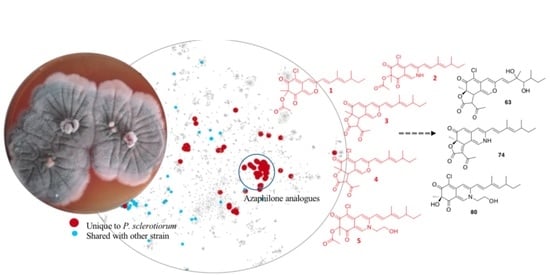
Dereplication, Annotation, and Characterization of 74 Potential Antimicrobial Metabolites from Penicillium Sclerotiorum Using t-SNE Molecular Networks
Abstract
:1. Introduction
2. Results
2.1. Penicillium Sclerotiorum Selection
2.2. Penicillium Sclerotiorum Metabolism Dereplication
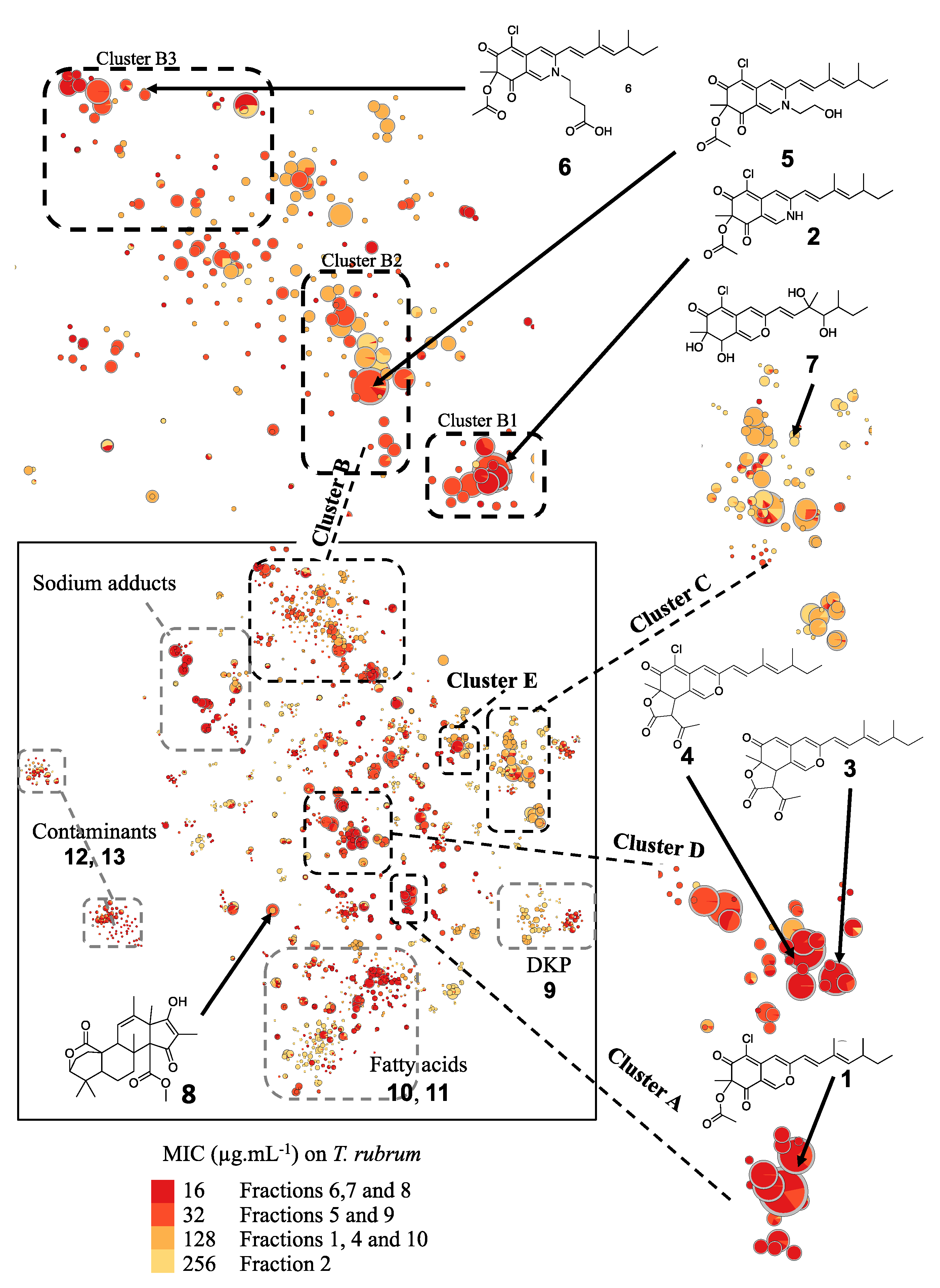
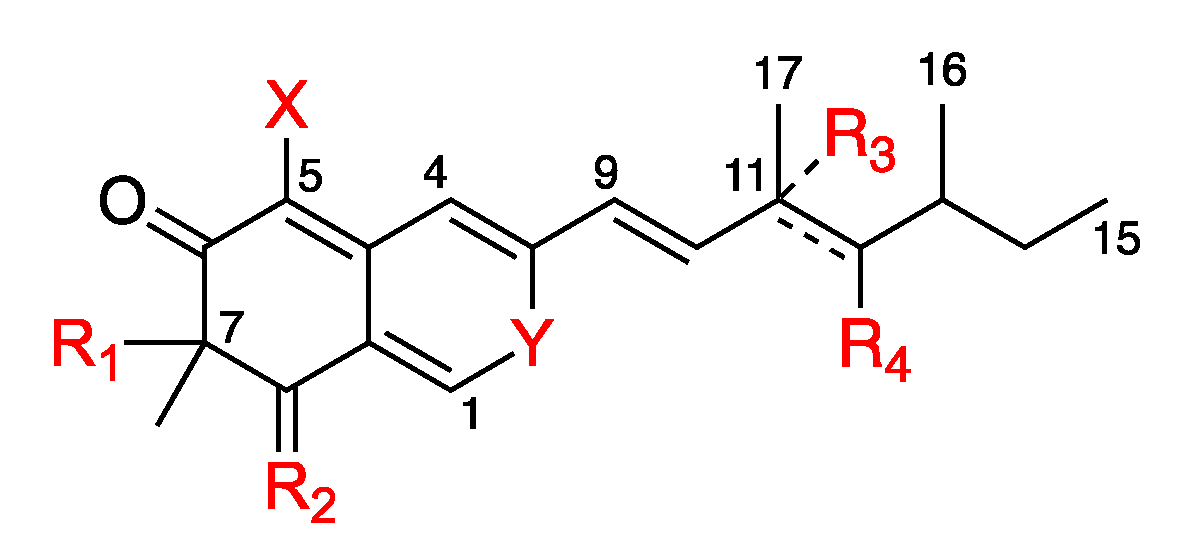
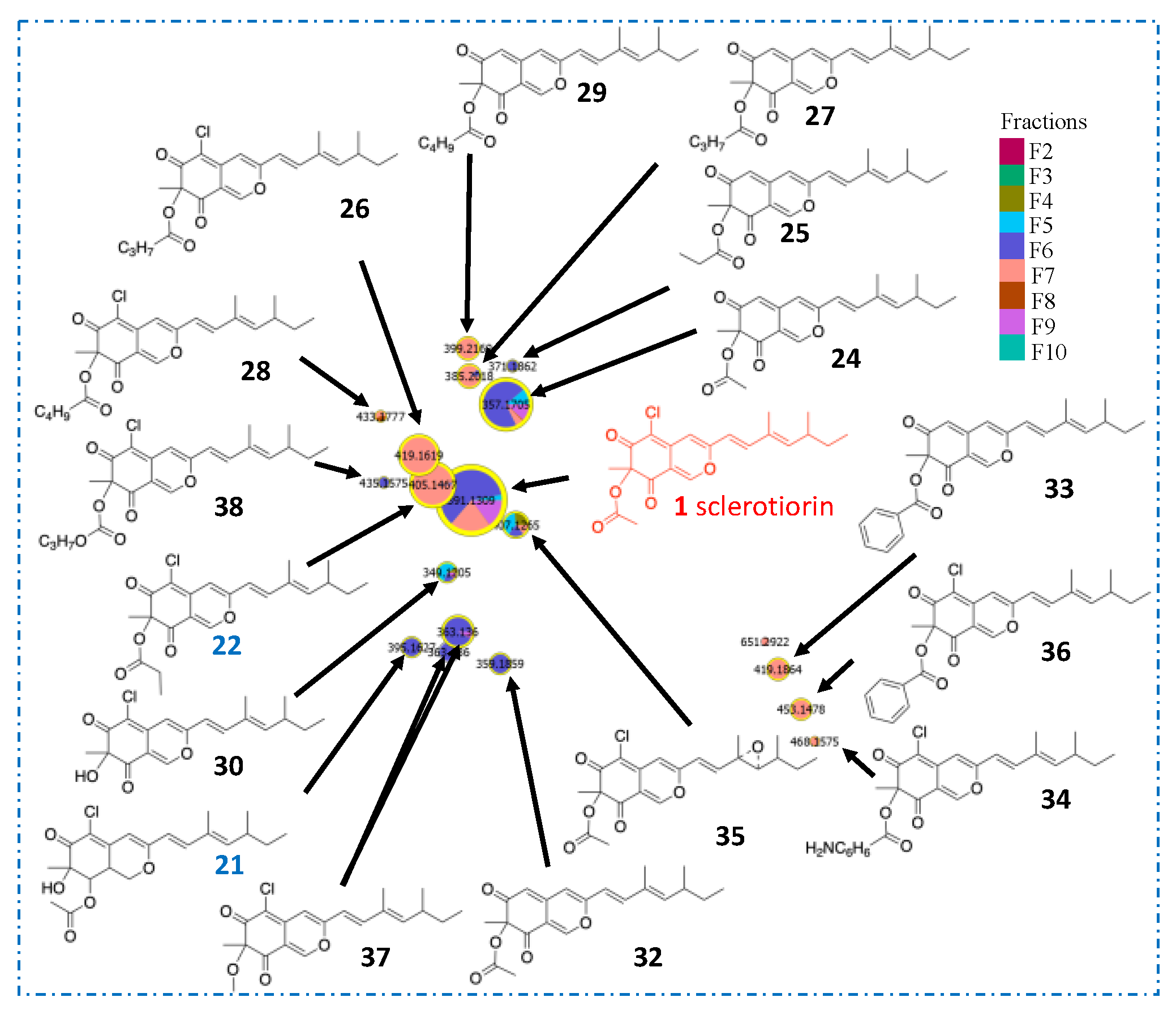
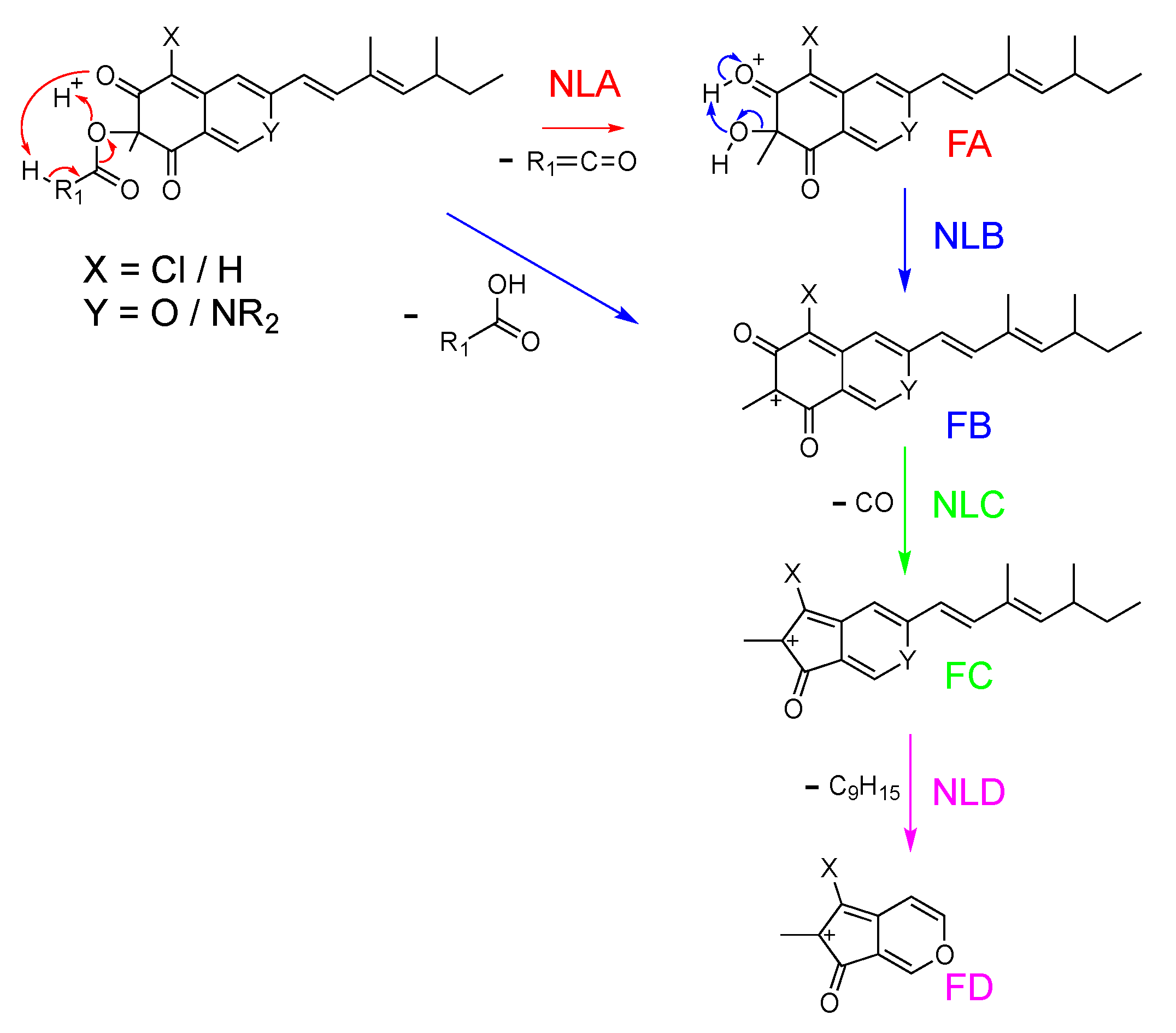
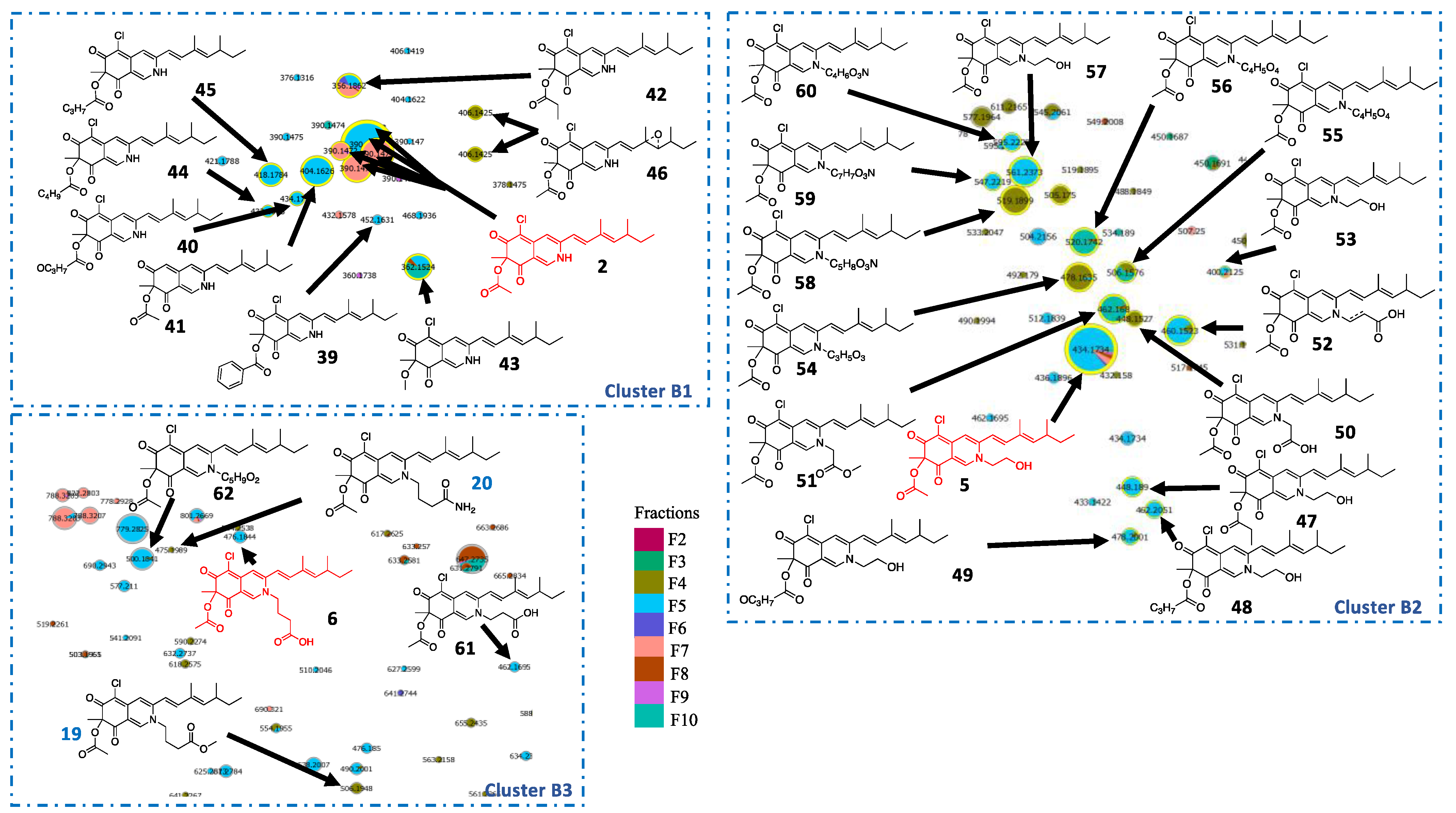
2.3. In Silico Azaphilone Structure Prediction Using MS/MS Data and a t-SNE Molecular Network
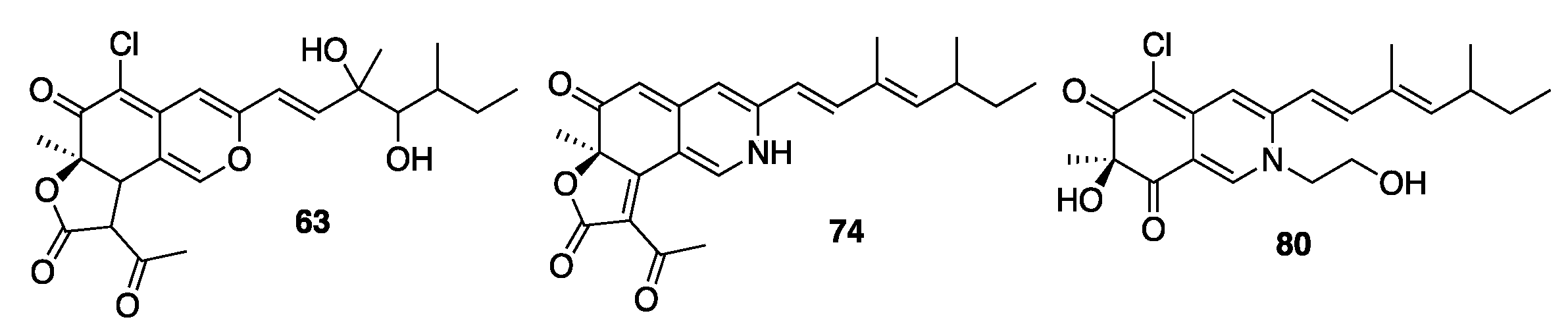
2.4. Isolation and Characterization of Compounds
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Identification of Termite Mutualistic Microorganisms
4.2.1. General Identification Procedure
4.2.2. Isolation and Identification of Penicillium Sclerotiorum SNB-CN111
4.3. Culture and Extraction of Microorganisms
4.3.1. General Cultivation and Extraction Procedure
4.3.2. Extraction of SNB-CN111
4.4. Isolation of Compounds
4.5. LC–MS/MS Analysis
4.6. Data Processing and Analysis
4.7. X-ray Structure Determination of Compounds 1 and 5
4.8. Biological Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knight, V.; Sanglier, J.-J.; DiTullio, D.; Braccili, S.; Bonner, P.; Waters, J.; Hughes, D.; Zhang, L. Diversifying Microbial Natural Products for Drug Discovery. Appl. Microbiol. Biotechnol. 2003, 62, 446–458. [Google Scholar] [CrossRef]
