Targeting Metabolic Reprogramming to Improve Breast Cancer Treatment: An In Vitro Evaluation of Selected Metabolic Inhibitors Using a Metabolomic Approach
Abstract
:1. Introduction
2. Results
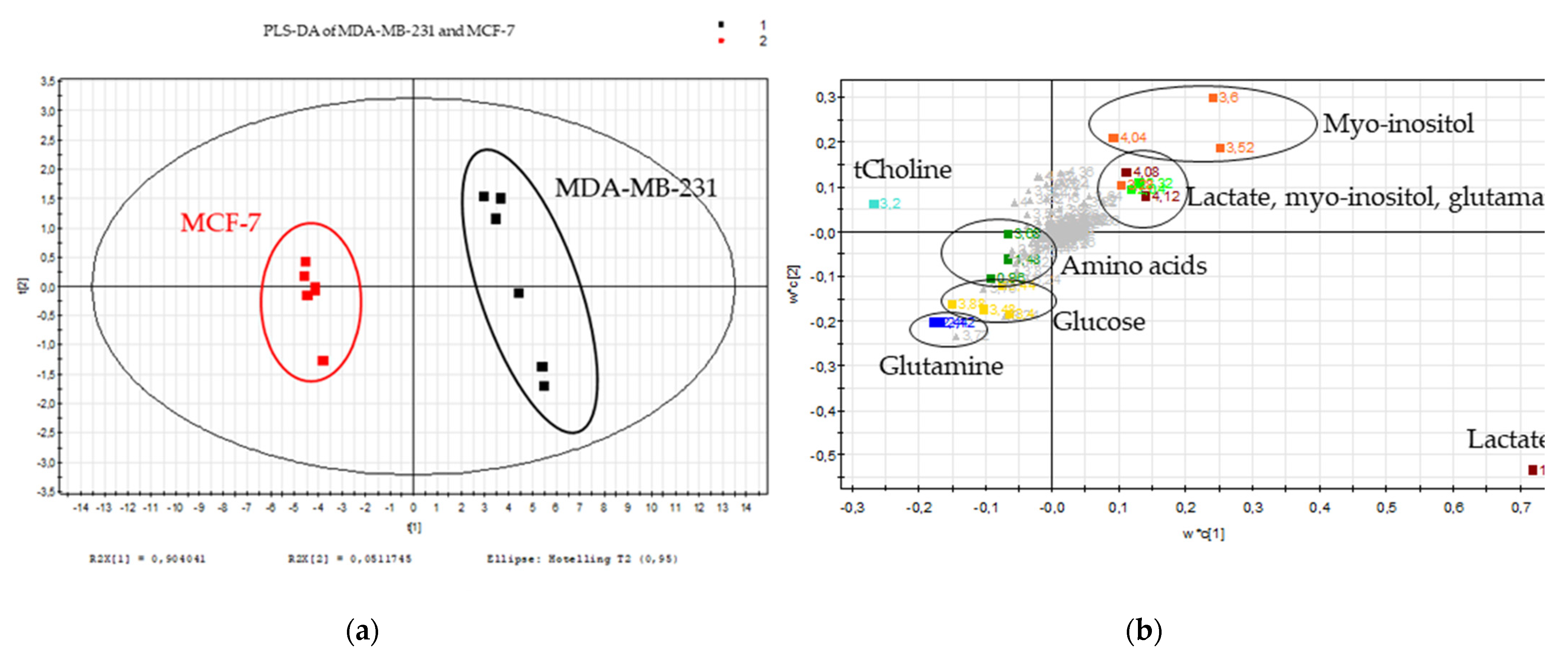
2.1. Metabolic Signature of Breast Cancer Tested Cell Lines
