Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. Metabolic Features Derived from Untargeted Metabolomics Analysis
2.3. Metabolic Differences in CRC Compared to Colorectal Adenomas
2.4. Metabolic Enrichment and Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Biospecimen Handling, Metabolomics Analysis, and Data Pre-Processing
4.3. Feature Identification
4.4. Metabolic Enrichment and Pathway Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Vargas, A.J.; Thompson, P.A. Diet and nutrient factors in colorectal cancer risk. Nutr. Clin. Pract. 2012, 27, 613–623. [Google Scholar] [CrossRef]
- Schmit, S.L.; Rennert, H.S.; Rennert, G.; Gruber, S.B. Coffee Consumption and the Risk of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 634–639. [Google Scholar] [CrossRef]
- Guertin, K.A.; Loftfield, E.; Boca, S.M.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Huang, W.Y.; Xiong, X.; Freedman, N.D.; Cross, A.J.; et al. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am. J. Clin. Nutr. 2015, 101, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Genet. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulou, E.; Bulusu, V.; Kamphorst, J.J. Metabolic scavenging by cancer cells: When the going gets tough, the tough keep eating. Br. J. Cancer 2016, 115, 635–640. [Google Scholar] [CrossRef]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism 2018, 87, A1–A9. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Bathe, O.F.; Farshidfar, F. From genotype to functional phenotype: Unraveling the metabolomic features of colorectal cancer. Genes 2014, 5, 536–560. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, J.; Li, R.; Fang, Z.M.; Zhu, X.H.; Yi, X.; Lan, H.; Wei, X.; Jiang, D.S. Disturbed energy and amino acid metabolism with their diagnostic potential in mitral valve disease revealed by untargeted plasma metabolic profiling. Metabolomics 2019, 15, 57. [Google Scholar] [CrossRef] [PubMed]
- Yusof, H.M.; Ab-Rahim, S.; Suddin, L.S.; Saman, M.S.A.; Mazlan, M. Metabolomics Profiling on Different Stages of Colorectal Cancer: A Systematic Review. Malays. J. Med. Sci. 2018, 25, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.A.; Hilsden, R.; McGregor, S.E.; Buie, W.D.; MacLean, A.; Vogel, H.J.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857. [Google Scholar] [CrossRef]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef]