- Harvey, A.L. Natural Products as a Screening Resource. Curr. Opin. Chem. Biol. 2007, 11, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Stork, N.E.; McBroom, J.; Gely, C.; Hamilton, A.J. New Approaches Narrow Global Species Estimates for Beetles, Insects, and Terrestrial Arthropods. Proc. Natl. Acad. Sci. USA 2015, 112, 7519–7523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brune, A. Symbiotic Digestion of Lignocellulose in Termite Guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Beemelmanns, C.; Guo, H.; Rischer, M.; Poulsen, M. Natural Products from Microbes Associated with Insects. J. Org. Chem. 2016, 12, 314–327. [Google Scholar] [CrossRef] [Green Version]
- Dillon, R.J.; Dillon, V.M. The Gut Bacteria of Insects: Nonpathogenic Interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Tanaka, J.; Namihira, T.; Shinzato, N. Antibiotics Production by an Actinomycete Isolated from the Termite Gut. J. Basic Microbiol. 2012, 52, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Jiang, D.; Kong, L.; Zhang, P.; Xu, J. Antifungal Activities of Metabolites Produced by a Termite-Associated Streptomyces canus BYB02. J. Agric. Food Chem. 2013, 61, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Nirma, C.; Eparvier, V.; Stien, D. Antifungal Agents from Pseudallescheria boydii SNB-CN73 Isolated from a Nasutitermes sp. Termite. J. Nat. Prod. 2013, 76, 988–991. [Google Scholar] [CrossRef]
- Nirma, C.; Eparvier, V.; Stien, D. Antibacterial Ilicicolinic Acids C and D and Ilicicolinal from Neonectria discophora SNB-CN63 Isolated from a Termite Nest. J. Nat. Prod. 2015, 78, 159–162. [Google Scholar] [CrossRef]
- Nirma, C.; Eparvier, V.; Stien, D. Reactivation of Antibiosis in the Entomogenous Fungus Chrysoporthe sp. SNB-CN74. J. Antibiot. 2015, 68, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Sorres, J.; Nirma, C.; Barthélemy, M.; Eparvier, V.; Stien, D. Tyroscherin and Tyroscherin Analogs from Pseudallescheria boydii SNB-CN85 Isolated from Termite Termes Cf. hispaniolae. Phytochem. Lett. 2017, 22, 142–144. [Google Scholar] [CrossRef]