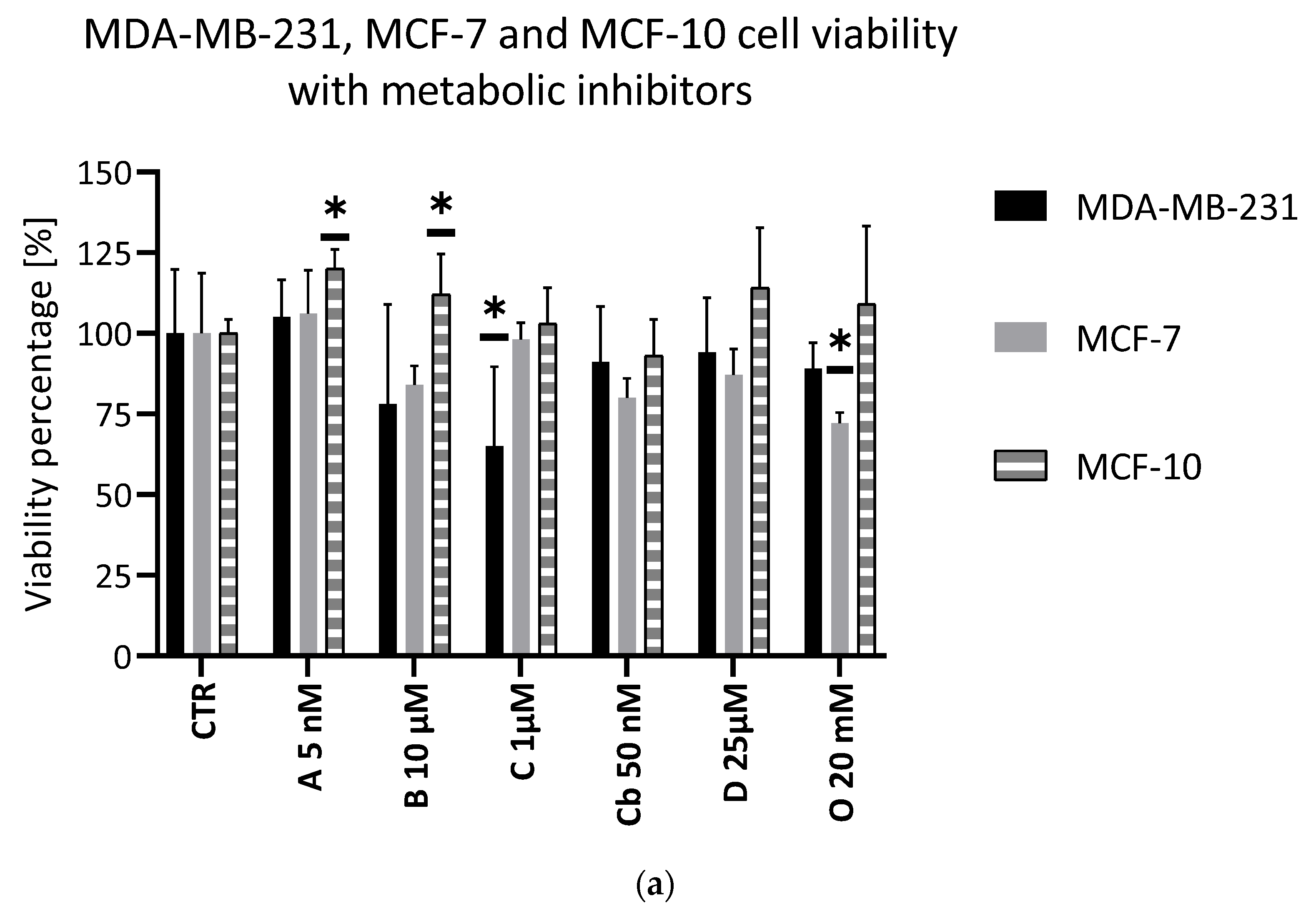
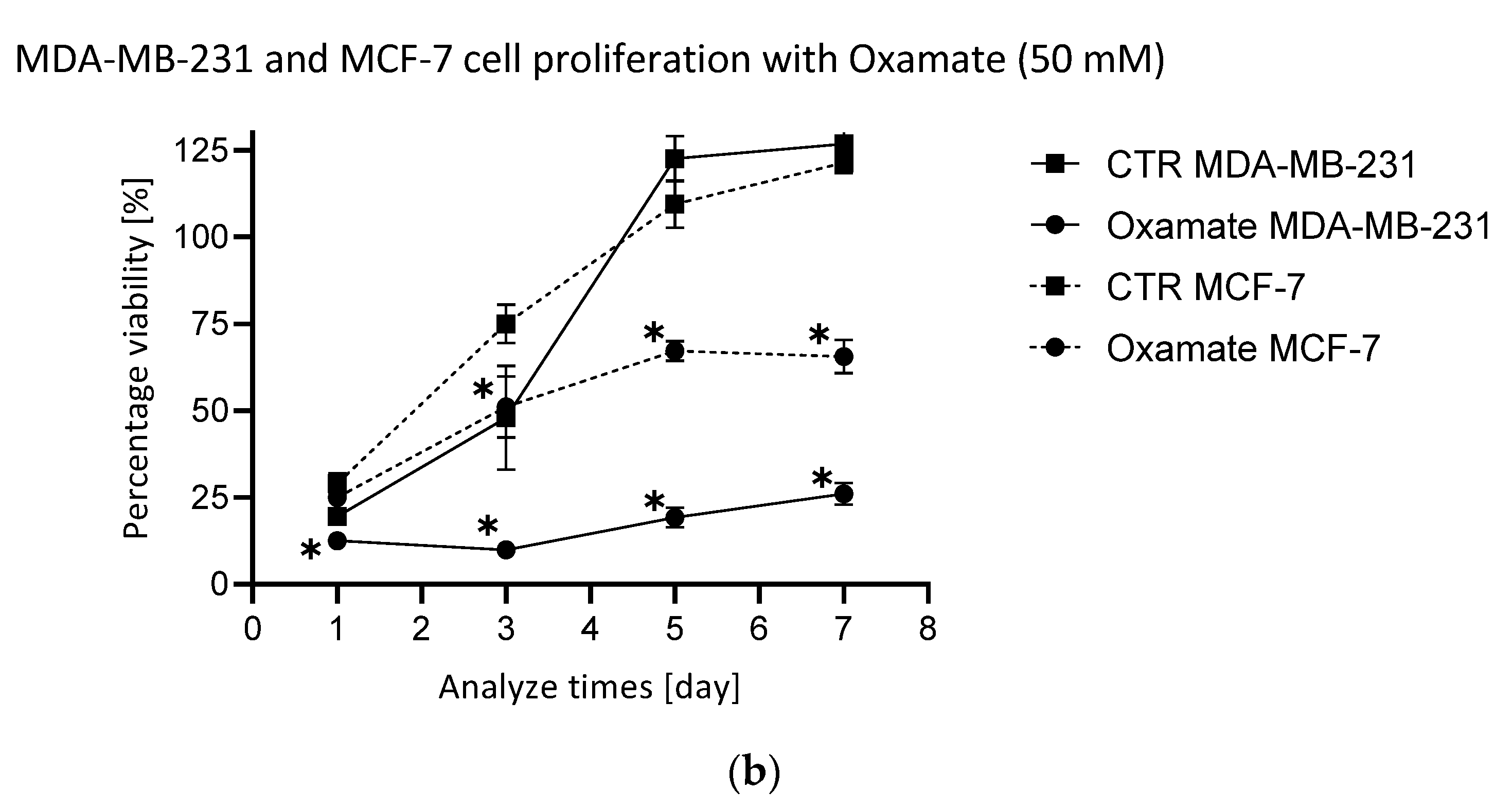
2.2. Assessment of the Efficacy of the Selected Metabolic Inhibitors
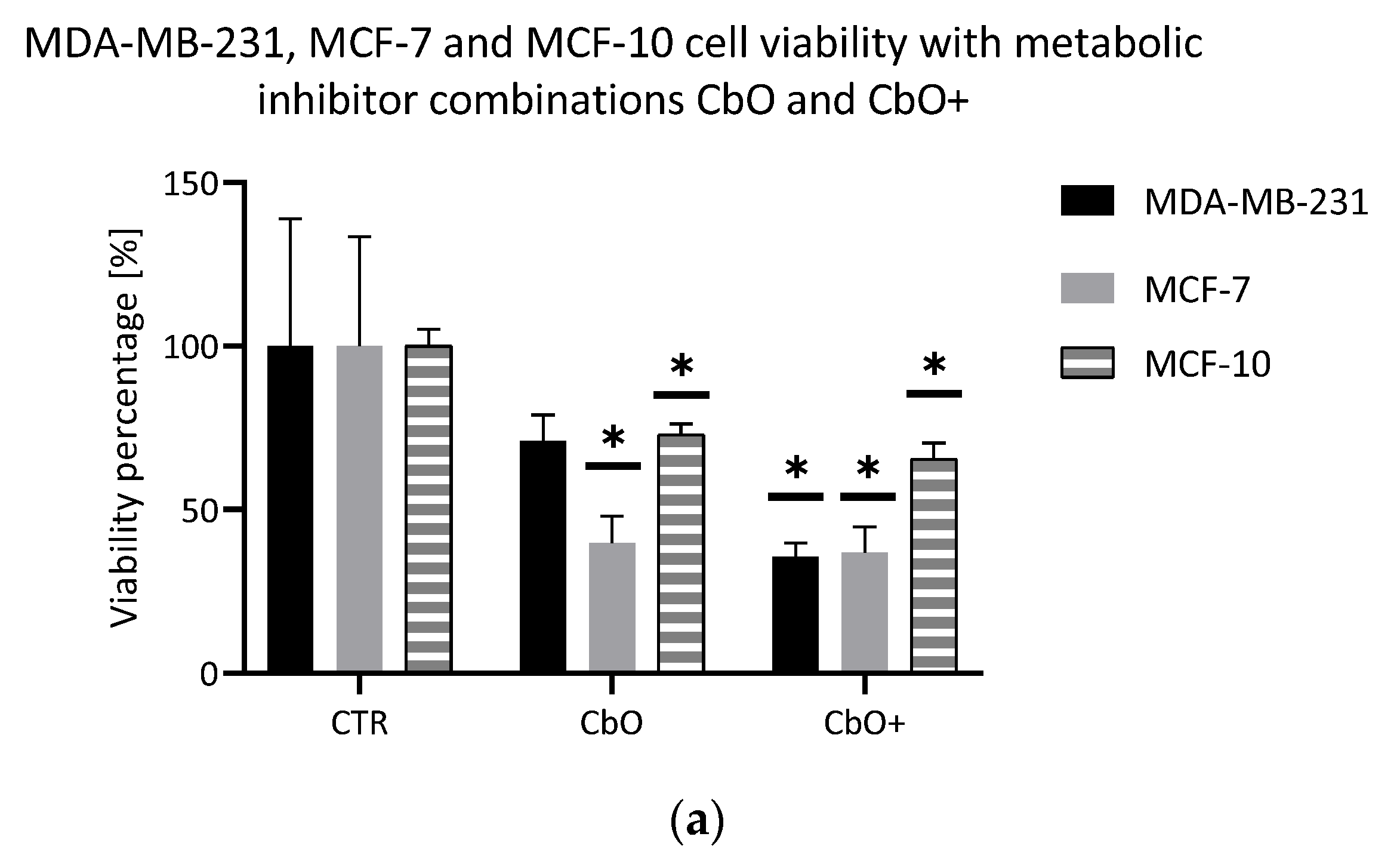
2.3. Evaluation of Double Combinations of Metabolic Inhibitors
2.4. Evaluation of Triple Combinations of Metabolic Inhibitors