- Ulrich, C.M.; Gigic, B.; Bohm, J.; Ose, J.; Viskochil, R.; Schneider, M.; Colditz, G.A.; Figueiredo, J.C.; Grady, W.M.; Li, C.I.; et al. The ColoCare Study: A Paradigm of Transdisciplinary Science in Colorectal Cancer Outcomes. Cancer Epidemiol. Biomark. Prev. 2019, 28, 591–601. [Google Scholar] [CrossRef]
- Pissios, P. Nicotinamide N-Methyltransferase: More Than a Vitamin B3 Clearance Enzyme. Trends Endocrinol. Metab. 2017, 28, 340–353. [Google Scholar] [CrossRef]
- Ghonimy, A.; Zhang, D.M.; Farouk, M.H.; Wang, Q. The Impact of Carnitine on Dietary Fiber and Gut Bacteria Metabolism and Their Mutual Interaction in Monogastrics. Int. J. Mol. Sci. 2018, 19, 1008. [Google Scholar] [CrossRef]
- Peng, Y.-F.; Goyal, H.; Xu, G.-D. Serum bilirubin has an important role in multiple clinical applications. J. Lab. Precis. Med. 2017, 2, 82. [Google Scholar] [CrossRef]
- Yang, L.; Ge, L.Y.; Yu, T.; Liang, Y.; Yin, Y.; Chen, H. The prognostic impact of serum bilirubin in stage IV colorectal cancer patients. J. Clin. Lab. Anal. 2018, 32, e22272. [Google Scholar] [CrossRef]
- Seyed Khoei, N.; Jenab, M.; Murphy, N.; Banbury, B.L.; Carreras-Torres, R.; Viallon, V.; Kuhn, T.; Bueno-de-Mesquita, B.; Aleksandrova, K.; Cross, A.J.; et al. Circulating bilirubin levels and risk of colorectal cancer: Serological and Mendelian randomization analyses. BMC Med. 2020, 18, 229. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Zhang, X.; Tu, S.; Wang, Y.; Xu, B.; Wan, F. Mechanism of taurine-induced apoptosis in human colon cancer cells. Acta Biochim. Biophys. Sin. 2014, 46, 261–272. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Galeone, C.; Turati, F.; La Vecchia, C.; Tavani, A. Coffee consumption and risk of colorectal cancer: A meta-analysis of case-control studies. Cancer Causes Control. 2010, 21, 1949–1959. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bao, Z.; Zou, J.; Dong, J. Coffee consumption and risk of cancers: A meta-analysis of cohort studies. BMC Cancer 2011, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Sanchez-Espiridion, B.; Lin, M.; White, L.; Mishra, L.; Raju, G.S.; Kopetz, S.; Eng, C.; Hildebrandt, M.A.T.; Chang, D.W.; et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer 2017, 123, 4066–4074. [Google Scholar] [CrossRef]
- Buldak, R.J.; Hejmo, T.; Osowski, M.; Buldak, L.; Kukla, M.; Polaniak, R.; Birkner, E. The Impact of Coffee and Its Selected Bioactive Compounds on the Development and Progression of Colorectal Cancer In Vivo and In Vitro. Molecules 2018, 23, 3309. [Google Scholar] [CrossRef]