- Wang, M. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Olivon, F.; Elie, N.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MetGem Software for the Generation of Molecular Networks Based on the T-SNE Algorithm. Anal. Chem. 2018, 90, 13900–13908. [Google Scholar] [CrossRef]
- Brel, O.; Touré, S.; Levasseur, M.; Lechat, C.; Pellissier, L.; Wolfender, J.-L.; Van-Elslande, E.; Litaudon, M.; Dusfour, I.; Stien, D.; et al. Paecilosetin Derivatives as Potent Antimicrobial Agents from Isaria farinosa. J. Nat. Prod. 2020, 83, 2915–2922. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MZmine 2 Data-Preprocessing to Enhance Molecular Networking Reliability. Anal. Chem. 2017, 89, 7836–7840. [Google Scholar] [CrossRef]
- Touré, S.; Desrat, S.; Pellissier, L.; Allard, P.-M.; Wolfender, J.-L.; Dusfour, I.; Stien, D.; Eparvier, V. Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. Int. J. Mol. Sci. 2019, 20, 1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfender, J.-L.; Nuzillard, J.-M. Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography–High-Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- MacCurtin, T.; Reilly, J. Sclerotiorine, a Chlorinated Metabolic Product of Penicillium sclerotiorum, Van Beyma. Nature 1940, 146, 335. [Google Scholar] [CrossRef]
- Osmanova, N.; Schultze, W.; Ayoub, N. Azaphilones: A Class of Fungal Metabolites with Diverse Biological Activities. Phytochem. Rev. 2010, 9, 315–34228. [Google Scholar] [CrossRef]
- Gao, J.-M.; Yang, S.-X.; Qin, J.-C. Azaphilones: Chemistry and Biology. Chem. Rev. 2013, 113, 4755–4811. [Google Scholar] [CrossRef] [PubMed]