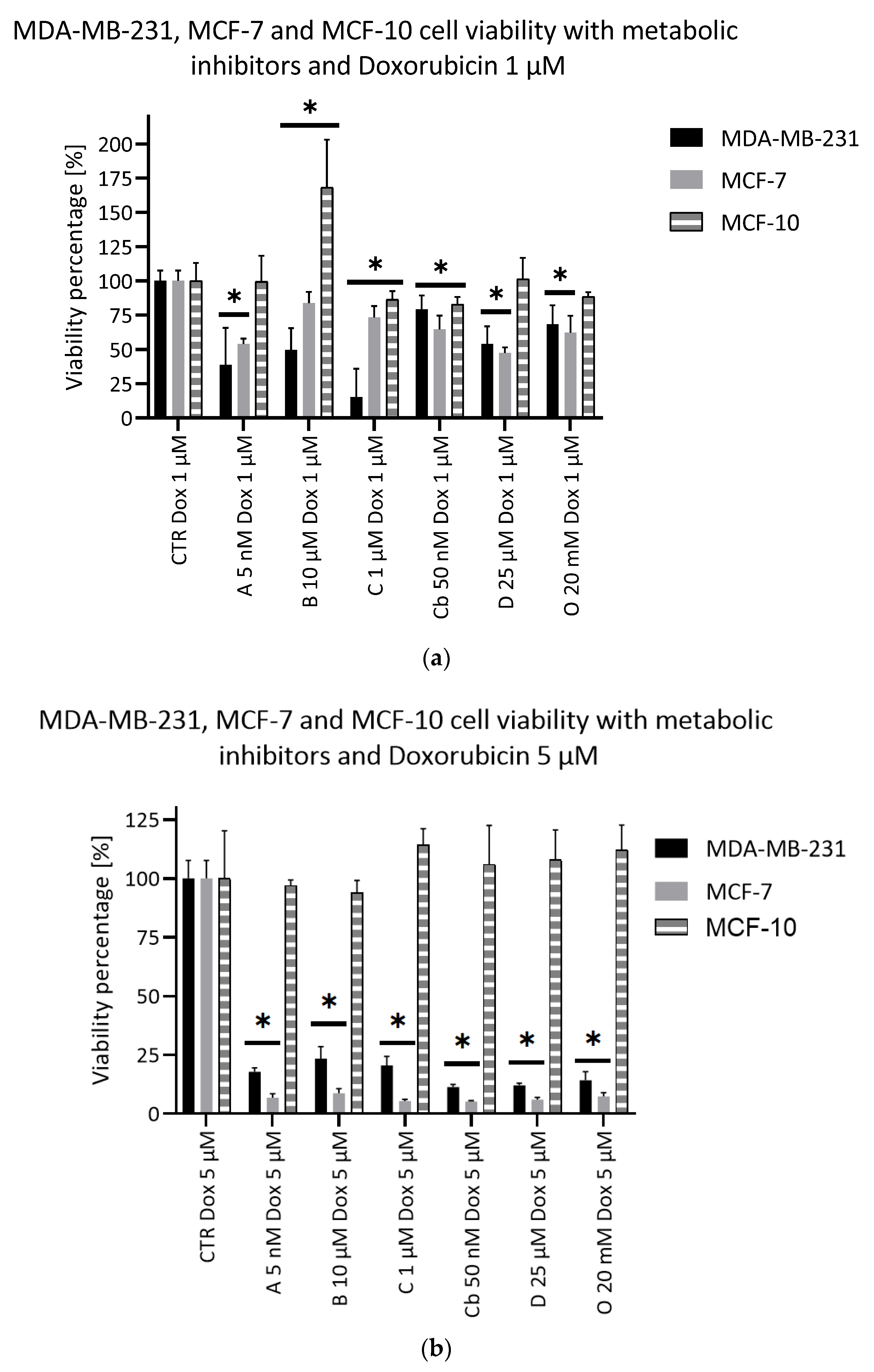
2.5. Potentiating Doxorubicin Efficacy with Metabolic Inhibitors
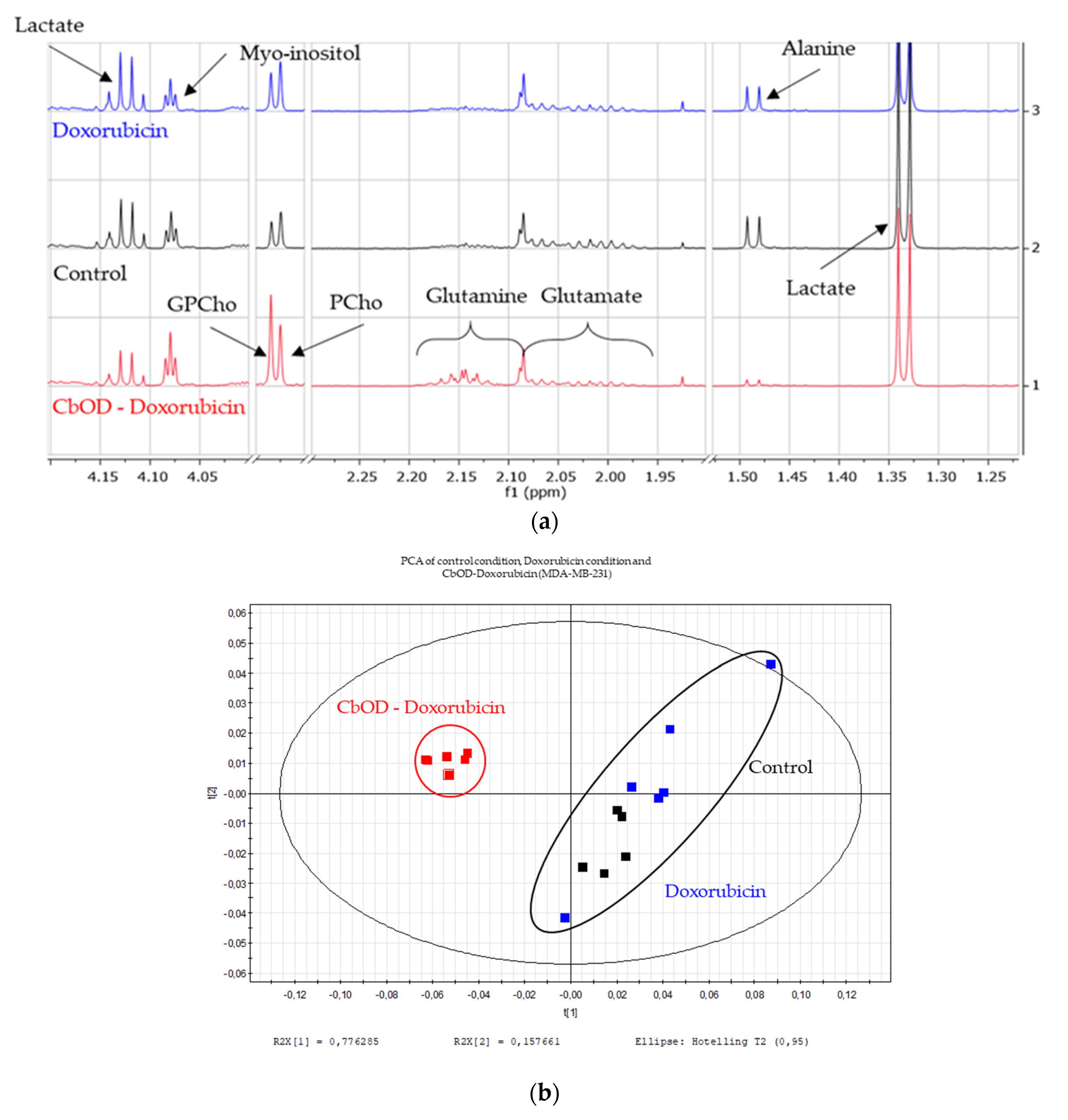
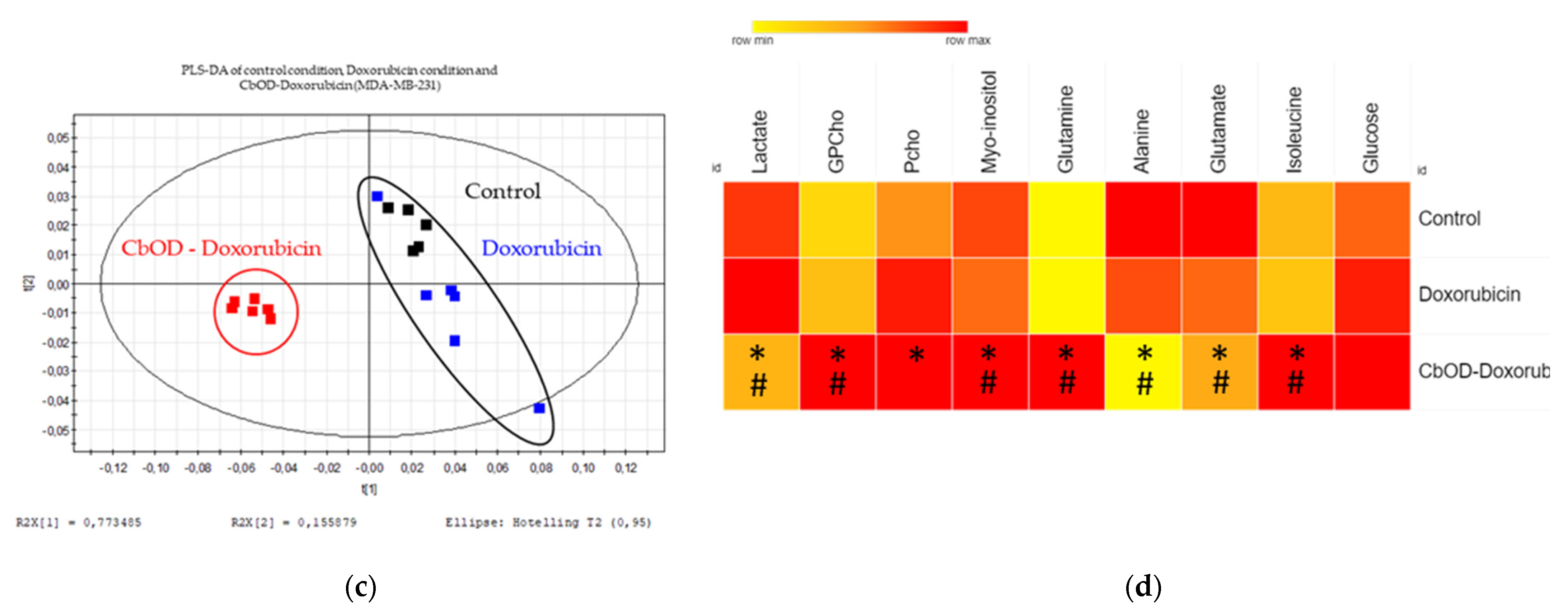
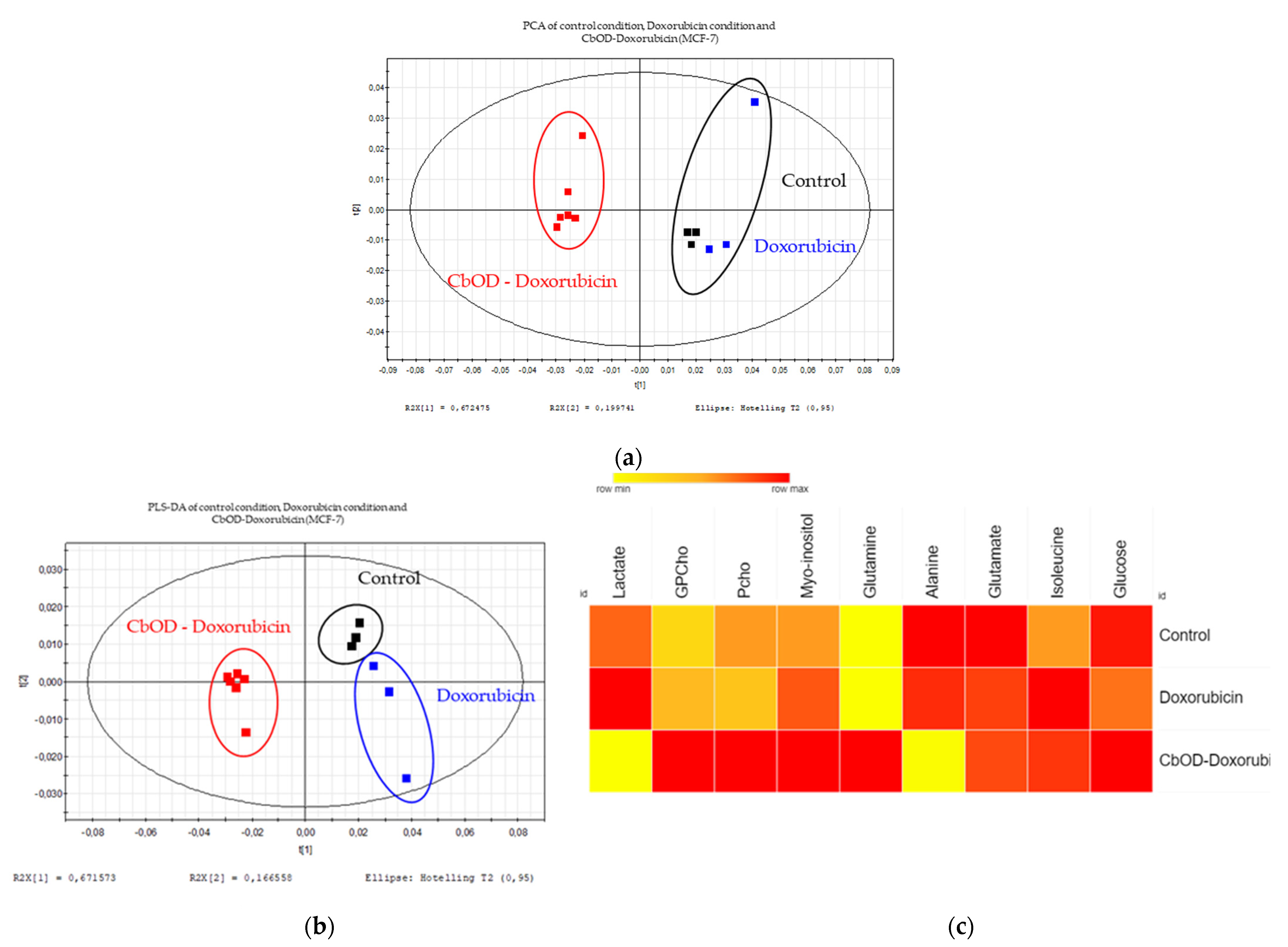
2.6. Impact of Doxorubicin and Combinations of Metabolic Inhibitors on Cell Metabolism
3. Discussion
3.1. Highlighting Potential Targetable Metabolic Pathway
3.2. Evaluation of the Efficacy of Metabolic Inhibitors and Their Ability to Potentiate Doxorubicin
3.3. Evaluation of the Combinations of Metabolic Inhibitors
3.3.1. CbO and CbOD Combinations
3.3.2. Other Triple Combinations
3.3.3. Conclusion about Metabolic Inhibitor Combinations
3.4. Impact of Doxorubicin and Inhibitor Combinations on Metabolism
4. Materials and Methods
4.1. Cell lines and Culture
4.2. Inhibitors
4.3. 1H-NMR Sample Collection and Extraction
4.4. Spectra Processing
4.5. Multivariate Data Analysis, Metabolic Signature, Heatmap
4.6. Viability Assay
4.7. Proliferation Assay
4.8. Statistical Method
5. General Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fondation Contre le Cancer le Cancer en Chiffres. Available online: https://www.cancer.be/le-cancer/le-cancer-en-chiffres%0D (accessed on 20 June 2020).