- Zhang, X.; Albanes, D.; Beeson, W.L.; van den Brandt, P.A.; Buring, J.E.; Flood, A.; Freudenheim, J.L.; Giovannucci, E.L.; Goldbohm, R.A.; Jaceldo-Siegl, K.; et al. Risk of colon cancer and coffee, tea, and sugar-sweetened soft drink intake: Pooled analysis of prospective cohort studies. J. Natl. Cancer Inst. 2010, 102, 771–783. [Google Scholar] [CrossRef]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Esmaeelian, B.; Abbott, C.A.; Le Leu, R.K.; Benkendorff, K. 6-bromoisatin found in muricid mollusc extracts inhibits colon cancer cell proliferation and induces apoptosis, preventing early stage tumor formation in a colorectal cancer rodent model. Mar. Drugs 2013, 12, 17–35. [Google Scholar] [CrossRef]
- Rudd, D.A.; Benkendorff, K.; Chahal, C.; Guinan, T.; Gustafsson, O.J.R.; Esmaeelian, B.; Krysinska, H.; Pogson, L.; Voelcker, N.H.; Abbott, C.A. Mapping insoluble indole metabolites in the gastrointestinal environment of a murine colorectal cancer model using desorption/ionisation on porous silicon imaging. Sci. Rep. 2019, 9, 12342. [Google Scholar] [CrossRef]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiao, Y.; Elson, P.; Tan, H.; Plummer, S.J.; Berk, M.; Aung, P.P.; Lavery, I.C.; Achkar, J.P.; Li, L.; et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J. Clin. Oncol. 2007, 25, 2696–2701. [Google Scholar] [CrossRef]
- Bae, S.; Ulrich, C.M.; Neuhouser, M.L.; Malysheva, O.; Bailey, L.B.; Xiao, L.; Brown, E.C.; Cushing-Haugen, K.L.; Zheng, Y.; Cheng, T.-Y.D.; et al. Plasma Choline Metabolites and Colorectal Cancer Risk in the Women’s Health Initiative Observational Study. Cancer Res. 2014, 74, 7442–7452. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Butler, L.M.; Yuan, J.-M.; Huang, J.Y.; Su, J.; Wang, R.; Koh, W.-P.; Ong, C.-N. Plasma fatty acids and risk of colon and rectal cancers in the Singapore Chinese Health Study. NPJ Precis. Oncol. 2017, 1, 38. [Google Scholar] [CrossRef]
- Volpato, M.; Hull, M.A. Omega-3 polyunsaturated fatty acids as adjuvant therapy of colorectal cancer. Cancer Metastasis Rev. 2018, 37, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Zhou, C.; Zhao, L.; Zhang, G.; Zhang, Y. Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer. J. Pharm. Biomed. Anal. 2016, 118, 349–355. [Google Scholar] [CrossRef]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by (1)H-NMR Spectrometry. Dis. Markers 2019, 2019, 3491852. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, P.; Wang, F.; Liu, W.; Yang, J.; Qin, H. An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann. Surg. 2012, 255, 720–730. [Google Scholar] [CrossRef]