- Mapari, S.A.S.; Thrane, U.; Meyer, A.S. Fungal Polyketide Azaphilone Pigments as Future Natural Food Colorants? Trends Biotechnol. 2010, 28, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Godzien, J.; Gil de la Fuente, A.; Otero, A.; Barbas, C. Metabolite Annotation and Identification. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2018; Volume 82, pp. 415–445. [Google Scholar] [CrossRef]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Son, S.; Ko, S.-K.; Kim, J.W.; Lee, J.K.; Jang, M.; Ryoo, I.-J.; Hwang, G.J.; Kwon, M.C.; Shin, K.-S.; Futamura, Y.; et al. Structures and Biological Activities of Azaphilones Produced by Penicillium sp. KCB11A109 from a Ginseng Field. Phytochemistry 2016, 122, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.-B. Azaphilone and Isocoumarin Derivatives from the Sponge-Derived Fungus Eupenicillium sp. 6A-9. Tetrahedron Lett. 2018, 59, 3345–3348. [Google Scholar] [CrossRef]
- Tang, J.-L.; Zhou, Z.-Y.; Yang, T.; Yao, C.; Wu, L.-W.; Li, G.-Y. Azaphilone Alkaloids with Anti-Inflammatory Activity from Fungus Penicillium sclerotiorum Cib-411. J. Agric. Food Chem. 2019, 67, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Long, Y.; Lei, X.; Xu, J.; Huang, Z.; She, Z.; Lin, Y.; Li, J.; Liu, L. Azaphilones Isolated from an Alga-Derived Fungus Penicillium sp. ZJ-27. Phytochem. Lett. 2016, 18, 180–186. [Google Scholar] [CrossRef]
- Michael, A.P.; Grace, E.J.; Kotiw, M.; Barrow, R.A. Isochromophilone IX, a Novel GABA-Containing Metabolite Isolated from a Cultured Fungus, Penicillium sp. Aust. J. Chem. 2003, 56, 13. [Google Scholar] [CrossRef]