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Azevedo-Silva, J.Ã.; Queirós, O.; Ribeiro, A.; Baltazar, F.; Young, K.H.; Pedersen, P.L.; Pretoand, A.; Casal, M. The cytotoxicity of 3-bromopyruvate in breast cancer cells depends on extracellular pH. Biochem. J. 2015, 467, 247–258. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Reid, M.; Sanderson, S.; Locasale, J. Cancer Metabolism, 6th ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9783319614014. [Google Scholar]
- Polanski, R.; Hodgkinson, C.L.; Fusi, A.; Nonaka, D.; Priest, L.; Kelly, P.; Trapani, F.; Bishop, P.W.; White, A.; Critchlow, S.E.; et al. Activity of the monocarboxylate transporter 1 inhibitor azd3965 in small cell lung cancer. Clin. Cancer Res. 2014, 20, 926–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beloueche-Babari, M.; Wantuch, S.; Galobart, T.C.; Koniordou, M.; Parkes, H.G.; Arunan, V.; Chung, Y.L.; Eykyn, T.R.; Smith, P.D.; Leach, M.O. MCT1 inhibitor AZD3965 increases mitochondrial metabolism, facilitating combination therapy and noninvasive magnetic resonance spectroscopy. Cancer Res. 2017, 77, 5913–5924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ackerstaff, E.; Glunde, K.; Bhujwalla, Z.M. Choline phospholipid metabolism: A target in cancer cells? J. Cell. Biochem. 2003, 90, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Andreasssen, T.; Jensen, L. Estrogen receptor a promotes breast cancer by reprogramming choline metabolism. Jpn. Cult. Stud. 2014, 167–181. [Google Scholar] [CrossRef] [Green Version]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010, 20, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Quintero, M.; Mackenzie, N.; Brennan, P.A. Hypoxia-inducible factor 1 (HIF-1) in cancer. Eur. J. Surg. Oncol. 2004, 30, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Xue, J.; Li, Z.; Shi, X.; Jiang, B.H.; Fang, J. Chrysin inhibits expression of hypoxia-inducible factor 1-α through reducing hypoxia-inducible factor-1α stability and inhibiting its protein synthesis. Mol. Cancer Ther. 2007, 6, 220–226. [Google Scholar] [CrossRef] [Green Version]
- OMS Cancer du Sein: Prévention et Lutte Contre la Maladie Facteurs de Risque du Cancer du Sein. Available online: https://www.who.int/topics/cancer/breastcancer/fr/index2.html (accessed on 28 January 2020).
- Key, T.; Verkasalo, P.; Banks, E. Epidemiology of breast cancer. Lancet Oncol. 2001, 2, 133–140. [Google Scholar] [CrossRef]
- Malhotra, G.K.; Zhao, X.; Band, H.; Band, V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. Ther. 2010, 10, 955–960. [Google Scholar] [CrossRef] [Green Version]
- Johnson-Arbor, K.; Dubey, R. Doxorubicin. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef]
- Dallons, M.; Schepkens, C.; Dupuis, A.; Tagliatti, V.; Colet, J.M. New Insights About Doxorubicin-Induced Toxicity to Cardiomyoblast-Derived H9C2 Cells and Dexrazoxane Cytoprotective Effect: Contribution of In Vitro 1H-NMR Metabonomics. Front. Pharmacol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Curtis, N.J.; Mooney, L.; Hopcroft, L.; Michopoulos, F.; Whalley, N.; Zhong, H.; Murray, C.; Logie, A.; Revill, M.; Byth, K.F.; et al. Pre-clinical pharmacology of AZD3965, a selective inhibitor of MCT1: DLBCL, NHL and Burkitt’s lymphoma anti-tumor activity. Oncotarget 2017, 8, 69219–69236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bean, J.F.; Qiu, Y.Y.; Yu, S.; Clark, S.; Chu, F.; Madonna, M.B. Glycolysis inhibition and its effect in doxorubicin resistance in neuroblastoma. J. Pediatr. Surg. 2014, 49, 981–984. [Google Scholar] [CrossRef] [PubMed]
- García-Castillo, V.; López-Urrutia, E.; Villanueva-Sánchez, O.; Ávila-Rodríguez, M.A.; Zentella-Dehesa, A.; Cortés-González, C.; López-Camarillo, C.; Jacobo-Herrera, N.J.; Pérez-Plasencia, C. Targeting metabolic remodeling in triple negative breast cancer in a murine model. J. Cancer 2017, 8, 178–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schepkens, C.; Dallons, M.; Dehairs, J.; Talebi, A.; Jeandriens, J.; Drossart, L.M.; Auquier, G.; Tagliatti, V.; Swinnen, J.V.; Colet, J.M. A new classification method of metastatic cancers using a1H-NMR-based approach: A study case of melanoma, breast, and prostate cancer cell lines. Metabolites 2019, 9, 281. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.A.; Mueller-Klieser, W. Lactate: A metabolic key player in cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [Green Version]
- Meric-Bernstam, F.; Tannir, N.M.; Mier, J.W.; DeMichele, A.; Telli, M.L.; Fan, A.C.; Munster, P.N.; Carvajal, R.D.; Orford, K.W.; Bennett, M.K.; et al. Phase 1 study of CB-839, a small molecule inhibitor of glutaminase (GLS), alone and in combination with everolimus (E) in patients (pts) with renal cell cancer (RCC). J. Clin. Oncol. 2016, 34, 4568. [Google Scholar] [CrossRef]