- Tsun, Z.Y.; Possemato, R. Amino acid management in cancer. Semin. Cell Dev. Biol. 2015, 43, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
| CRC | HR a | LR b | |
|---|---|---|---|
| Number of Participants | 88 | 200 | 200 |
| Gender | |||
| Male n(%) | 60 (68.2) | 132 (66.0) | 132 (66.0) |
| Age (years) | |||
| Median (IQR) | 70.0 (60.0–76.0) | 65.4 (56.4–72.6) | 66.0 (55.3–72.9) |
| Body Mass Index (kg/m2) | |||
| Median (IQR) | 26.1 (23.8–29.4) | 27.3 (24.3–30.0) | 27.2 (24.6–30.9) |
| Underweight < 18.5 n(%) | 0 (0) | 3 (1.5) | 1 (0.5) |
| Normal weight 18.5–24.9 n(%) | 26 (29.5) | 57 (28.5) | 51 (25.5) |
| Overweight 25–29.9 n(%) | 38 (43.2) | 82 (41.0) | 81 (40.5) |
| Obese ≥ 30 n(%) | 15 (17.0) | 48 (24.0) | 61 (30.5) |
| Missing | 9 (10.2) | 10 (5.0) | 6 (3.0) |
| Smoking status n(%) | |||
| Current | 20 (22.7) | 50 (25.0) | 35 (17.5) |
| Former | 30 (34.2) | 55 (27.5) | 60 (30.0) |
| Never | 35 (39.7) | 91 (45.5) | 97 (48.5) |
| Missing | 3 (3.4) | 4 (2.0) | 8 (4.0) |
| Site n(%) c | |||
| Colon - distal | 21 (23.9) | - | - |
| Colon - proximal | 33 (37.5) | - | - |
| Rectum | 34 (38.6) | - | - |
| CRC stage n(%) d | |||
| I | 30 (34.1) | - | - |
| II | 17 (19.3) | - | - |
| III | 18 (20.5) | - | - |
| IV | 12 (13.6) | - | - |
| Unspecified | 3 (3.4) | - | - |
| Missing | 8 (9.1) | - | - |
| Histopathology of polyps n(%) e | |||
| Hyperplastic | - | - | 11 (5.5) |
| Tubular < 1 cm | - | - | 189 (94.5) |
| Tubular > 1 cm | - | 64 (32.0) | - |
| Tubulo-villous | - | 128 (64.0) | - |
| Villous | - | 8 (4.0) | - |
| CRC vs. HR + LR | CRC vs. HR | CRC vs. LR | |
|---|---|---|---|
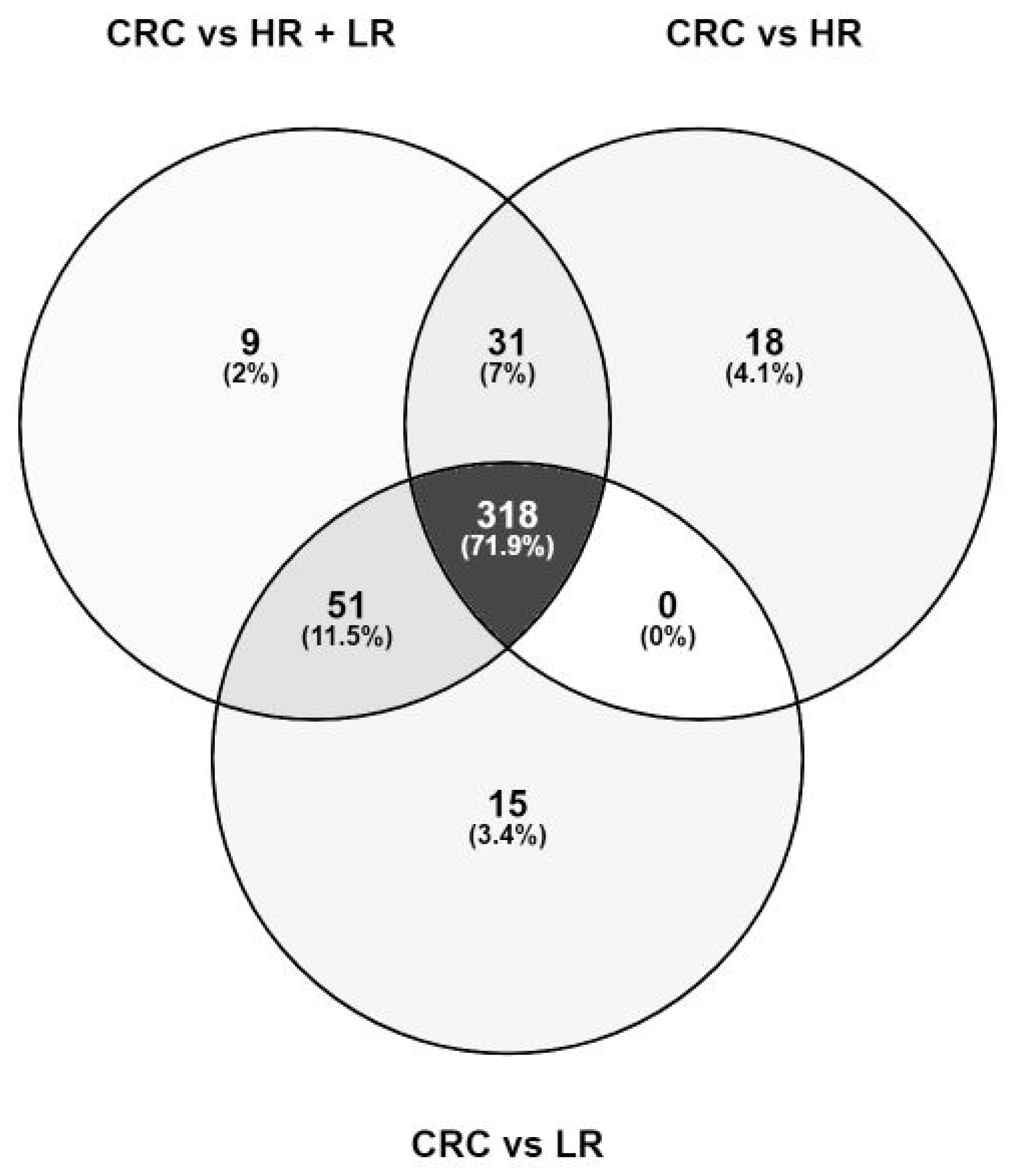
| Sum of Statistically Significant Features | 409 | 367 | 384 |
| q-value a | |||
| 5.0 × 10−2–1.0 × 10−2 | 103 | 101 | 85 |
| 1.0 × 10−2–1.0 × 10−3 | 78 | 86 | 97 |
| 1.0 × 10−3–1.0 × 10−5 | 122 | 131 | 99 |
| 1.0 × 10−5–1.0 × 10−10 | 92 | 43 | 88 |
| 1.0 × 10−10–1.0 × 10−20 | 11 | 6 | 15 |
| <1.0 × 10−20 | 3 | 0 | 0 |
| Pathway and Metabolite Name | RT a | m/zb | ID Level c | q-Value d | OR [CI.Low; CI.Up] e |
|---|---|---|---|---|---|
| Nicotinate and nicotinamide metabolism | |||||
| 1-methylnicotinamide | 0.59 | 137.0711 | 1 | 9.46 × 10−9 | 0.20 [0.12; 0.34] |
| Carnitine pathway | |||||
| Carnitine | 0.59 | 162.1132 | 1 | 1.15 × 10−2 | 0.22 [0.08; 0.61] |
| Tetradecanoylcarnitine (C14:0) | 5.99 | 372.3109 | 1 | 1.16 × 10−4 | 0.25 [0.13; 0.48] |
| Tetradecenoylcarnitine (C14:1) | 5.83 | 370.2959 | 2 | 1.46 × 10−5 | 0.38 [0.25; 0.57] |
| Tetradecadiencarnitine (C14:2) | 5.62 | 368.2799 | 2 | 5.02 × 10−5 | 0.39 [0.25; 0.59] |