- Arai, N.; Shiomi, K.; Tomoda, H.; Tabata, N.; Yang, D.J.; Masuma, R.; Kawakubo, T.; Omura, S. Isochromophilones III-VI, Inhibitors of Acyl-CoA: Cholesterol Acyltransferase Produced by Penicillium multicolor FO-3216. J. Antibiot. 1995, 48, 696–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Qiu, P.; Liu, H.; Li, J.; Shao, C.; Yan, T.; Cao, W.; She, Z. Identification of Anti-Inflammatory Polyketides from the Coral-Derived Fungus Penicillium sclerotiorin: In Vitro Approaches and Molecular-Modeling. Bioorg. Chem. 2019, 88, 102973. [Google Scholar] [CrossRef] [PubMed]
- Pairet, L.; Wrigley, S.K.; Reynolds, I.E.; Hayes, M.A. Taxonomy, Fermentation, Isolation, Structure Elucidation and Biological Activity. J. Antibiot. 1995, 48, 913–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niessen, W.M.A.; Ceballos, R.A.C. Fragmentation of Drugs and Pesticides. In Interpretation of MS-MS Mass Spectra of Drugs and Pesticides; John Wiley & Sons, Inc.: Hoboken, NY, USA, 2017; pp. 129–349. [Google Scholar] [CrossRef]
- Niessen, W.M.A.; Ceballos, R.A.C. Fragmentation of Even-Electron Ions. In Interpretation of MS-MS Mass Spectra of Drugs and Pesticides; John Wiley & Sons, Inc.: Hoboken, NY, USA, 2017; pp. 71–128. [Google Scholar] [CrossRef]
- Svilar, L.; Stankov-Jovanovic, V.; Lesage, D.; Dossmann, H.; Tabet, J.C. High-resolution mass spectrometry and hydrogen/deuterium exchange study of mitorubrin azaphilones and nitrogenized analogues. J. Mass Spectrom. 2012, 47, 969–977. [Google Scholar] [CrossRef]
- Williams, J.P.; Nibbering, N.M.M.; Green, B.N.; Patel, V.J.; Scrivens, J.H. Collision-Induced Fragmentation Pathways Including Odd-Electron Ion Formation from Desorption Electrospray Ionisation Generated Protonated and Deprotonated Drugs Derived from Tandem Accurate Mass Spectrometry. J. Mass Spectrom. 2006, 41, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B.; de Visser, S.P.; Shaik, S. Mechanism of Oxidation Reactions Catalyzed by Cytochrome P450 Enzymes. Chem. Rev. 2004, 104, 3947–3980. [Google Scholar] [CrossRef] [PubMed]