- Meric-Bernstam, F. Phase 1 study of CB-839, a first-in-class, orally administered small molecule inhibitor of glutaminase in patients with refractory solid tumors. Mol. Cancer Ther. 2015, 126, 2566. [Google Scholar] [CrossRef]
- Hong, C.S.; Graham, N.A.; Gu, W.; Espindola Camacho, C.; Mah, V.; Maresh, E.L.; Alavi, M.; Bagryanova, L.; Krotee, P.A.L.; Gardner, B.K.; et al. MCT1 Modulates Cancer Cell Pyruvate Export and Growth of Tumors that Co-express MCT1 and MCT4. Cell Rep. 2016, 14, 1590–1601. [Google Scholar] [CrossRef] [Green Version]
- Kwiatkowska, E.; Wojtala, M.; Gajewska, A.; Soszyński, M.; Bartosz, G.; Sadowska-Bartosz, I. Effect of 3-bromopyruvate acid on the redox equilibrium in non-invasive MCF-7 and invasive MDA-MB-231 breast cancer cells. J. Bioenerg. Biomembr. 2016, 48, 23–32. [Google Scholar] [CrossRef]
- Chong, D.; Ma, L.; Liu, F.; Zhang, Z.; Zhao, S.; Huo, Q.; Zhang, P.; Zheng, H.; Liu, H. Synergistic antitumor effect of 3-bromopyruvate and 5-fluorouracil against human colorectal cancer through cell cycle arrest and induction of apoptosis. Anticancer. Drugs 2017, 28, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Mag, P.; Samarghandian, S.; Azimi-nezhad, M.; Borji, A.; Hasanzadeh, M.; Jabbari, F. Inhibitory and Cytotoxic Activities of Chrysin on Human Breast Adenocarcinoma Cells by Induction of Apoptosis. Pharmacogn. Mag. 2016, 12, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Khoo, B.Y.; Chua, S.L.; Balaram, P. Apoptotic Effects of Chrysin in Human Cancer Cell Lines. Int. J. Mol. Sci. 2010, 11, 2188–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasouli, S.; Zarghami, N. Synergistic growth inhibitory effects of Chrysin and Metformin combination on breast cancer cells through hTERT and cyclin D1 suppression. Asian Pacific J. Cancer Prev. 2018, 19, 977–982. [Google Scholar] [CrossRef]
- Paris, L.; Cecchetti, S.; Spadaro, F.; Abalsamo, L.; Lugini, L.; Pisanu, M.E.; Iorio, E.; Natali, P.G.; Ramoni, C.; Podo, F. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. Breast Cancer Res. 2010, 12, R27. [Google Scholar] [CrossRef] [Green Version]
- Miskimins, W.K.; Ahn, H.J.; Kim, J.Y.; Ryu, S.; Jung, Y.S.; Choi, J.Y. Synergistic anti-cancer effect of phenformin and oxamate. PLoS ONE 2014, 9, e85576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, G.; Liu, Y.; Li, X.; Xu, P.; Luo, Y. Targeting glucose metabolism in chondrosarcoma cells enhances the sensitivity to doxorubicin through the inhibition of lactate dehydrogenase-A. Oncol. Rep. 2014, 31, 2727–2734. [Google Scholar] [CrossRef] [Green Version]
- Abalsamo, L.; Spadaro, F.; Bozzuto, G.; Paris, L.; Cecchetti, S.; Lugini, L.; Iorio, E.; Molinari, A.; Ramoni, C.; Podo, F. Inhibition of phosphatidylcholine-specific phospholipase C results in loss of mesenchymal traits in metastatic breast cancer cells. Breast Cancer Res. 2012, 14, R50. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, Y.; Nakayama, Y.; Fukusaki, E.; Irino, Y. Glutamate production from ammonia via glutamate dehydrogenase 2 activity supports cancer cell proliferation under glutamine depletion. Biochem. Biophys. Res. Commun. 2018, 495, 761–767. [Google Scholar] [CrossRef] [Green Version]


| Inhibitors | Low Concentration | High Concentration |
|---|---|---|
| Oxamate | 20 mM | 50 mM |
| Chrysin | 1 µM | 5 µM |
| AZD3965 | 5 nM | 50 nM |
| D609 | 25 µM | 75 µM |
| 3-bromo-pyruvate | 10 µM | 50 µM |
| CB-839 | 50 nM | 1 µM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Draguet, A.; Tagliatti, V.; Colet, J.-M. Targeting Metabolic Reprogramming to Improve Breast Cancer Treatment: An In Vitro Evaluation of Selected Metabolic Inhibitors Using a Metabolomic Approach. Metabolites 2021, 11, 556. https://doi.org/10.3390/metabo11080556
Draguet A, Tagliatti V, Colet J-M. Targeting Metabolic Reprogramming to Improve Breast Cancer Treatment: An In Vitro Evaluation of Selected Metabolic Inhibitors Using a Metabolomic Approach. Metabolites. 2021; 11(8):556. https://doi.org/10.3390/metabo11080556
Chicago/Turabian StyleDraguet, Anaïs, Vanessa Tagliatti, and Jean-Marie Colet. 2021. "Targeting Metabolic Reprogramming to Improve Breast Cancer Treatment: An In Vitro Evaluation of Selected Metabolic Inhibitors Using a Metabolomic Approach" Metabolites 11, no. 8: 556. https://doi.org/10.3390/metabo11080556
APA StyleDraguet, A., Tagliatti, V., & Colet, J.-M. (2021). Targeting Metabolic Reprogramming to Improve Breast Cancer Treatment: An In Vitro Evaluation of Selected Metabolic Inhibitors Using a Metabolomic Approach. Metabolites, 11(8), 556. https://doi.org/10.3390/metabo11080556