| Hexanoylcarnitine (C6:0) | 3.33 | 260.1855 | 2 | 1.86 × 10−3 | 0.42 [0.25; 0.68] |
| Hexadecenoylcarnitine (C16:1) | 6.10 | 398.3263 | 2 | 9.27 × 10−7 | 0.22 [0.12; 0.39] |
| Hexadecadienoylcarnitine (C16:2) | 5.93 | 396.3105 | 2 | 7.41 × 10−5 | 0.29 [0.16; 0.51] |
| Octanoylcarnitine (C8:0) | 4.42 | 288.2177 | 2 | 9.86 × 10−4 | 0.46 [0.3; 0.7] |
| Decanoylcarnitine (C10:0) | 5.13 | 316.2495 | 1 | 7.29 × 10−3 | 0.53 [0.35; 0.8] |
| Decenoylcarnitine (C10:1) isomer 2 | 4.96 | 314.2327 | 2 | 7.58 × 10−4 | 0.35 [0.2; 0.61] |
| Decenoylcarnitine (C10:1) isomer 1 | 4.87 | 314.2328 | 2 | 1.61 × 10−4 | 0.44 [0.27; 0.7] |
| Dodecanoylcarnitine (C12:0) | 5.64 | 344.2804 | 1 | 9.34 × 10−5 | 0.37 [0.23; 0.58] |
| Dodecenoylcarnitine (C12:1) | 5.50 | 342.2638 | 2 | 4.40 × 10−4 | 0.33 [0.19; 0.58] |
| Propionylcarnitine (C3:0) | 1.32 | 218.1382 | 1 | 2.15 × 10−5 | 5.14 [2.56; 10.68] |
| Bilirubin pathway | |||||
| Bilirubin | 7.93 | 583.2554 | 1 | 2.53 × 10−4 | 0.46 [0.31; 0.67] |
| Bilirubin isomer 2 | 5.11 | 585.2696 | 2 | 4.90 × 10−7 | 0.33 [0.21; 0.5] |
| Bilirubin isomer 1 | 4.31 | 585.2685 | 2 | 8.43 × 10−3 | 0.46 [0.27; 0.77] |
| Bile acid metabolism | |||||
| Taurine | 0.63 | 126.0219 | 1 | 6.10 × 10−13 | 16.17 [7.81; 35.24] |
| Glycochenodeoxycholic acid | 6.44 | 450.3216 | 1 | 1.37 × 10−2 | 1.46 [1.13; 1.9] |
| Caffeine pathway | |||||
| Caffeine | 3.19 | 195.0884 | 1 | 2.14 × 10−3 | 1.28 [1.11; 1.49] |
| Theobromine | 2.38 | 181.0721 | 1 | 8.42 × 10−3 | 1.46 [1.14; 1.89] |
| Theophylline | 2.81 | 181.0723 | 1 | 4.20 × 10−2 | 1.33 [1.05; 1.71] |
| Phenolic acid metabolism | |||||
| Hippuric acid | 3.07 | 180.0657 | 1 | 6.52 × 10−21 | 3.15 [2.46; 4.13] |
| Nucleotide metabolism | |||||
| Hypoxanthine | 1.16 | 137.0456 | 1 | 7.60 × 10−3 | 2.14 [1.3; 3.59] |
| Tryptophan pathway | |||||
| Indoleacetic acid | 4.13 | 176.0716 | 1 | 1.17 × 10−10 | 4.23 [2.77; 6.68] |
| Indole-3-propionic acid | 4.56 | 190.0870 | 1 | 1.19 × 10−12 | 2.57 [1.99; 3.37] |
| Indolelactic acid | 3.83 | 206.0823 | 1 | 2.70 × 10−3 | 3.06 [1.59; 5.98] |
| Indole | |||||
| Isatin | 3.31 | 148.0394 | 1 | 7.34 × 10−12 | 5.01 [3.2; 8.09] |
| Linoleic acid and glycerophospholipid metabolism | |||||
| LysoPC (14:0) isomer 2 | 6.73 | 468.3076 | 2 | 3.02 × 10−2 | 0.55 [0.34; 0.88] |
| LysoPC (15:0) | 6.88 | 482.3230 | 2 | 1.98 × 10−2 | 0.47 [0.27; 0.82] |
| LysoPC (16:0) | 7.00 | 496.3400 | 2 | 1.07 × 10−7 | 0.04 [0.01; 0.12] |
| LysoPC (16:1) | 6.82 | 494.3243 | 2 | 3.06 × 10−5 | 0.32 [0.19; 0.52] |