- Arand, M.; Cronin, A.; Oesch, F.; Mowbray, S.L.; Alwyn Jones, T. The Telltale Structures of Epoxide Hydrolases. Drug Metab. Rev. 2003, 35, 365–383. [Google Scholar] [CrossRef]
- Hemtasin, C.; Kanokmedhakul, S.; Moosophon, P.; Soytong, K.; Kanokmedhakul, K. Bioactive Azaphilones from the Fungus Penicillium multicolor CM01. Phytochem. Lett. 2016, 16, 56–60. [Google Scholar] [CrossRef]
- Wang, X.; Sena Filho, J.G.; Hoover, A.R.; King, J.B.; Ellis, T.K.; Powell, D.R.; Cichewicz, R.H. Chemical Epigenetics Alters the Secondary Metabolite Composition of Guttate Excreted by an Atlantic-Forest-Soil-Derived Penicillium citreonigrum. J. Nat. Prod. 2010, 73, 942–948. [Google Scholar] [CrossRef] [Green Version]
- Seto, H.; Tanabe, M. Utilization of 13C-13C Coupling in Structural and Biosynthetic Studies. III. Ochrephilone—A New Fungal Metabolite. Tetrahedron Lett. 1974, 15, 651–654. [Google Scholar] [CrossRef]
- Kanokmedhakul, S.; Kanokmedhakul, K.; Nasomjai, P.; Louangsysouphanh, S.; Soytong, K.; Isobe, M.; Kongsaeree, P.; Prabpai, S.; Suksamrarn, A. Antifungal Azaphilones from the Fungus Chaetomium cupreum CC3003. J. Nat. Prod. 2006, 69, 891–895. [Google Scholar] [CrossRef]
- Beauxis, Y.; Genta-Jouve, G. MetWork: A Web Server for Natural Products Anticipation. Bioinformatics 2019, 35, 1795–1796. [Google Scholar] [CrossRef]
