backlim | 41 | 2 | 6 | 7 | 73.2 |
| Cumulus | 3 | 14 | 3 | 3 | 60.9 |
| Gijnlim | 9 | 1 | 31 | 1 | 73.8 |
| Grolim | 1 | 4 | 3 | 19 | 65.5 |
| Specificity [%] | 73.2 | 66.7 | 72.1 | 63.3 |
| Origin | 2016 | 2017 | 2018 |
|---|---|---|---|
| Germany | 105 | 77 | 31 |
| Greece | 14 | 7 | 4 |
| Netherlands | 10 | 10 | 11 |
| Peru | 7 | 4 | 2 |
| Poland | 16 | 11 | 8 |
| Variety | 2016 | 2017 |
|---|---|---|
| Backlim | 33 | 23 |
| Cumulus | 12 | 11 |
| Gijnlim | 22 | 20 |
| Grolim | 18 | 11 |
| Approach | Parameter | Description | Value |
|---|---|---|---|
| RF | ntree | number of trees | 10,000 |
| min.node.size | number of samples in terminal node | 1 | |
| mtry | number of candidate features | 138 (p3/4) 1 | |
| case.weights | weights for sampling of training observations | chosen according to the size of the respective class | |
| Boruta | importance | applied importance measure | impurity_corrected |
| pValue | confidence level | 0.01 | |
| maxRuns | maximum number of importance source runs | 100 | |
| SMD | s | predefined number of surrogate splits | 35 (p ∙ 0.05) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wenck, S.; Creydt, M.; Hansen, J.; Gärber, F.; Fischer, M.; Seifert, S. Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites 2022, 12, 5. https://doi.org/10.3390/metabo12010005
Wenck S, Creydt M, Hansen J, Gärber F, Fischer M, Seifert S. Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites. 2022; 12(1):5. https://doi.org/10.3390/metabo12010005
Chicago/Turabian StyleWenck, Soeren, Marina Creydt, Jule Hansen, Florian Gärber, Markus Fischer, and Stephan Seifert. 2022. "Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth" Metabolites 12, no. 1: 5. https://doi.org/10.3390/metabo12010005
APA StyleWenck, S., Creydt, M., Hansen, J., Gärber, F., Fischer, M., & Seifert, S. (2022). Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites, 12(1), 5. https://doi.org/10.3390/metabo12010005