| LysoPC (17:0) | 7.13 | 510.3539 | 2 | 2.64 × 10−3 | 0.4 [0.23; 0.68] |
| LysoPC (18:0) | 7.24 | 524.3713 | 2 | 2.89 × 10−7 | 0.15 [0.07; 0.29] |
| LysoPC (18:1) | 7.06 | 522.3557 | 2 | 1.62 × 10−2 | 0.34 [0.16; 0.73] |
| LysoPC (20:4) | 6.90 | 544.3402 | 2 | 1.09 × 10−4 | 0.22 [0.11; 0.44] |
| LysoPC (22:5) | 6.97 | 570.3538 | 2 | 1.15 × 10−2 | 0.35 [0.17; 0.71] |
| LysoPC (22:6) | 6.89 | 568.3390 | 2 | 4.92 × 10−2 | 0.48 [0.25; 0.89] |
| LysoPC (P-16:0) | 7.11 | 480.3475 | 2 | 6.50 × 10−5 | 0.23 [0.12; 0.45] |
| PC (36:4) | 8.65 | 782.5728 | 2 | 5.87 × 10−3 | 0.3 [0.14; 0.64] |
| PC (38:4) | 9.21 | 810.6029 | 2 | 1.40 × 10−3 | 0.16 [0.05; 0.44] |
| Fatty acid metabolism | |||||
| Docosahexaenoic acid (DHA) | 7.23 | 329.2475 | 1 | 7.37 × 10−3 | 0.45 [0.27; 0.75] |
| Choline metabolism | |||||
| Choline | 0.58 | 104.108 | 1 | 4.67 × 10−2 | 0.26 [0.09; 0.8] |
| Valine, leucine and isoleucine biosynthesis | |||||
| Proline | 0.69 | 116.0712 | 1 | 5.02 × 10−3 | 3.87 [1.69; 9.12] |
| Valine | 0.80 | 118.0866 | 1 | 2.89 × 10−2 | 0.21 [0.06; 0.69] |
| Vitamin E pathway | |||||
| γ-carboxyethyl hydroxychroman | 5.27 | 265.1430 | 1 | 3.25 × 10−2 | 2.49 [1.22; 5.1] |
| Phenylacetate metabolism | |||||
| Phenylacetylglutamine | 3.11 | 265.1190 | 1 | 3.15 × 10−24 | 3.51 [2.71; 4.67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumpenberger, T.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Robinot, N.; Leeb, G.; Habermann, N.; Kok, D.E.G.; Scalbert, A.; Ueland, P.-M.; et al. Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas. Metabolites 2021, 11, 119. https://doi.org/10.3390/metabo11020119
Gumpenberger T, Brezina S, Keski-Rahkonen P, Baierl A, Robinot N, Leeb G, Habermann N, Kok DEG, Scalbert A, Ueland P-M, et al. Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas. Metabolites. 2021; 11(2):119. https://doi.org/10.3390/metabo11020119
Chicago/Turabian StyleGumpenberger, Tanja, Stefanie Brezina, Pekka Keski-Rahkonen, Andreas Baierl, Nivonirina Robinot, Gernot Leeb, Nina Habermann, Dieuwertje E G Kok, Augustin Scalbert, Per-Magne Ueland, and et al. 2021. "Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas" Metabolites 11, no. 2: 119. https://doi.org/10.3390/metabo11020119
APA StyleGumpenberger, T., Brezina, S., Keski-Rahkonen, P., Baierl, A., Robinot, N., Leeb, G., Habermann, N., Kok, D. E. G., Scalbert, A., Ueland, P.-M., Ulrich, C. M., & Gsur, A. (2021). Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas. Metabolites, 11(2), 119. https://doi.org/10.3390/metabo11020119