- da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating Annotations of Molecular Networks Using in Silico Fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying Metabolites by Integrating Metabolome Databases with Mass Spectrometry Cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Torbašinović, H.; Obrenović, S.; Mehta, S.S.; Tsugawa, H.; Wermuth, T.; Schauer, N.; Jahn, M.; Biedendieck, R.; et al. Comprehensive Comparison of in Silico MS/MS Fragmentation Tools of the CASMI Contest: Database Boosting Is Needed to Achieve 93% Accuracy. J. Cheminform. 2017, 9, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, J.M.; Sato, M.; Sugimoto, S.; Chiou, G.; Garg, N.K.; Tang, Y.; Watanabe, K. Identification and Characterization of the Chaetoviridin and Chaetomugilin Gene Cluster in Chaetomium globosum Reveal Dual Functions of an Iterative Highly-Reducing Polyketide Synthase. J. Am. Chem. Soc. 2012, 134, 17900–17903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, J.M.; Cascio, D.; Dietrich, D.; Sato, M.; Watanabe, K.; Sawaya, M.R.; Vederas, J.C.; Tang, Y. Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis. J. Am. Chem. Soc. 2015, 137, 9885–9893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chidananda, C.; Sattur, A.P. Sclerotiorin, a Novel Inhibitor of Lipoxygenase from Penicillium frequentans. J. Agric. Food Chem. 2007, 55, 2879–2883. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Flack, H.D. On Enantiomorph-Polarity Estimation. Acta Crystallogr. A 1983, 39, 876–881. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Determination of Absolute Structure Using Bayesian Statistics on Bijvoet Differences. J. Appl. Crystallogr. 2008, 41, 96–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and Analysis of Crystal Structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing: EUCAST. Available online: https://eucast.org/ (accessed on 11 April 2016).
- Tempête, C.; Werner, G.; Favre, F.; Rojas, A.; Langlois, N. In Vitro Cytostatic Activity of 9-Demethoxyporothramycin B. Eur. J. Med. Chem. 1995, 30, 647–650. [Google Scholar] [CrossRef]


| Compounds/Fractions | C. Albicans | T. Rubrum |
|---|---|---|
| 1 | >64 | 32 |
| 2 | >64 | 64 |
| 5 | >64 | >64 |
| 23 | 64 | 32 |
| 63 | >64 | >64 |
| 74 | >64 | >64 |
| 75 | >64 | >64 |
| 80 | >64 | >64 |
| F5 | 256 | 32 |
| F6 | 128 | 16 |
| F7 | 128 | 16 |
| Control | 1 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hebra, T.; Elie, N.; Poyer, S.; Van Elslande, E.; Touboul, D.; Eparvier, V. Dereplication, Annotation, and Characterization of 74 Potential Antimicrobial Metabolites from Penicillium Sclerotiorum Using t-SNE Molecular Networks. Metabolites 2021, 11, 444. https://doi.org/10.3390/metabo11070444
Hebra T, Elie N, Poyer S, Van Elslande E, Touboul D, Eparvier V. Dereplication, Annotation, and Characterization of 74 Potential Antimicrobial Metabolites from Penicillium Sclerotiorum Using t-SNE Molecular Networks. Metabolites. 2021; 11(7):444. https://doi.org/10.3390/metabo11070444
Chicago/Turabian StyleHebra, Téo, Nicolas Elie, Salomé Poyer, Elsa Van Elslande, David Touboul, and Véronique Eparvier. 2021. "Dereplication, Annotation, and Characterization of 74 Potential Antimicrobial Metabolites from Penicillium Sclerotiorum Using t-SNE Molecular Networks" Metabolites 11, no. 7: 444. https://doi.org/10.3390/metabo11070444
APA StyleHebra, T., Elie, N., Poyer, S., Van Elslande, E., Touboul, D., & Eparvier, V. (2021). Dereplication, Annotation, and Characterization of 74 Potential Antimicrobial Metabolites from Penicillium Sclerotiorum Using t-SNE Molecular Networks. Metabolites, 11(7), 444. https://doi.org/10.3390/metabo11070444